Abstract
Monocyte subsets have been shown to differ in the pattern of chemokine receptor expression and their migratory properties, both in human and mouse. Previously we have characterized in the swine several monocyte subpopulations, based on the expression of CD163, Tük4 and SLA-II, which share features with the populations described in human and mouse. Here, we have analysed the expression of different chemokine receptors in the CD163−Tük4+SLA-II− and CD163+Tük4−SLA-II+ populations of porcine monocytes. CD163+Tük4−SLA-II+ monocytes expressed higher CX3CR1 but lower CCR2 and CXCR4 mRNA levels than CD163−Tük4+SLA-II− monocytes. Moreover, porcine CCL2 binding on Tük4+SLA-II− but not on Tük4−SLA-II+ cells was detected by using a CCL2-green fluorescence protein (pCCL2-GFP) fusion protein. Finally, flow cytometric analyses of monocytes recovered after chemotaxis assays show a clear increase in the proportion of Tük4+SLA-II− cells in the fraction migrating toward CCL2, consistent with the polarized CCR2 expression in this monocyte population. The pattern of expression of these chemokine receptors reinforces the similarities of these porcine subsets with their human and mouse counterparts.
Keywords: porcine monocyte, chemokine receptor, chemotaxis
1. INTRODUCTION
Monocytes play critical roles in innate and adaptive immunity and can be considered potential targets for vaccine delivery, as they are circulating precursors of macrophages and dendritic cells (DC). However, the successful development of such vaccination strategies would require a better understanding of the mechanisms that control the traffic and recruitment of these cells to specific tissues and their potential for further functional differentiation in vivo.
Monocytes constitute a heterogeneous population comprising several subsets of cells with distinct phenotypes and functional properties. In humans and mice, two major monocyte populations differing in phenotype and migratory and differentiation capacities have been characterized [12, 19]. Human CD14hiCD16− classical monocytes express high levels of CCR2 and low levels of CX3CR1 and are CD62L+, whereas CD14loCD16+ monocytes express low levels of CCR2 and high levels of CX3CR1, and are CD62L− [10, 21]. Their counterparts in mice are Ly-6ChiCCR2+CD62L+CX3CR1lo and Ly-6C−/loCCR2−CD62L−CX3CR1hi, respectively. Ly-6Chi monocytes are recruited to inflammatory sites and give rise to macrophages and DC, whereas Ly-6C−/lo monocytes have been proposed as precursors for steady state DC [3, 10, 21].
In the swine we have characterized two major monocyte populations based on the expression of CD163. Porcine CD163− and CD163+ monocytes differ in the expression of several surface markers, the pattern of cytokine production, the accessory cell capacity and the permissiveness to virus infection [6, 7, 17, 18]. Adhesion molecules, such as VLA-4 (CD49d/CD29) and LFA-1 (CD11a/CD18), and co-stimulatory molecules CD80/CD86 are also expressed at higher levels in CD163+ monocytes [8]. Like human CD14+CD16+ monocytes, swine CD163+ monocytes can produce high amounts of TNF-α but not IL-10, and have a strong antigen presentation capacity [8, 17]. These CD163− and CD163+ monocytes can be further subdivided into four subsets, according to the expression of SLA-II and Tük4 [22], that would correspond to different developmental stages of monocytes: CD163−Tük4+SLA-II−, CD163−Tük4+SLA-II+, CD163+Tük4+SLA-II+ and CD163+Tük4−SLA-II+ [8].
The aim of this study has been to analyse the expression of CCR2 and other chemokine receptors in the CD163−Tük4+SLA-II− and CD163+Tük4−SLA-II+ populations and their in vitro migratory properties in response to CCL2/MCP-1.
2. MATERIALS AND METHODS
2.1. Animals and cells
Blood samples were obtained from 12-month-old outbred Large-White pigs. The reported experiments have been executed in full compliance with guidelines by the ethical committee of the Institute.
Peripheral blood mononuclear cells (PBMC) were isolated on Percoll discontinuous gradients after blood sedimentation through dextran, as previously described [11]. Cells were resuspended in RPMI 1640 medium (Bio-Whittaker, Verviers, Belgium), containing 10% foetal calf serum (FCS, Bio-Whittaker), 2 mM L-glutamine, 5 × 10−5 M 2-mercaptoethanol and 30 μg/mL gentamicin (Lonza, Walkersville, MD, USA).
2.2. Monoclonal antibodies
Monoclonal antibodies (mAb) to porcine CD3 (BB23-8E6, IgG2b) and CD8α (76-2-11, IgG2a) were kindly provided by M. Pescovitz (Indiana University, Indianapolis, IN, USA) and J. Lunney (USDA, Beltsville, MD, USA), respectively. Crossreactive anti-human CD14 mAb Tük4 (IgG2a) was purchased from Dako (Glostrup, Denmark). MAb to porcine CD45RA (3C3/9, IgG1) [4], CD163 (2A10/11, IgG1) [17], swine major histocompatibility complex class-II antigen SLA-II DR (1F12, IgG2b) [5] and CD172a/SWC3 (BL1H7, IgG1) [1] were produced in our laboratory.
For multi-colour immunofluorescence assays, anti-SLA-II mAb 1F12 was purified by affinity chromatography on Protein A-Sepharose CL-4B (GE Healthcare, Uppsala, Sweden) and labelled with either biotin or Alexa 488 (Molecular Probes, Eugene, OR, USA).
2.3. Flow cytometry
For single-colour staining, cells (2–5 × 105/well) were incubated with 50 μL of unlabelled mAb for 30 min at 4 °C. After washed in PBS containing 0.1% bovine serum albumin (BSA) and 0.01% sodium azide (FACS buffer), cells were incubated with phycoerythrin (PE)-conjugated rabbit (Fab’)2 anti-mouse immunoglobulin (Dako). Then they were washed in FACS buffer and fixed in 0.1% formaldehyde prior to analysis in a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA).
For two-colour staining, cells were incubated with Tük4 followed by Allophycocyanin (APC)-conjugated goat anti-mouse immunoglobulin (Becton Dickinson). After washing, free binding sites were blocked with 5% normal mouse serum. Then Alexa 488- or biotin-conjugated anti-SLA-II mAb was added and biotinylated mAb were detected with streptavidin-PerCP (Pharmingen, San Jose, CA, USA). Subsequently, cells were washed and fixed in formaldehyde prior to analysis by flow cytometry.
2.4. Chemokine binding assay
PBMC or monocytes (2.5 × 105/well) were washed with FACS buffer and incubated for 30 min on ice with various dilutions of supernatant from a transfected CHO cell line stably expressing CCL2-GFP. Then cells were incubated with Alexa 488-conjugated rabbit anti-GFP IgG (Molecular Probes, Eugene, OR, USA) for 30 min. After thorough washing with FACS buffer, cells were fixed with 0.1% paraformaldehyde in PBS and analysed by flow cytometry. To assess CCL2 binding to different monocyte subsets, PBMC were incubated for 30 min on ice with 50 μL/well of supernatant of the CHO/CCL2-GFP cell line, followed by Alexa 488-conjugated rabbit anti-GFP IgG. Then, selected combinations of Tük4, CD172a and biotin-labelled SLA-II specific mAb were added and revealed with secondary antibodies labelled with APC, PE or streptavidin-PerCP, respectively, as above described. Isotype matched unlabelled and biotinylated mAb were used as controls.
2.5. Sorting of monocyte subpopulations
Monocytes were magnetically isolated from PBMC using the autoMACS cell-sorting technique (Miltenyi Biotec, Bergisch-Gladbach, Germany). PBMC were first depleted of lymphocytes using a cocktail of mAb to CD3 (T cells), CD8α (T subpopulation and natural killer cells) and CD45RA (B cells and a T-cell subset). More than 95% of the resultant cells were CD172a+. For sorting of the CD163−SLA-II− monocyte subpopulation, the monocyte-enriched fraction was incubated on ice with a mixture of mAb to CD163 and SLA-II for 30 min, washed with PBS containing 5% FCS, and 2.5 mM EDTA and 0.1% sodium azide (MACS buffer) and then incubated with goat anti-mouse IgG magnetic microbeads for 15 min. After washing with MACS buffer, the cell suspension was passed through the autoMACS separation column to deplete monocytes expressing these markers. The effluent negative fraction was analysed by flow cytometry and contained more than 95% of CD163−Tük4+SLA-II− monocytes.
For sorting of the CD163+Tük4−SLA-II+ monocyte subpopulation, the monocyte-enriched fraction was incubated with Tük4 and passed through the autoMACS separation column. The effluent negative fraction was then incubated with the anti-CD163 mAb and magnetic microbeads as described above. Cells were passed through the autoMACS column and magnetically labelled cells (CD163+Tük4−) were collected. Its analysis by flow cytometry determined that this fraction contained more than 95% of CD163+Tük4−SLA-II+ monocytes.
2.6. RT-PCR analysis of chemokine receptor mRNA expression
Total RNA was isolated from cells using the Tripure Isolation Reagent (Roche, Mannheim, Germany) [9]. First-strand cDNA was obtained from 5 μg of total RNA, previously denatured by heating for 2 min at 65 °C and immediately placed on ice, in a total volume of 50 μL, containing 50 mM Tris-HCl (pH 8.6), 75 mM KCl, 1 mM MnSO4, 10 mM DTT, 50 μM of each dNTP, 0.5 mM oligo(dT), 20 U of RNasin ribonuclease inhibitor (Promega, Madison, WI, USA) and 12.5 U of Moloney murine leukaemia virus RT (Epicentre, Madison, WI, USA). Reaction mixtures were incubated for 1 h at 37 °C. For PCR, a variable amount of the cDNA (typically 1 μL) was used in a total volume of 25 μL of a PCR mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.005% Tween-20, 0.005% Nonidet P-40, 0.5 to 2 mM MgCl2 (depending on the oligonucleotide pair), 50 μM of each deoxynucleoside triphosphate, 10 pmol of each specific oligonucleotide (forward and reverse primers) and 1 U of DyNAzyme II DNA polymerase (Finnzymes Oy, Espoo, Finland).
Primers for amplification of chemokine receptors (Tab. I) were selected from the nucleotide sequences of Sus scrofa CCR2 (GenBank accession number AB119271), CCR5 (AB119272), CCR6 (DQ991099), CCR7 (AB090356), CXCR1 (NM001001622) and CXCR4 (AB116560) and of the conserved regions of Homo sapiens CX3CR1 (BC028078). PCR amplifications were performed by 40 cycles consisting of template denaturation (94 °C, 45 s), primer annealing (52–62 °C, depending on the oligonucleotide pair, 30 s) and extension (72 °C, 1 min). PCR products were run on 1.5% agarose gels. Expected amplified fragment sizes are shown in Table I. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta-actin expression were used for quantitative and qualitative control of RNA. Specificity of the PCR products was verified by DNA sequencing.
Table I.
Primers and PCR conditions used in this study.
| Molecule | Primers | Sequence | Product | Tan |
|---|---|---|---|---|
| CCR7 | CCR7F | GGC TCA AGA CCA TGA CCG AC | 712 | |
| CCR7R | CCG ATG AAG GCG TAC AAG | |||
| CCR4 | CCR4F | CTC CCT GAG AGC AAG GAC | 364 | |
| CCR4R | CTC CAG GAA GAG CAC CAC | |||
| CXCR3 | CXCR3F | GCC CTC TTC AAC ATC AAC TTC | 612 | 52 |
| CXCR3R | CAG CAT CCA CAT CCG CTC | |||
| CX3CR1 | CX3CR1F | CAA CAG CAA GAA GCC CAA G | 444 | |
| CX3CR1R | AGC AGG GGG AGT AGG AAG | |||
| CCR2 | CCR2F | TTG TGT GAC CCA AGA GAG ACT TAC G | 529 | 58 |
| CCR2R | GTT ACA GCC AAA CCA TCC TAA AGC | |||
| CCR5 | CCR5F | GCA ATG AGA AGA AGA AGC ACA AGG | 493 | 55 |
| CCR5R | GGA GCA GAT GCC AAA CGG ATA G | |||
| CXCR4 | CXCR4F | TTA CAC AGA GGA TGA CTT GGG CTC | 707 | 57 |
| CXCR4R | GGA TGA GGA TAA CGG TGG TTT TG | |||
| CCR1 | CCR1F | AAGGGCTATGATGCAACCAC | 504 | 53 |
| CCR1R | CTGGGTCTTGGAAAAGTGGA | |||
| CCR6 | CCR6F | CTGGTGACGGCCGTCAACCTG | 191 | 60 |
| CCR6R | TTCACACACCACAGGTCTTTCATGATC | |||
| CCL2 | CCL2F | CCTCATCCTCCAGCATGAAGGTCTCTGC | 328 | 62 |
| CCL2R | GGTGGAGTCAGGCTTCAAGGCTTCGG | |||
| CCL2-GFP | CCL2GFPR | GAGGCTTCGGAGTTTGGTTTTTCTTGTCCAGG | 313 | 62 |
| CCL2-V5 | CCL2V5R | AGGCTTCGGAGTTTGGTTTTTCTTGTCCAGG | 312 | 62 |
| GAPDH | GAPDHF | CCATCACCATCTTCCAGGAGCGAG | 285 | 55 |
| GAPDHR | AAGTTGTCATGGATGACCTTGGCCA | |||
| B-Actin | B-Actin-F | TGCTGTCCCTGTACGCCTCTG | 220 | 55 |
| B-Actin-R | ATGTCCCGCACGATCTCCC |
2.7. Cloning and expression of recombinant porcine CCL2/MCP-1
Total RNA was isolated from CD172+ cells and the corresponding cDNA obtained as above described. Based on the complete sequence of porcine CCL2 (GeneBank accession number NM214214), two primers were designed for PCR amplification of full-length coding cDNA (Tab. I). The template cDNA (2 μL) was used in a total volume of 25 μL of a PCR mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.005% Tween 20, 0.005% Nonidet P-40, 1 mM MgCl2, 50 μM each dNTP, 10 pmol each of specific oligonucleotides, and 1 U of DyNAzyme II DNA polymerase. After denaturation of cDNA at 94 °C for 5 min, amplification was performed for 35 cycles at 94 °C for 30 s, 62 °C for 30 s and 72 °C for 45 s followed by an extra-extension time of 10 min at 72 °C. After agarose-gel electrophoresis of PCR-amplified products, a band of approximately 328 base pairs was purified from the gel and cloned into the pcDNA3.1 plasmid following the manufacturer instructions (Invitrogen, San Diego, CA, USA). Several clones were sequenced to confirm the orientation and the integrity of the inserts.
Two new anti-sense primers were designed to clone the full-length CCL2 ORF as fusion proteins to GFP and V5-His. PCR amplifications were carried out using DyNAzyme II DNA polymerase with CCL2F primer and the corresponding anti-sense primer (CCL2-GFPR or CCL2-V5HR) (Tab. I). The respective PCR products were cloned into pcDNA3.1/CT-GFP-TOPO or pcDNA3.1/V5-His-TOPO, according to the manufacturer protocol (Invitrogen). Several clones of each construct (pCCL2-GFP or pCCL2-V5-His) were sequenced to confirm the orientation and the integrity of the inserts.
2.8. Recombinant fusion proteins production
CHO cells were transfected with plasmid pCCL2-GFP, encoding CCL2-GFP fusion protein, by using the Lipofectamine Plus Reagent (Invitrogen). The day prior to transfection 3 × 105 CHO cells were placed in M6 plates (Nunc, Rochester, USA) and grown in Dulbecco’s modified Eagle’s minimal medium (DEMEM) (BioWhittaker) supplemented with 30 μg/mL gentamicin, 2 mM L-glutamine and 10% FCS. Immediately before transfection, cells were rinsed with medium without serum or antibiotics (transfection medium, TM). For each well, 500 ng of plasmid DNA and 6 μL of PLUS reagent were mixed, diluted in 100 μL of TM and incubated for 15 min at room temperature. After that, 4 μL of Lipofectamine diluted in 900 μL of TM was added to the transfection mixture. After 15 min of incubation at room temperature, DNA-Lipofectamine complexes were added to the cells and incubated at 37 °C in a 5% CO2 atmosphere for 4 h. Then, 1 mL of fresh growth medium was added and cells cultured for 24 h. Monensin (2 μg/mL), a protein secretion inhibitor, was added 4 h before harvesting. Cells were washed and analysed in a flow cytometer. Non-transfected cells and cells incubated with Lipofectamine were used as negative controls.
Cells were then selected for stable plasmid integration using geneticin (Gibco G-418, Grand Island, NY, USA) at a concentration of 800 μg/mL. After the initial selection period of 10 days, two rounds of limiting dilution cloning were performed. Isolated clones were expanded and screened by flow cytometry and Western analysis for the production of the fusion protein.
2.9. Western blot analysis
Transfected CHO cells (2 × 105) were treated with 2 μg/mL Monensin (Sigma, St. Louis, MO, USA), washed with PBS and solubilized in 0.2 mL of lysis buffer (10 mM Tris-HCl pH 7.4, 1 mM EDTA, 1% NP-40, 1 mg/mL BSA, 10 μg/mL of aprotinin and 1 mM PMSF). After centrifugation, the supernatant was mixed with electrophoresis sample buffer, boiled and run on a 15% SDS-PAGE gel under reducing conditions. Separated proteins were transferred to nitrocellulose and fusion proteins analysed by Western blotting with a biotin-conjugated goat anti-GFP polyclonal antibody (Rockland Inmunochemicals, Gilbertsville, PA, USA) and streptavidin-HRP (Thermo Fisher Scientific, Rockford, IL, USA). Peroxidase activity was visualized using the ECL detection assay (GE Healthcare).
2.10. Chemotaxis assays
Monocyte subset migration was measured by chemotaxis through a 5 μm-pore polycarbonate filter in Transwell chambers (Corning, Lowell, MA, USA). Enriched blood monocytes (3.5 × 105 cells) were resuspended in RPMI with 1% BSA and 25 mM HEPES and placed in the top chamber. Serial dilutions of pCCL2-GFP were added to the lower chamber. Two or more replicate wells were used for each point. After 2 h incubation, migrated cells were analysed by flow cytometry. The migration index was determined as the ratio between cells migrated in response to the chemokine and to the inverted construction used as control.
3. RESULTS
3.1. Differential expression of chemokine receptor mRNA transcripts in monocyte subsets
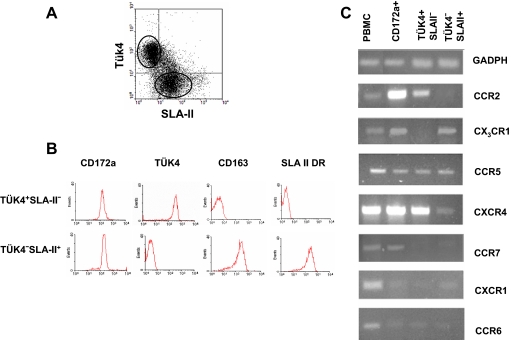
Enriched peripheral blood monocytes from healthy pigs were magnetically sorted based on the expression of SLA-II, Tük4 and CD163. Two discrete populations were selected, one expressing high levels of Tük4 but neither SLA-II or CD163, and other expressing SLA-II and CD163, but not Tük4 (Figs. 1A and 1B).
Figure 1.
Chemokine receptor mRNA expression in monocyte subsets. (A) Dot plot of Tük4 versus SLA-II expression in monocytes obtained from PBMC (20 000 cells were acquired). Three subsets can be identified: Tük4+SLA-II−, Tük4+SLA-II+, and Tük4−SLA-II+. (B) The Tük4+SLA-II− and Tük4−SLA-II+ monocyte subsets were magnetically isolated as described in Materials and methods section, and the expression of the indicated markers was analysed by flow cytometry (5 000 cells acquired). (C) Total RNA isolated from PBMC, the whole monocyte population (CD172a+) or the Tük4+SLA-II− and Tük4−SLA-II+ monocyte subsets shown in (B) were reverse transcribed, and the cDNA amplified by PCR with specific primers for GAPDH or different chemokine receptors. PCR products were analysed by 1.5% agarose gel electrophoresis. Data are representative from three independent experiments using different donors. (A color version of this figure is available at www.vetres.org.)
The expression of CCR2, CX3CR1, CCR5, CXCR4, CCR7, CXCR1 and CCR6 transcripts in these monocyte subsets was determined by RT-PCR (Fig. 1C). GAPDH is shown as quantitative and qualitative control of RNA, this housekeeping gene was chosen by its stability in many tissues [24], parallel amplification of a fragment of beta-actin gene as a second control (not shown) gave identical results. CCR2, CXCR4 and CX3CR1 expression differed between these cell populations. CCR2 and CXCR4 mRNA were clearly detected in CD163−Tük4+SLA-II− but not detected or detected at low levels in CD163+Tük4−SLA-II+ cells; whereas CX3CR1 expression was negative in CD163−Tük4+SLA-II− cells but positive in CD163+Tük4−SLA-II+ cells. Both subsets expressed CCR5 mRNA at similar levels. CCR7 was clearly detected in the whole monocyte population, but its expression was mostly negative in these two subsets. CXCR1 and CCR6 mRNA were very weak or undetectable in the whole monocyte population as well as in the sorted monocyte subsets.
3.2. Differential responsiveness of Tük4+SLA II− and Tük4−SLA-II+ monocytes to CCL2
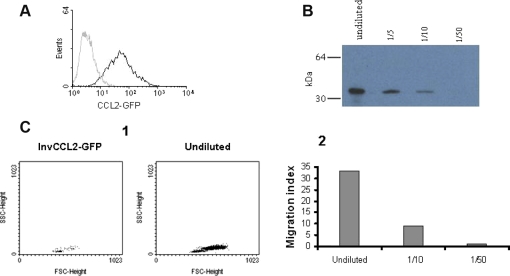
We next tested whether the different expression of CCR2 in these two monocyte subsets was associated with a distinct response to CCL2 in chemotaxis assays. To clone the porcine CCL2, primers CCL2F and CCL2R that flank the CCL2 translation start and stop codons were designed based on the sequence available in databases and used for PCR amplification of the full-length coding sequence of porcine CCL2 (Tab. I). Since no specific antibodies were available at that time for detecting its expression, we made a GFP tagged construct by subcloning in the plasmid pcDNA3.1/CT-GFP-TOPO the porcine CCL2 full-length coding sequence in frame with that of GFP (pCCL2-GFP). Another plasmid containing the CCL2 sequence in an inverted orientation fused with GFP (pInvCCL2-GFP) was also constructed. CHO cells were transfected with plasmids pCCL2-GFP or pInvCCL2-GFP and the expression of GFP-fused proteins was analysed by flow cytometry. After geneticin selection of transfectants and two rounds of limiting dilution, several cell clones stably expressing each of these constructs were obtained (Fig. 2A).
Figure 2.
Expression of recombinant porcine CCL2. (A) CHO cell line stably expressing the porcine CCL2 fused to GFP. The expression of GFP fusion protein was directly analysed by flow cytometry. Non transfected CHO cells were used as negative control (grey histogram). 5 000 cells were acquired. (B) Western blot of CCL2-GFP produced by transfected CHO cells. Different dilutions of supernatant were resolved by 15% SDS-PAGE under reducing conditions and revealed with biotinylated anti-GFP and streptavidin-HRP. Numbers on the left indicate the position of MW markers. (C) Chemotactic activity of CCL2-GFP on porcine blood monocytes. Chemotaxis was assessed with the Transwell cell migration system and subsequent flow cytometry counting of migrated cells by a 45 s acquisition. (1) FSC versus SSC dot plot of migrated cells in response to supernatants from CHO cells expressing CCL2-GFP or the inverted sequence of pCCL2 fused to GFP (InvCCL2-GFP, negative control). (2) Results expressed as migration index, calculated as the ratio of the number of cells migrating to the chemokine and the number of cells in the negative control. Results from one representative experiment out of three performed are shown. (A color version of this figure is available at www.vetres.org.)
The expression of GFP-fused proteins in these clones was confirmed by Western blot using a biotin-conjugated goat anti-GFP polyclonal antibody and streptavidin-HRP. As shown in Figure 2B, a specific band with an apparent molecular weight of 39 kDa, the expected size for the CCL2-GFP fusion protein, was clearly detected in supernatants from the CHO clones expressing pCCL2-GFP.
The functional activity of GFP-fused CCL2 was evaluated in a chemotaxis assay using purified monocytes. Supernatants from CHO cells transfected with the porcine CCL2-GFP expression plasmid showed a dose-dependent chemoattractant activity, whereas no effect was observed with control supernatants obtained from CHO cells transfected with pInvCCL2-GFP (Fig. 2C).
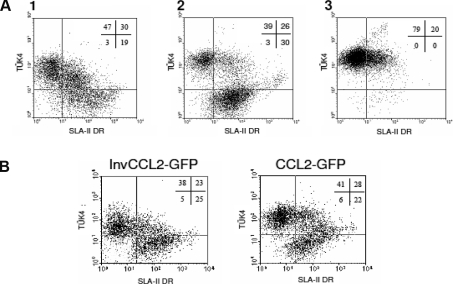
Two-colour flow cytometric analyses of monocytes recovered after chemotaxis revealed that cells migrating were mostly of the Tük4+SLA-II− phenotype while Tük4−SLA-II+ cells did not migrate (Fig. 3A). To rule out that the increase in the proportion of Tük4+SLA-II− cells in the migrating population was due to changes in the phenotype induced by CCL2, we analysed the effect of this chemokine on the expression of Tük4 and SLA-II markers in monocytes cultured in Teflon containers without observing significant changes in the expression of these markers (Fig. 3B).
Figure 3.
Differential chemotactic responsiveness of monocyte subsets to pCCL2. (A) Isolated monocytes were subjected to chemotaxis induced by CCL2 or kept under identical conditions (input) for 3 h. Input cells (1), cells that did not migrate (2) or cells that migrated (3) were recovered and the expression of Tük4 and SLA II analysed by two-colour flow cytometry. Dot plots correspond to a representative experiment out of four performed. (B) Isolated monocytes were cultured for 3 h in Teflon containers in the presence of supernatants of CHO cells transfected with pCCL2-GFP or InvCCL2-GFP, and the expression of Tük4 and SLA-II analysed by two colour flow cytometry. 10 000 cells were acquired.
3.3. Binding of pCCL2 to monocyte subsets
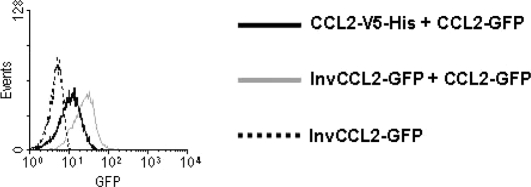
CCL2 and other chemokines bind to their receptors with enough affinity to allow their detection by flow cytometry. Therefore we tested whether the CCL2-GFP fusion protein could bind to porcine PBMC. A clear binding of CCL2-GFP to monocytes was observed. This binding was specific since it was competed by unlabeled CCL2. Moreover, no binding was detected when supernatants from cells transfected with the InvCCL2-GFP were used (Fig. 4).
Figure 4.
Inhibitory effect of unlabeled CCL2 on the binding of CCL2-GFP to porcine monocytes. PBMC were incubated with 25 μL of supernatant containing CCL2-GFP and 25 μL of supernatant from CHO cells transfected with a construct with the sequence of pCCL2 fused to V5-His epitope (CCL2-V5-His) or with InvCCL2-GFP. GFP bound to the cells was detected with Alexa 488- conjugated rabbit anti-GFP IgG. Histograms shown were obtained on gates corresponding to monocytes (CD172a+). 6 000 cells were acquired.
To study the CCL2 binding in different monocyte subsets, freshly isolated PBMC were incubated with CCL2-GFP and mAb to CD172a, Tük4 and SLA-II. CD172a+ monocytes were gated according to their FSC/SSC characteristics and the binding of CCL2-GFP to monocyte subsets, defined based on the expression of Tük4 and SLA-II, was analysed. Tük4+SLA-II− and Tük4+SLA-II+ monocytes showed comparable CCL2 binding ability, whereas no CCL2 binding was detected within the Tük4−SLA-II+ population (Fig. 5).
Figure 5.
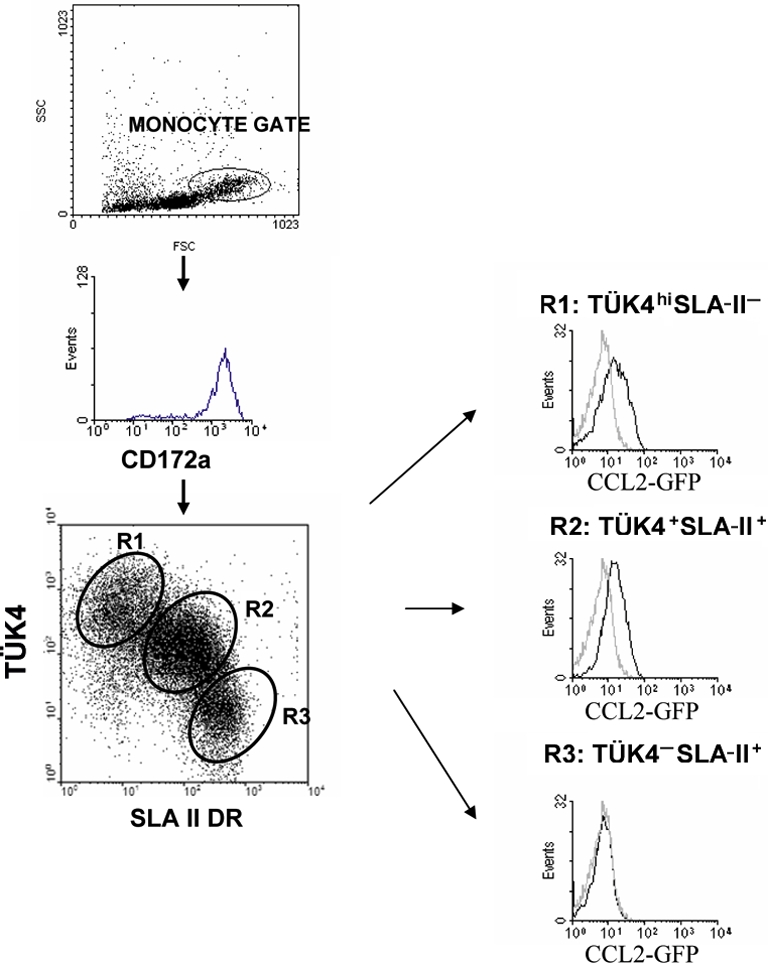
Differential binding of CCL2-GFP to monocyte subsets. Porcine PBMC were incubated with CCL2-GFP (CHO cell supernatant) followed by Alexa 488-conjugated rabbit anti-GFP IgG, and then double stained for Tük4 and SLA II. The histograms are gated on the appropriate monocyte subpopulation regions as defined in the dot-plots. Grey histograms show the background fluorescence of monocytes incubated with supernatants from CHO cells transfected with the InvCCL2-GFP construct. 25 000 cells were acquired. (A color version of this figure is available at www.vetres.org.)
4. DISCUSSION
Circulating monocytes exhibit a marked phenotypic and functional heterogeneity, a common feature in every mammalian species studied, which appears to be related with their maturation stage [12]. Human monocytes have been divided in two major subsets according to the expression of CD16 and CD14 [28]. Monocyte subsets have also been identified in mouse and rat, based on the expression of Ly-6C and CD43, respectively, and on that of the chemokine receptors CCR2 and CX3CR1 [10, 16, 20, 27]. These monocyte subsets may play different functional roles in immunity and inflammation; they have distinct migration patterns by which they are recruited into tissues and they can give rise to distinct DC and macrophages.
In previous studies we have characterized four subsets of porcine monocytes which differ in the levels of expression of Tük4, an anti-human CD14 mAb crossreacting in pig [22], CD163 and SLA-II antigens, and appear to represent sequential stages of the monocyte maturation [8]. Within this maturation pathway, the CD163−Tük4hiSLA-II− subset would correspond to the most immature stage, and the CD163+Tük4−SLA-II+ to the most mature. Like human CD16+ monocytes, porcine CD163+Tük4−SLA-II+ monocytes exhibit a phenotype more closely related to macrophages, produce higher levels of TNF and have higher antigen presenting potential [6, 8].
In this study we have analysed in the swine the expression of chemokine receptors in two blood monocyte subsets, corresponding to the first and late stages of their maturation. RT-PCR analyses showed that CD163−Tük4+SLA-II− and CD163+Tük4−SLA-II+ monocytes differ in the mRNA expression of CCR2 and CX3CR1. Consistent with these data, Tük4+SLA-II− but not Tük4−SLA-II+ monocytes bind CCL2-GFP and migrate in response to this chemokine.
The expression of other chemokine receptors, namely CCR5 and CCR7, in these porcine monocyte subsets also compares with that seen in human and murine monocytes. Thus, like human CD14hiCD16− and CD14loCD16+ [25], porcine Tük4+SLA-II− and Tük4−SLA-II+ monocytes express similar levels of mRNA CCR5. By using mAb and flow cytometry Ancuta et al. also reported similar low levels of CCR5 expression in human CD14hiCD16− and CD14loCD16+ monocyte subsets [2]. With respect to the CCR7 expression, it was mostly negative in the two porcine monocyte subsets studied, but was clearly detected in the whole monocyte population, suggesting that it is expressed by an intermediate population. This pattern is consistent with studies in the mouse, where this receptor has been found selectively expressed in the intermediate Ly-6Cme population, but not in Ly-6Chi or Ly-6Clo cells [16]. However, we cannot rule out a minor contamination of DC in the whole monocyte population (CD172a+) that may contribute to CCR7.
On the other hand, differences between swine and human were observed in the expression of CXCR4 that may reflect species-specific differences. Whereas CXCR4 is expressed at higher levels in porcine Tük4+SLA-II− monocytes than in Tük4−SLA-II+, human CD16+ monocytes express higher or similar CXCR4 levels than CD16− and migrate more efficiently in response to SDF-1 [2].
Biotin- or fluorochrome-labelled chemokines can be an alternative or a complement to the use of antibodies for characterization of chemokine receptors. pCCL2, expressed as a fusion protein linked to GFP, maintains its chemoattractant activity and can be used, in combination with antibodies against cell surface markers to analyse the CCL2 binding capacity of different leukocyte populations. The binding seems specific, as fluorescent and unlabeled CCL2 compete for binding to their receptor.
In addition to Tük4+SLA-II− cells, CCL2-GFP also binds to Tük4+SLA-II+, but not to Tük4−SLA-II+ cells. Consistent with this finding, a fraction of Tük4+SLA-II+ cells responded to CCL2-GFP in chemotaxis assays. Although monocyte phenotype could change upon stimulation with CCL2 [13], an effect on the phenotype of Tük4+SLA-II− cells following stimulation with CCL2 seems unlikely, since no changes were seen by flow cytometry in cells cultured in Teflon in the presence of CCL2.
The CCL2 binding capacity of the distinct monocyte subsets correlates well with CCR2 expression, as determined by RT-PCR, suggesting that CCR2 is the main receptor for this chemokine in monocytes. Although CCL2 can also bind to D6 and Duffy/DARC “decoy” receptors [29], the expression of these receptors in monocytes is low or undetectable [14, 15]. This is consistent with previous studies which identify a single class of high affinity receptor for CCL2 in human monocytes [23, 26].
In summary, the Tük4+SLA-II− and Tük4−SLA-II+ porcine monocyte subsets differ in the expression of CCR2 and CX3CR1 and in their response to CCL2, reinforcing their similarities with their human and mouse counterparts. Characterization of the migratory properties of these monocyte subsets will contribute to a better understanding of their functional roles in immune and inflammatory responses, providing relevant information for the design of more effective vaccines. Moreover, the conservation among species of migration patterns of monocyte subsets makes the swine a useful model for studying pathogenesis of inflammatory diseases in which monocytes are involved.
Acknowledgments
This work was supported by grants AGL-2005-07073, CSD2006-0007 and RTA2008-00038-00-00 from the Spanish Ministry of Science and Innovation.
References
- 1.Alvarez B., Sanchez C., Bullido R., Marina A., Lunney J., Alonso F., et al., A porcine cell surface receptor identified by monoclonal antibodies to SWC3 is a member of the signal regulatory protein family and associates with protein-tyrosine phosphatase SHP-1, Tissue Antigens (2000) 55:342–351 [DOI] [PubMed] [Google Scholar]
- 2.Ancuta P., Rao R., Moses A., Mehle A., Shaw S.K., Luscinskas F.W., Gabuzda D., Fractalkine preferentially mediates arrest and migration of CD16+ monocytes, J. Exp. Med. (2003) 197:1701–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auffray C., Sieweke M.H., Geissmann F., Blood monocytes: Development, heterogeneity and relationships with dendritic cells, Annu. Rev. Immunol. (2009) 27:669–692 [DOI] [PubMed] [Google Scholar]
- 4.Bullido R., Gomez del Moral M., Doménech N., Alonso F., Ezquerra A., Domínguez J., Monoclonal antibodies to a high molecular weight isoform of porcine CD45: biochemical and tissue distribution analyses, Vet. Immunol. Immunopathol. (1997) 56:151–162 [DOI] [PubMed] [Google Scholar]
- 5.Bullido R., Doménech N., Alvarez B., Alonso F., Babin M., Ezquerra A., et al., Characterization of five monoclonal antibodies specific for swine class II major histocompatibility antigens and crossreactivity studies with leukocytes of domestic animals, Dev. Comp. Immunol. (1997) 21:311–322 [DOI] [PubMed] [Google Scholar]
- 6.Chamorro S., Revilla C., Alvarez B., Lopez-Fuertes L., Ezquerra A., Dominguez J., Phenotypic characterization of monocyte subsets in the pig, Immunobiology (2000) 202:82–93 [DOI] [PubMed] [Google Scholar]
- 7.Chamorro S., Revilla C., Gomez N., Alvarez B., Alonso F., Ezquerra A., Dominguez J., In vitro differentiation of porcine blood CD163− and CD163+ monocytes into functional dendritic cells, Immunobiology (2004) 209:57–65 [DOI] [PubMed] [Google Scholar]
- 8.Chamorro S., Revilla C., Alvarez B., Alonso F., Ezquerra A., Dominguez J., Phenotypic and functional heterogeneity of porcine blood monocytes and its relation with maturation, Immunology (2005) 114:63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski P., Sacchi N., Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction, Anal. Biochem. (1987) 162:156–159 [DOI] [PubMed] [Google Scholar]
- 10.Geissmann F., Jung S., Littman D.R., Blood monocytes consist in two principal subsets with distinct migratory properties, Immunity (2003) 19:71–82 [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez S., Mendoza C., Sanchez-Vizcaino J.M., Alonso F., Inhibitory effect of African swine fever virus on lectin-dependent swine lymphocyte proliferation, Vet. Immunol. Immunopathol. (1990) 26:71–80 [DOI] [PubMed] [Google Scholar]
- 12.Gordon S., Taylor P.R., Monocyte and macrophage heterogeneity, Nat. Rev. Immunol. (2005) 5:953–964 [DOI] [PubMed] [Google Scholar]
- 13.Green S.R., Han K.H., Chen Y., Almazan F., Charo I.F., Miller Y.I., Quehenberger O., The CC chemokine MCP-1 Stimulates surface expression of CX3CR1 and enhances the adhesion of monocytes to fractalkine/CX3CL 1via p38 MAPK, J. Immunol. (2006) 176:7412–7420 [DOI] [PubMed] [Google Scholar]
- 14.McKimmie C.S., Fraser A.R., Hansell C., Gutierrez L., Philipsen S., Connell L., et al., Hemopoietic cell expression of the chemokine decoy receptor D6 is dynamic and regulated by GATA1, J. Immunol. (2008) 181:3353–3363 [DOI] [PubMed] [Google Scholar]
- 15.Pogo A.O., Chaudhuri A., The duffy protein: a malarial and chemokine receptor, Semin. Hematol. (2000) 37:122–129 [DOI] [PubMed] [Google Scholar]
- 16.Qu C., Edwards E.W., Tacke F., Angeli V., Llodra J., Sanchez-Schmitz G., et al., Role of CCR8 and other chemokine pathways in the migration of monocyte-derived dendritic cells to lymph nodes, J. Exp. Med. (2004) 200:1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez C., Domenech N., Vazquez J., Alonso F., Ezquerra A., Dominguez J., The porcine 2A10 antigen is homologous to human CD163 and related to macrophage differentiation, J. Immunol. (1999) 162:5230–5237 [PubMed] [Google Scholar]
- 18.Sanchez-Torres C., Gomez-Puertas P., Gomez del Moral M., Alonso F., Escribano J.M., Ezquerra A., Domınguez J., Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection, Arch. Virol. (2003) 148:2307–2323 [DOI] [PubMed] [Google Scholar]
- 19.Serbina N.V., Jia T., Hohl T.M., Pamer E.G., Monocyte- mediated defense against microbial pathogens, Annu. Rev. Immunol. (2008) 26:421–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunderkötter C., Nikolic T., Dillon M.J., van Rooijen N., Stehling M., Drevets D.A., Leenen P.J.M., Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response, J. Immunol. (2004) 172:4410–4417 [DOI] [PubMed] [Google Scholar]
- 21.Tacke F., Randolph G., Migratory fate and differentiation of blood monocyte subsets, Immunobiology (2006) 211:609–618 [DOI] [PubMed] [Google Scholar]
- 22.Thacker E., Summerfield A., McCullough K., Ezquerra A., Dominguez J., Alonso F., et al., Summary of workshop findings for porcine myelomonocytic markers, Vet. Immunol. Immunopathol. (2001) 80:93–109 [DOI] [PubMed] [Google Scholar]
- 23.Valente A.J., Rozek M.M., Schwartz C.J., Graves D.T., Characterization of monocyte chemotactic protein-1 binding to human monocytes, Biochem. Biophys. Res. Commun. (1991) 176:309–314 [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al., Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes, Genome Biol. (2002) 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber C., Belge K.U., von Hundelshausen P., Draude G., Steppich B., Mack M., et al., Differential chemokine receptor expression and function in human monocytes subpopulations, J. Leukoc. Biol. (2000) 67:699–704 [DOI] [PubMed] [Google Scholar]
- 26.Yoshimura T., Leonard E.J., Identification of high affinity receptors for human monocyte chemoattractant protein-1 on human monocytes, J. Immunol. (1990) 145:292–297 [PubMed] [Google Scholar]
- 27.Yrlid U., Jenkins C.D., MacPherson G.G., Relationships between distinct blood monocyte subsets and migrating intestinal lymph dendritic cells in vivo under steady-state conditions, J. Immunol. (2006) 176:4155–4162 [DOI] [PubMed] [Google Scholar]
- 28.Ziegler-Heitbrock H.W., Fingerle G., Strobel M., Schraut W., Stelter F., Schutt C., et al., The novel subset of CD14+/CD16+ blood monocytes exhibits features of tissue macrophages, Eur. J. Immunol. (1993) 23:2053–2058 [DOI] [PubMed] [Google Scholar]
- 29.Zlotnik A., Yoshie O., Nomiyama H., The chemokine and chemokine receptor superfamilies and their molecular evolution, Genome Biol. (2006) 7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]