Abstract
Background
In the United States, clade B is the predominant human immunodeficiency virus (HIV) subtype, whereas in sub-Saharan Africa, clades A, C, and D are the predominant subtypes. HIV subtype may have an impact on HIV disease progression. The effect of HIV subtype on the risk of dementia has, to our knowledge, not been examined. The objective of this study was to examine the relationship between HIV subtype and the severity of HIV-associated cognitive impairment among individuals initiating antiretroviral therapy in Uganda.
Methods
Sixty antiretroviral-naive HIV-infected individuals with advanced immunosuppression who were at risk of HIV-associated cognitive impairment underwent neurological, neuropsychological, and functional assessments, and gag and gp41 regions were subtyped. Subtype assignments were generated by sequence analysis using a portion of the gag and gp41 regions.
Results
Thirty-three HIV-infected individuals were infected with subtype A, 2 with subtype C, 9 with subtype D, and 16 with A/D recombinants. Eight (89%) of 9 HIV-infected individuals with subtype D had dementia, compared with 7 (24%) of 33 HIV-infected individuals with subtype A (P = .004).
Conclusions
These results suggest that, in untreated HIV-infected individuals with advanced immunosuppression who are at risk of developing HIV-associated cognitive impairment, HIV dementia may be more common among patients infected with subtype D virus than among those infected with subtype A virus. These findings provide the first evidence, to our knowledge, to demonstrate that HIV subtypes may have a pathogenetic factor with respect to their capacity to cause cognitive impairment. Additional studies are needed to confirm this observation and to define the mechanism by which subtype D leads to an increased risk of neuropathogenesis.
Human immunodeficiency virus (HIV)–associated neurocognitive disorders (HANDs) are characterized by disabling cognitive, behavioral, and motor dysfunction and are a common neurological manifestation of advanced HIV infection. The prevalence of HIV-associated dementia, the most severe form of HAND, in resource-limited settings such as countries in sub-Saharan Africa is largely unknown, but a recent study suggests that the prevalence in Uganda may be as high as 31% [1].
HIV type 1 (HIV-1) is characterized by extensive genetic diversity and can be divided into 3 classes: group M (major), group O (outlier), and group N (new, non-M, and non-O). Group M is responsible for >90% of cases of HIV infection globally and is represented by 9 major subtypes or clades (A–D, F–H, J, and K) [2]. The HIV-1 subtypes in the United States differ from the subtypes seen elsewhere in the world. In the United States, clade B is the predominant subtype, whereas in sub-Saharan Africa, clades A, C, and D are the predominant subtypes [2].
HIV subtype has an impact on HIV disease progression. Studies from Uganda, Kenya, and Tanzania have demonstrated that HIV-infected individuals infected with subtype D virus have a faster progression to AIDS and a higher mortality rate than do HIV-infected individuals infected with subtype A virus [3–6]. These data demonstrate a virus-specific component, which is subtype specific and impacts virulence. The effect of HIV subtype on the risk of HIV dementia has, to our knowledge, not been examined in well-characterized HIV-infected individuals in Africa. The objective of this study was to characterize the HIV subtype among HIV-infected individuals initiating highly active antiretroviral therapy (HAART) in Uganda and to examine the relationship between HIV subtype and the severity of HIV-associated cognitive impairment.
METHODS
Participants
The study enrolled 60 antiretroviral-naive HIV-infected individuals from a larger project evaluating the effect of HAART on HIV-infected individuals who were at risk of developing cognitive impairment at an infectious diseases clinic in Kampala, Uganda, from September 2005 through January 2007 [7]. HIV-infected individuals were chosen to receive HAART using the following inclusion criteria: advanced HIV infection, with a CD4 lymphocyte count <200 cells/μL; attendance of at least 2 clinic visits in the prior 6 months; residence within a 20-km radius of Kampala; performance on a screening test for HIV dementia (International HIV Dementia Scale [IHDS], ≤10) suggestive of HAND [8]; and provision of written informed consent. Exclusion criteria included age <18 years, an active or known past opportunistic infection of the central nervous system, fever (temperature, >37.5°C), and a history of a chronic neurologic disorder, active psychiatric disorder, alcoholism, physical deficit (eg, amputation), severe functional impairment (Karnofsky performance scale, <50) [9], or severe medical illness that would interfere with the ability to perform the study evaluations. The evaluations were translated into the local language, Luganda.
Clinical assessments
HIV-infected individuals received standardized questionnaires for assessment of demographic information and medical, psychiatric, and neurologic history and underwent a neurologic examination [1], including assessments of cranial nerves, limb strength and coordination, limb vibration and pin sensation, gait, and deep tendon reflexes in all extremities [1]; patients were also evaluated for fever, headache, neck stiffness, and focal abnormalities. HIV-infected patients with suspected opportunistic infection of the central nervous system or neoplasm were excluded from the study.
The neurocognitive assessment included a screening test for HIV dementia, the IHDS [8], and a full neuropsychological test battery. Participants were antiretroviral naive prior to the neurocognitive assessment. The IHDS consists of 3 subtests (timed finger tapping, a timed alternating hand sequence test, and recall of 4 items at 2 min) and has been validated as a sensitive screening test for HIV dementia in Uganda [8]. The neuropsychological testing battery included the World Health Organization–University of California–Los Angeles Auditory Verbal Learning test for verbal memory [10], the Timed Gait and Grooved Pegboard Tests to assess motor performance, the Symbol Digit Test [11] and Color Trails Test [10] to assess psychomotor speed performance, the Digit Span Forward and Backward to assess attention, the finger tapping test to assess motor performance, and the category naming test to assess verbal fluency. The functional assessment included the Karnofsky performance scale [9]. These assessments were used to assign a Memorial Sloan Kettering (MSK) dementia stage of 0 (normal neurocognitive function), 0.5 (equivocal symptoms or signs without impairment in capacity to perform activities of daily living), or ≥1 (HIV dementia; ie, abnormal neuropsychological test findings and mild-to-moderate functional impairment) [12]. A diagnosis of HIV dementia required impairment in ≥2 neuropsychological tests in which the subject scored <1.5 standard deviations below the locally determined mean for his or her normative age and education group and had symptomatic and/or functional complaints consistent with dementia or findings from a neurologic examination consistent with HIV dementia (eg, extrapyramidal signs) [1].
CD4 lymphocyte counts and plasma HIV loads were determined for all HIV-infected subjects on the same day as the neurocognitive assessment. Analysis of cerebrospinal fluid specimens and neuroimaging were not performed in this study.
RNA extraction and amplification of gag and gp41 fragments
Viral RNA was extracted from 140 μL of serum using QlAmp Viral RNA Mini Kit (Qiagen) with the modifications listed below. The final elution was performed with 50 μL of deionized diethylpyrocarbonate–treated water into a tube that contained 1 μL of 100 U of RNAse inhibitor (RNAsin; Promega). The eluted RNA was used for reverse-transcription polymerase chain reaction in 2 separate reactions with HIV-1–specific primers in the gag and gp41 env regions. Information about the primer sequences and amplification protocols have been described in detail elsewhere [13, 14].
The purified nested polymerase chain reaction products from the gag and gp41 regions were used for automated sequencing with a BigDye terminator cycle-sequencing ready reaction kit. The sequencing reactions were then run in a 377 DNA sequencer (PE Applied Biosystems). These sequences, along with reference sequences from the HIV sequence database, were aligned using the CLUSTALW multiple-sequence alignment program [15] and were optimized by hand using BIOEdit, version 5.09 [16].
Phylogenetic analysis and subtype assignment
Phylogenetic trees were generated using Nimble Tree (http://sray.med.som.jhmi.edu/SCRoftware/), which incorporates PHYLIP, version 3.572c [15]. DNADIST was used to calculate the genetic distance matrix using a maximum likelihood model with a transition-to-transversion ratio of 2.0 [17]. Trees were generated from the distance matrix using the neighbor-joining algorithm in NEIGHBOR [15]. Bootstrap confidence intervals were calculated by randomly permuting the sequence alignment 100 times with SEQBOOT [15]. Consensus topology was derived by the use of CONSENSE [16]. Bootstrap values >75% were considered significant. Nucleotide positions in relation to HXB2 were determined using the HIV numbering engine and reference sequences for different HIV-1 group M subtypes obtained from Los Alamos (http://web.lanl.gov/seq-db.html). Reference sequences used included fragments of the full-length sequences previously generated from this region (accession numbers AF-484502 to AF-484520 and HXB2 (accession number K03455). Sequence subtype classification for both the gag and gp41 fragments was based on their relation to the closest reference sequence. The reference sequences (accession numbers) that were used were A1 (U51190), A2 (AF286238), B (K03455), C (U46016), D (U88824), F1 (AJ249238), F2 (AJ249237), G (U88826), H (AF005496), J (AF082394), and K (AJ249239). The sequence analysis was based on a portion of the gp41 (HXB2 nucleotide [nt] 7867–8283) and gag (HXB2 nt 1240–1907) regions. Sequences generated for this study have been submitted to GenBank (accession numbers EU841418–EU841480 and EU850290–EU850351). Subjects were considered infected with a specific subtype if both regions analyzed were from the same subtype. If there was a discordance between the gag and gp41 subtype assignments, the subject was considered to be infected with a recombinant strain. Subtype assignments were confirmed by using the Web-based REGA subtyping tool (http://www.bioafrica.net/subtypetool/html). Sequence fragments that demonstrated any evidence of incorporating a recombinant breakpoint based on their position on the tree or from the REGA subtyping tool were further analyzed using SimPlot [18]. Potential break points were confirmed by generating a phylogenetic tree on each portion on both sides of the putative break point. If the 2 portions clustered significantly to 2 different subtypes, the sequence fragment was considered to have incorporated a recombination break point.
Data analysis
For each neuropsychological test, a Z score was calculated using age- and education-adjusted normative data obtained from 100 HIV-uninfected individuals in Uganda [1]. Distributional tests have confirmed that the resultant Z scores follow a normal distribution, and scores are summarized as mean ± standard deviation. Baseline differences in demographics were examined using the t test and the χ2 test among HIV-infected individuals infected with subtype A and D, the 2 predominant subtypes. A logistic regression model was used to examine for a difference between the frequency of HIV dementia among HIV-infected individuals infected with subtypes D and A. The association between subtype and MSK score was tested using a χ2 test of association.
RESULTS
The demographic characteristics of the 60 HIV-infected individuals for whom subtypes of the gp41 and gag regions were determined are summarized in Table 1, stratified by infecting HIV subtype. There were no differences in age, education, sex, CD4 lymphocyte count, or log plasma HIV RNA level, as stratified by infecting HIV subtype using clade assignment by phylogenetic analysis of gag only, gp41 only, or gag and gp41. There was also no difference in CD4 lymphocyte count or log plasma HIV RNA level among HIV-infected individuals with and without dementia.
Table 1.
Demographic Characteristics of Human Immunodeficiency Virus (HlV)–Infected Individuals, Stratified by HIV Subtype
| HIV subtype |
||||
|---|---|---|---|---|
| Characteristic | A | C | D | A/D recombinant |
| Patients for whom the gag gene fragment was analyzed | ||||
| No. of patients | 45 | 2 | 20 | |
| Age, years | 34.0 ± 0.9 | 37.0 ± 5.0 | 33.5 ± 1.2 | ... |
| Education, years | 9.0 ± 0.7 | 13.0 ± 0.0 | 10.5 ± 0.9 | ... |
| Percentage of male subjects | 27 | 50 | 30 | ... |
| CD4 lymphocyte count, cells/μL | 105.0 ± 10.8 | 51.0 ± 5.0 | 108.5 ± 14.7 | ... |
| Log plasma HIV RNA level, copies/mL | 5.3 ± 0.1 | 5.2 ± 0.6 | 5.2 ± 0.2 | ... |
| Patients for whom the gp41 gene fragment was analyzed | ||||
| No. of patients | 41 | 2 | 20 | ... |
| Age, years | 33.0 ± 0.9 | 37.0 ± 5.0 | 35.5 ± 1.4 | ... |
| Education, years | 8.0 ± 0.8 | 13.0 ± 0.0 | 9.5 ± 0.9 | ... |
| Percentage of male subjects | 27 | 50 | 35 | ... |
| CD4 lymphocyte count, cells/μL | 105.0 ± 11.3 | 51.0 ± 5.0 | 95.5 ± 17.9 | ... |
| Log plasma HIV RNA level, copies/mL | 5.3 ± 0.1 | 5.2 ± 0.6 | 5.2 ± 0.2 | ... |
| Patients for whom the gag and gp41 gene fragments were analyzed | ||||
| No. of patients | 33 | 2 | 9 | 16 |
| Age, years | 34.0 ± 1.0 | 37.0 ± 5.0 | 35.0 ± 1.9 | 32.5 ± 1.5 |
| Education, years | 8.0 ± 0.8 | 13.0 ± 0.0 | 10.0 ± 0.9 | 10.0 ± 1.2 |
| Percentage of male subjects | 27 | 50 | 44 | 18 |
| CD4 lymphocyte count, cells/μL | 90.0 ± 12.6 | 51.0 ± 5.0 | 66.0 ± 18.5 | 148.0 ± 18.4 |
| Log plasma HIV RNA level, copies/mL | 5.3 ± 0.2 | 5.2 ± 0.6 | 5.1 ± 0.3 | 5.3 ± 0.2 |
NOTE. Data are median ± standard error, unless otherwise indicated.
Analysis of the gag gene fragment
On the basis of phylogenetic analysis of the gag sequence fragment, 45 individuals were classified as being infected with subtype A1, 2 individuals were infected with subtype C, and 20 individuals were infected with subtype D. No sequences had any evidence of recombination. On the basis of the gag data only, there were no differences in the frequency of HIV dementia between individuals infected with subtype A (9, 24, and 12 individuals had MSK ratings of 0, 0.5, and ≥1, respectively) and subtype D (1, 9, and 10 individuals had ratings of 0, 0.5, and ≥1, respectively), although we noted a trend toward a greater frequency of HIV dementia among HIV subtype D–infected individuals (P = .15).
Analysis of the gp41 gene fragment
Forty-one individuals were classified as having subtype A infection, 2 individuals were classified as having subtype C infection, and 20 were classified as having subtype D infection on the basis of phylogenetic analysis of this fragment. No sequences demonstrated any evidence of recombination. For this fragment, differences in sub-types A1 and A2 could not be inferred and were all classified as subtype A. Individuals infected with subtype D were more likely to have HIV dementia than were those infected with subtype A (P = .02).
HIV subtype using phylogenetic analysis of the gag and gp41 gene
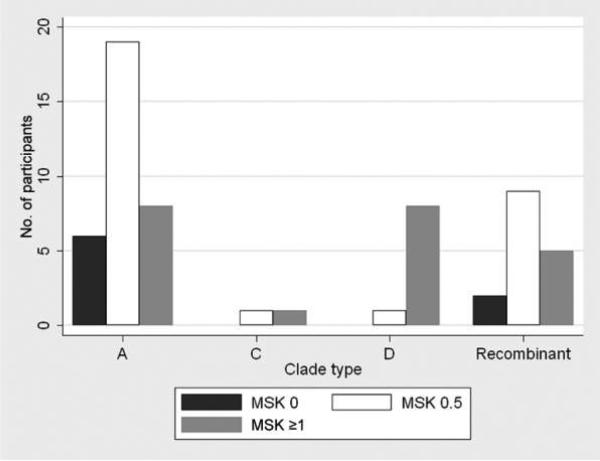
There were 60 subjects who had sequence data from both the gag and gp41 regions. If the sequence fragments had a concordant subtype for both regions, patients were considered to be infected with that subtype. Thirty-three HIV-infected individuals were classified as being infected with subtype A, 2 were classified as being infected with subtype C, 9 were classified as being infected with subtype D, and 16 were classified as being infected with recombinants with a discordant subtype assignment for gag and gp41. One-half the recombinants had a subtype A gag and D gp41, and the other half had a subtype D gag and A gp41. As shown in Figure 1, individuals with HIV subtype D infection had an increased frequency (89%) of HIV dementia (MSK stage, ≥1), compared with HIV subtype A–infected individuals (24%; P = .004). There was no difference in the frequency of HIV dementia between persons infected with A/D recombinants and persons infected with subtype A or between persons infected with A/D recombinants and persons infected with subtype D. HIV subtype D–infected individuals had more-impaired digit span forward performance, compared with HIV subtype A–infected individuals (P = .04) (Table 2). There were no differences between HIV subtype D–infected and HIV subtype A–infected individuals in any of the other individual neuropsychological tests.
Figure 1.
Frequency of human immunodeficiency virus (HIV)–associated cognitive impairment among antiretroviral-naive individuals in Uganda infected with HIV subtype A (n = 33), C (n = 2), D (n = 9), and A/ D recombinant (n = 16). HIV dementia stage was defined using the Memorial Sloan Kettering (MSK) stages of 0 (normal; ie, normal neurocognitive function), 0.5 (equivocal or subclinical; ie, equivocal symptoms or signs without impairment in capacity to perform activities of daily living; equivalent to asymptomatic neurocognitive impairment or mild neurocognitive disorder [19]), and ≥1 (HIV dementia; ie, unequivocal evidence [symptoms or signs including performance on neuropsychological tests] and mild-moderate functional impairment) [12].
Table 2.
Neuropsychological Test Differences Stratified by Subtype Using Analysis of gag and gp41 Genes
| Median score ± standard error, by HIV subtype |
||||
|---|---|---|---|---|
| Test | A (n = 33) | C (n = 2) | D (n = 9) | A/D recombinant (n = 16) |
| WHO UCLA AVLT trial 5 | 38.0 ± 1.2 | 39.5 ± 10.0 | 35.0 ± 1.8 | 36.5 ± 1.6 |
| Timed Gait | 9.0 ± 0.4 | 9.0 ± 0.4 | 9.7 ± 0.6 | 8.2 ± 0.3 |
| Grooved Pegboard Test | ||||
| Dominant hand | 85.0 ± 4.2 | 146.5 ± 79.5 | 98.0 ± 11.1 | 72.0 ± 5.4 |
| Nondominant hand | 102.0 ± 13.7 | 158.5 ± 77.5 | 121.0 ± 12.5 | 100.0 ± 6.3 |
| Symbol Digit Modalities Test | 23.0 ± 1.7 | 31.0 ± 4.0 | 20.0 ± 2.6 | 20.5 ± 2.2 |
| Color Trails Test | ||||
| Part 1 | 94.0 ± 11.0 | 75.5 ± 12.5 | 125.0 ± 12.8 | 88.0 ± 10.8 |
| Part 2 | 180.0 ± 16.4 | 270.5 ± 116.5 | 215.0 ± 24.7 | 174.0 ± 21.5 |
| Digit Span | ||||
| Forward | 6.0 ± 0.2 | 5.5 ± 0.5 | 5.0 ± 0.2a | 5.0 ± 0.2 |
| Backward | 3.0 ± 0.1 | 3.5 ± 0.5 | 3.0 ± 0.3 | 3.0 ± 0.2 |
| Finger tapping test | ||||
| Dominant hand | 165.0 ± 7.3 | 181.0 ± 21.0 | 163.0 ± 15.8 | 200.0 ± 12.0 |
| Nondominant hand | 172.0 ± 8.8 | NA | 192.5 ± 37.5 | 170.5 ± 10.0 |
| Category naming | 13.0 ± 0.8 | 15.0 ± 4.0 | 12.0 ± 1.1 | 13.0 ± 0.5 |
NOTE. NA, not available; WHO UCLA AVLT, World Health Organization–University of California-Los Angeles Auditory Verbal Learning test.
P = .04 for subtype D vs subtype A.
DISCUSSION
The results of our study suggest that, among HIV-infected individuals with advanced immunosuppression at risk of developing cognitive impairment, HIV dementia is more common among subtype D–infected persons than among subtype A–infected persons, as determined by phylogenetic analysis of both the gag and gp41 regions. These findings provide, to our knowledge, the first direct evidence in well-characterized HIV-infected individuals at a similar stage of HIV disease that HIV subtypes may have a different biological impact on neurological complications of HIV infection—namely, HAND. These results are consistent with previous data suggesting that HIV subtype D may be associated with more-rapid HIV disease progression—specifically, a lower CD4 cell count during follow-up and a faster progression to death—compared with subtype A. In a study of 140 HIV-infected seroconverters in Rakai, Uganda, 59% of subtype D–infected subjects experienced progression to AIDS or died, whereas 29% of subtype A–infected subjects experienced progression to AIDS, and none died [3]. Similar results were seen in another study of 1045 HIV-infected individuals in Uganda in which subtype D infection was associated with a lower CD4 cell count during follow-up and a faster progression to death [4]. Also, in a study from Senegal, the incidence of AIDS was 14.5 cases per 100 person-years for subtype D–infected individuals, 16.0 cases per 100 person-years for subtype C–infected individuals, and 3.5 cases per 100 person-years for subtype A–infected individuals [2, 20]. Another study of 428 HIV-infected women in Tanzania also found that subtype D–infected individuals experienced more-rapid progression to a CD4 cell count ≤200 cells/μL and World Health Organization stage 4 illness and a higher mortality rate, compared with subtype A–infected individuals [2, 5]. These data suggest that HIV subtypes may differ with respect to virulence and the risk of HIV-associated complications from advanced immunodeficiency.
A limitation to the study is that HIV-infected individuals at risk of developing cognitive impairment were involved in the study. They had abnormal performances on a screening test for HIV dementia (IHDS, ≤10), suggestive of HAND, and CD4 lymphocyte counts <200 cells/μL. However, dementia is primarily observed in patients with advanced immunosuppression (CD4 lymphocyte count, <200 cells/μL). In addition, our subtype distribution for the subjects studied in Kampala is remarkably similar to the subtype distribution of the polymerase gene among 279 women who attended a prenatal clinic in Kampala (subtype A, 53%; subtype D, 35%; subtype C, 2%; and recombinants, 10%) [21]. Nevertheless, the finding that 89% of subjects infected with subtype D had evidence of dementia, compared to 24% of those infected with subtype A, suggests that subtype D is associated with an increased risk of dementia. Additional studies need to be performed to determine whether the risk of HIV dementia is increased for subtype D versus subtype A among all HIV-infected individuals in Uganda.
One hypothesis for this differential rate of progression for HIV subtypes relates to differences in coreceptor use. HIV subtype A and subtype C favor the chemokine receptor CCR5 for viral entry throughout the course of infection [2, 22]. The use of CCR5 receptors seen in subtype A and C correlates with a nonsyncytium inducing macrophage-tropic version of HIV, which is associated with slower viral growth and replication [2, 22, 23]. In contrast, HIV subtype D displays a tropism for the syncytium-inducing chemokine receptor observed principally in T cells [2, 23] and possibly a dual tropism for both coreceptors [2, 5, 24]. CXCR4 T cell tropic strains are associated with more-rapid viral growth and replication, which could allow subtype D to infect more cells per unit of time than other subtypes, allowing for more-rapid HIV disease progression [2, 5].
With respect to potential mechanisms for a differential rate of neurovirulence by subtype, one possible mechanism relates to the tat gene. Recent data suggest that the tat gene in subtype C is associated with increased cell survival in rat hippocampal neuron cultures, compared with in subtype B [25], but the relative neurotoxicity of other clades, such as A and D, requires further study. The neurotoxicity of tat may occur through binding of the NMDA receptor, allowing for calcium-induced excitotoxicity, increased apoptosis, neuronal injury, and death. Another possible mechanism for an increased risk of neuro-pathogenesis from subtype D relates to differences in the V3 loop of the envelope gene [2, 26–29]. The hypervariable V3 loop of the envelope glycoprotein gp120 is involved in HIV entry into the CD4 cell, and clade D strains have been identified with a more-variable pattern of V3 loop amino acids than other subtypes [2, 27, 30]. Viral envelope diversity can influence the progression of neurological disease in several other retroviruses [2, 31, 32], and similar effects may be seen with the HIV envelope gene [2]. HIV dementia–associated strains differ predominantly in the V1 and V3 region of the gp120 envelope protein, which are the same regions that account for HIV subtype diversity [2, 28, 29, 32, 33]. Of note, in the phylogenetic analyses for subtype determination in this study, subtype D was associated with an increased risk of HIV dementia in the gp41 envelope gene, whereas only a trend was seen for an increased association of subtype D for HIV dementia in the gag gene. These results suggest that the envelope gene may be more critical for conferring an increased risk of HIV dementia than is the gag gene in subtype D–infected individuals. Differences in coreceptor use and syncytium inducibility as described above could also account for differences in neurovirulence.
The genetic diversity of the HIV virus is due to many factors, including its high replication rates, lack of a proofreading capacity with mismatch errors, and a propensity for recombination [2, 34, 35]. The HIV virus has more genetic diversity and interclade recombination occurs more frequently in Africa than in the United States [2, 36]. Thus, Africa is an ideal setting to examine issues related to subtype and the risk of HIV dementia. In Uganda, where 2 HIV subtypes predominate, our results suggest that subtype D is associated with an increased risk of HIV dementia. Additional studies are needed to confirm this observation. The precise mechanism by which subtype D leads to an increased risk of HIV dementia remains to be elucidated, but differences in the tat gene, envelope gene, coreceptor use, and syncytium inducibility may be involved. Additional studies are needed globally to define the frequency of HIV dementia in all subtypes and to define the rate of progression of HAND within specific HIV subtypes. The effect of HIV subtype on other neurological complications of HIV infection, such as sensory neuropathy and opportunistic infection of the central nervous system, also should be examined.
Acknowledgments
We thank Dr Andrew Kambugu and the staff of the Infectious Disease Clinic at Mulago Hospital for their assistance and participation. We also thank the 2 research assistants for the study, Alice Namudde and Julian Nkarayija, and Ms. Marie Sonderman for administrative assistance. In addition, we thank all of the HIV-infected patients who participated in the study at the Infectious Disease Institute in Kampala, Uganda.
Financial support. The Neurologic AIDS Research Consortium, which receives support from National Institute of Neurological Disorders and Stroke (NS 32228), National Institute of Mental Health (MH71150); and the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases.
Footnotes
Potential conflicts of interest. All authors: no conflicts.
References
- 1.Wong M, Robertson K, Nakasujja N, et al. Frequency of and risk factors for HIV dementia in an HIV clinic in sub-Saharan Africa. Neurology. 2007;68:350–5. doi: 10.1212/01.wnl.0000252811.48891.6d. [DOI] [PubMed] [Google Scholar]
- 2.Liner KJ, Hall CD, Robertson KR. Impact of human immunodeficiency virus (HIV) subtypes on HIV-associated neurological disease. J Neurovirol. 2007;13:291–304. doi: 10.1080/13550280701422383. [DOI] [PubMed] [Google Scholar]
- 3.Kiwanuka N, Laeyendecker O, Robb M, et al. Effect of Human Immunodeficiency Virus Type 1 (HIV-1) Subtype on Disease Progression in Persons from Rakai, Uganda, with Incident HIV-1 Infection. J Infect Dis. 2008;197:707–13. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 4.Kaleebu P, French N, Mahe C, et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J Infect Dis. 2002;185:1244–50. doi: 10.1086/340130. [DOI] [PubMed] [Google Scholar]
- 5.Vasan A, Renjifo B, Hertzmark E, et al. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis. 2006;42:843–52. doi: 10.1086/499952. [DOI] [PubMed] [Google Scholar]
- 6.Baeten JM, Chohan B, Lavreys L, et al. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195:1177–80. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 7.Sacktor N, Nakasujja N, Skolasky R, et al. Benefits and risks of stavudine therapy for HIV-associated neurologic complications in Uganda. Neurology. 2009;72:165–70. doi: 10.1212/01.wnl.0000339042.96109.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacktor NC, Wong M, Nakasujja N, et al. The International HIV Dementia Scale: a new rapid screening test for HIV dementia. AIDS. 2005;19:1367–74. [PubMed] [Google Scholar]
- 9.Karnofsky DA, Abelman WH, Craver LF, Burchenal J. The use of nitrogen mustards in the palliative treatment of carcinoma. Cancer. 1948;1:634–56. [Google Scholar]
- 10.Maj M, Satz P, Janssen R, et al. WHO Neuropsychiatric AIDS Study, Cross-sectional Phase II. Neuropsychological and neurological findings. Arch Gen Psychiatry. 1994;51:51–61. doi: 10.1001/archpsyc.1994.03950010051007. [DOI] [PubMed] [Google Scholar]
- 11.Smith A. Symbol digit modalities test. Western Psychological Service; Los Angeles: 1982. [Google Scholar]
- 12.Price RW, Brew BJ. The AIDS dementia complex. J Infect Dis. 1988;158:1079–83. doi: 10.1093/infdis/158.5.1079. [DOI] [PubMed] [Google Scholar]
- 13.Yang C, Dash BC, Simon F, et al. Detection of diverse variants of human immunodeficiency virus-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–5. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]
- 14.Yang C, Dash B, Hanna SL, et al. Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2001;17:361–5. doi: 10.1089/08892220150503726. [DOI] [PubMed] [Google Scholar]
- 15.Hall TA. BioEdit: a user-friendly biological sequencealignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 16.Felsentein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Carr JK, Salminen MO, Koch C, et al. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–43. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lole KS, Bollinger RC, Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antinori A, Arendt G, Becker JT, et al. Updated research nosology for HIV-associated neurocognitive disorders (HAND). Neurology. 2007;69:1789–99. doi: 10.1212/01.WNL.0000287431.88658.8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanki PJ, Hamel DJ, Sankale JL, et al. Human immunodeficiency virus type 1 subtypes differ in disease progression. J Infect Dis. 1999;179:68–73. doi: 10.1086/314557. [DOI] [PubMed] [Google Scholar]
- 21.Eshleman SH, Guay LA, Mwatha A, et al. Comparison of mother-to-child transmission rates in Ugandan women with subtype A versus D HIV-1 who received single-dose nevirapine prophylaxis: HIV Network For Prevention Trials 012. J Acquir Immune Defic Syndr. 2005;39:593–7. [PubMed] [Google Scholar]
- 22.Peeters M, Vincent R, Perret JL, et al. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–21. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 23.Tscherning C, Alaeus A, Fredriksson R, et al. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology. 1998;241:181–8. doi: 10.1006/viro.1997.8980. [DOI] [PubMed] [Google Scholar]
- 24.Kaleebu P, Nankya IL, Yirrell DL, et al. Relation between chemokine receptor use, disease stage, and HIV-1 subtypes A and D: results from a rural Ugandan cohort. J Acquir Immune Defic Syndr. 2007;45:28–33. doi: 10.1097/QAI.0b013e3180385aa0. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Huang Y, Reid R, et al. NMDA receptor activation by HIV-Tat protein is clade dependent. J Neurosci. 2008;28:12190–8. doi: 10.1523/JNEUROSCI.3019-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Jong JJ, De Ronde A, Keulen W, et al. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–80. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong P, Peeters M, Janssens W, et al. Correlation between genetic and biological properties of biologically cloned HIV type 1 viruses representing subtypes A, B, and D. AIDS Res Hum Retroviruses. 1995;11:239–48. doi: 10.1089/aid.1995.11.239. [DOI] [PubMed] [Google Scholar]
- 28.Dunfee RL, Thomas ER, Gorry PR, et al. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A. 2006;103:15160–5. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pillai SK, Pond SL, Liu Y, et al. Genetic attributes of cerebrospinal fluid-derived HIV-1 env. Brain. 2006;129:1872–83. doi: 10.1093/brain/awl136. [DOI] [PubMed] [Google Scholar]
- 30.de Wolf F, Hogervorst E, Goudsmit J, et al. Syncytium-inducing and non-syncytium-inducing capacity of human immunodeficiency virus type 1 subtypes other than B: phenotypic and genotypic characteristics. WHO Network for HIV Isolation and Characterization. AIDS Res Hum Retroviruses. 1994;10:1387–400. doi: 10.1089/aid.1994.10.1387. [DOI] [PubMed] [Google Scholar]
- 31.Johnston JB, Jiang Y, van Marle G, et al. Lentivirus infection in the brain induces matrix metalloproteinase expression: role of envelope diversity. J Virol. 2000;74:7211–20. doi: 10.1128/jvi.74.16.7211-7220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mankowski JL, Flaherty MT, Spelman JP, et al. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–60. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritola K, Robertson K, Fiscus SA, et al. Increased human immunodeficiency virus type 1 (HIV-1) env compartmentalization in the presence of HIV-1-associated dementia. J Virol. 2005;79:10830–4. doi: 10.1128/JVI.79.16.10830-10834.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liesch GR, DeStefano JJ. Analysis of mutations made during active synthesis or extension of mismatched substrates further define the mechanism of HIV-RT mutagenesis. Biochemistry. 2003;42:5925–36. doi: 10.1021/bi026998n. [DOI] [PubMed] [Google Scholar]
- 35.Simon V, Ho DD. HIV-1 dynamics in vivo: implications for therapy. Nat Rev Microbiol. 2003;1:181–90. doi: 10.1038/nrmicro772. [DOI] [PubMed] [Google Scholar]
- 36.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 2006. 20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]