This article reports the association of IGF-1 with human myopia and supports previous studies in animal models on IGF-1's role in axial ocular growth during myopia development.
Abstract
Purpose.
Evidence from human myopia genetic mapping studies (MYP3 locus), modulated animal models, and observations of glycemic control in humans suggests that insulin-like growth factor (IGF)-1 plays a role in the control of eye growth. This study was conducted to determine whether IGF-1 polymorphisms are associated with myopia in a large, international dataset of Caucasian high-grade myopia pedigrees.
Methods.
Two hundred sixty-five multiplex families with 1391 subjects participated in the study. IGF-1 genotyping was performed with 13 selected tag single nucleotide polymorphisms (SNPs) using allelic discrimination assays. A family-based pedigree disequilibrium test (PDT) was performed to test for association. Myopia status was defined using sphere (SPH) or spherical equivalent (SE), and analyses assessed the association of (1) high-grade myopia (≤ −5.00 D), and (2) any myopia (≤ −0.50 D) with IGF-1 markers. Results were declared significant at P ≤ 0.0038 after Bonferroni correction. Q values that take into account multiple testing were also obtained.
Results.
In all, three SNPs—rs10860860, rs2946834, and rs6214—were present at P < 0.05. SNP rs6214 showed positive association with both the high-grade– and any-myopia groups (P = 2 × 10−3 and P = 2 × 10−3, respectively) after correction for multiple testing.
Conclusions.
The study supports a genetic association between IGF-1 and high-grade myopia. These findings are in line with recent evidence in an experimental myopia model showing that IGF-1 promotes ocular growth and axial myopia. IGF-1 may be a myopia candidate gene for further investigation.
Myopia is the most common eye disorder, affecting approximately one third of the population older than 12 years in the United States.1 The public health impact, along with the associated costs of optical correction, is substantial.2,3 High-grade myopia can increase the risk of cataract, glaucoma, retinal detachment, and chorioretinal degeneration.4 The etiology of myopia is not fully understood, but both genetic and environmental factors are believed to be involved in its development and progression. Several genetic mapping studies have been performed to identify cytogenetic susceptibility regions for myopia,5 and research is ongoing to identify causative genes involved in myopia development and progression.
Insulin-like growth factor (IGF)-1 is a polypeptide that plays an important role in cell proliferation, differentiation, and apoptosis. Evidence from human genetic mapping studies, experimental myopia induction in animal models, and observations of an association between poor glycemic control and myopia in humans suggests that IGF-1 also has a role in controlling eye growth.
Before the present study, we had mapped a novel autosomal dominant high-grade myopia locus (MYP3; OMIM 603221; Online Mendelian Inheritance in Man; http://www.ncbi.nlm.nih.gov/Omim/ provided in the public domain by the National Center for Biotechnology Information, Bethesda, MD) to the long arm of chromosome 12, region 21-23, in a large German/Italian family.6 Linkage analysis in a cohort of 51 U.K. families suggested that the MYP3 locus could be responsible for high myopia in approximately 25% of the families.7 More recently, the MYP3 locus was replicated by our group with a whole-genome linkage scan involving 254 families with high-grade myopia from the Myopia International Consortium8 and by Nurnberg et al.9 in a large, six-generation German kindred. IGF-1 maps to 12q23.2 and lies within the MYP3 locus.
In two independent studies published recently, the chick model of experimental myopia showed accelerated ocular growth after intravitreal injections of insulin or IGF-1, suggesting that these molecules are powerful stimulators of axial myopia.10,11 Furthermore, the receptor expression levels for the IGF-1 gene in the chick model were upregulated in the retinal pigment epithelial layer during experimental myopia induction (−10-D lens treatment; Zhang Z, et al. IOVS 2007;48:ARVO E-Abstract 4417).
Cordain et al.12 speculated that high-glycemic-load carbohydrate diets may contribute to the development and progression of myopia, perhaps by affecting sensitivity to insulin or increasing free-circulating IGF-1 levels. This hypothesis is supported by a retrospective study that revealed poor metabolic control of glucose to be a risk factor for myopia.13 Also, in patients with primary growth hormone insensitivity, IGF-1 is an important regulator of ocular growth.14 It should be noted that the specific role of IGF-1 in myopigenesis in the context of glycemic control is currently unknown and remains to be investigated in epidemiologic studies.
The role of IGF-1 gene polymorphisms in association with human myopia has not been investigated to date. Therefore, we ascertained a large Caucasian cohort biased toward high-grade myopia and performed genetic association analyses of polymorphisms of IGF-1 with myopia refractive error phenotypes.
Materials and Methods
Subject Selection
Informed consent was acquired from all subjects before they entered the study, and the principles of the Declaration of Helsinki were followed. The study was approved by the Institutional Review Board at the Duke University Medical Center (Durham, NC). High-grade myopia status was assigned to sphere (SPH) or spherical equivalent (SE) refractive error [defined by sphere + (cylinder/2)] of −5.00 D or more in at least one eye. Families were eligible for inclusion if they had one member with high-grade myopia, and all available family members were recruited. Complete ophthalmic examinations were performed, including cycloplegic retinoscopy and/or autorefraction (model RM-8800; Topcon Medical Systems, Inc., Paramus, NJ). Refractions were recorded in positive cylinder format, and manifest refraction data were used in the analyses. Patients with systemic conditions, syndromic disorders, or ophthalmic conditions that could predispose to high myopia were excluded. A total of 265 multiplex high-grade myopia Caucasian families that included 1391 subjects participated in the study; the sample encompassed all families from the International Myopia Consortium.8 Of these, 153 families were ascertained in the United States (Duke University), 46 in the United Kingdom (Cardiff University), 25 in Denmark (National Eye Clinic, Kennedy Institute), 1 in Australia (University of Melbourne), and 40 in France (Toulouse University).
Marker Selection and Genotyping
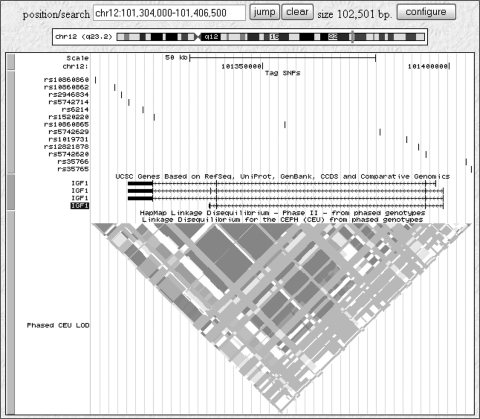
The SNPSelector program15 was used to choose tagging SNPs that met the criteria of a Pearson-squared correlation (r2) threshold of 0.67 in the linkage disequilibrium bins and a minor allele frequency (MAF) greater than 5% in the HapMap CEU population. Thirteen tag SNPs were selected, to provide full gene coverage. The SNPs chosen for the candidate gene are shown in Figure 1.
Figure 1.
Representation of the 13 SNPs selected for the IGF-1 gene (source; Human Genome Browser (http://genome.ucsc.edu/ provided in the public domain by UCSC Genome Bioinformatics, University of California at Santa Cruz). The HapMap linkage disequilibrium map for the region for European samples from the CEPH (30 trios) is also displayed.
After venous blood sample collection, total genomic DNA was extracted (AutoPure LS DNA Extractor and Puregene reagents; Gentra Systems Inc; Minneapolis, MN). For genotyping reactions, custom allelic discrimination assays were performed (TaqMan; Applied Biosystems [ABI], Foster City, CA), consisting of unlabeled polymerase chain reaction (PCR) primers and the minor groove binding (MGB) group probes (TaqMan FAM and VIC dye-labeled; ABI). The assays comprised two unlabeled PCR primers and two allele-specific probes. PCR reactions were then performed (Taqman Universal PCR Master Mix, GeneAmp PCR System 9700; ABI), and the allelic discrimination calls were read (7900HT Fast PCR System; ABI).
All 1391 subjects who participated in the study were genotyped. For quality control purposes, two CEPH (Centre d'Etude du Polymorphisme Humain) DNA standards were included in each 96-well plate, and two blind duplicate samples were used per plate. Genotype submission to the analysis database required 100% matching quality control genotypes within and across plates and with at least 95% genotyping efficiency. All genotypes were in Hardy-Weinberg equilibrium (HWE). Two datasets with unrelated samples were formed in which one affected individual sample per family was randomly selected to cluster within a designated affected group, and one unaffected individual sample per family was selected to add to a designated unaffected group. An exact test implemented in the Genetic Data Analysis (GDA) program was used to test HWE in which 3200 permutations were performed to estimate the empiric P value for each marker.16
Statistical Analyses
Analyses assessed the following myopic refractive error phenotypes (1) high myopia (≤−5.0D), and (2) any myopia (≤−0.5D). The phenotypes were based on either SPH or SE. The dataset was stratified in this manner to investigate the effect of genetic background on common myopia within this cohort. It is notable that more than 85% of the cohort was older than 20 years (an age by which refractive state becomes relatively stable). The family-based pedigree disequilibrium test (PDT)17 was performed to test the association of IGF-1 polymorphisms with each myopia phenotype state. After Bonferroni correction for multiple testing, statistical significance was defined as P ≤ 0.0038 (0.05/13). Although we did not account for the multiple phenotypes tested to set this threshold, the somewhat less-stringent Bonferroni correction is justified, because the phenotypes studied are nonindependent. We also used a less conservative multiple-correction method for a comparison, for which we obtained q-values for all markers with the Q-Value program (http://faculty.washington.edu/jstorey/qvalue/).18 This program converts the P value to a q value at a marker, where q value is the estimate of the proportion of false positives incurred (false-discovery rate) when the marker is called significant.
Results
The number of affected and unaffected subjects for each classification is described in detail in Table 1. Table 2 depicts clinical information for the dataset based on SPH classifications. Among 13 IGF-1 SNPs tested, three SNPs—rs10860860, rs2946834, and rs6214—showed P < 0.05. The SNP rs6214 showed significant association with the high-grade– and any-myopia groups (P = 2 × 10−3, q = 0.08) after correction for multiple testing. The positive association was consistent with analyses performed on SPH and SE phenotypes. Table 3 provides the P values (PDT) for all the polymorphisms tested.
Table 1.
Summary of Number of Subjects across Various Phenotype Classifications in the Cohort
| Group | Definition Based on Myopia Severity | Affected (A) | Unaffected (N) | Unknown (U) | Total |
|---|---|---|---|---|---|
| High-grade myopia | SPH Data: A ≤ −5 D, N ≥ −0.49 D, U others | 628 | 368 | 395 | 1391 |
| SE Data: A ≤ −5 D, N ≥ −0.49 D, U others | 580 | 433 | 378 | 1391 | |
| Any myopia | SPH Data: A ≤ −0.5 D, N ≥ 0 D, −0.5 D < U < 0 D | 1023 | 341 | 27 | 1391 |
| SE Data: A ≤ −0.5 D, N ≥ 0 D, −0.5 D < U < 0 D | 958 | 391 | 42 | 1391 |
Table 2.
Clinical Information Data
| High-Grade Myopia |
Any Myopia |
|||||
|---|---|---|---|---|---|---|
| Affected (A) | Unaffected (N) | Unknown (U) | Affected (A) | Unaffected (N) | Unknown (U) | |
| Sample size by SPH | 628 | 368 | 395 | 1023 | 341 | 27 |
| Age at examination, y | 43.47 ± 20.43 (93) | 45 ± 20.47 (55) | 43.54 ± 21.49 (69) | 43.50 ± 20.83 (162) | 44.56 ± 20.11 (48) | 48 ± 24.34 (7) |
| Sphere OD, D | −9.59 ± 4.6 (624) | 0.74 ± 1.29 (242) | −2.04 ± 1.42 (394) | −6.67 ± 5.22 (1018) | 0.85 ± 1.33 (215) | −0.13 ± 0.188 (27) |
| Sphere OS, D | −9.54 ± 4.37 (625) | 0.76 ± 1.58 (241) | −2.04 ± 1.42 (389) | −6.66 ± 5.07 (1014) | 0.81 ± 1.44 (215) | 0.33 ± 2.44 (26) |
| Cylinder OD, D | 1.02 ± 1.06 (550) | 0.77 ± 0.98 (284) | 0.81 ± 0.94 (344) | 0.94 ± 1.02 (894) | 0.79 ± 1.0 (262) | 0.55 ± 0.62 (22) |
| Cylinder OS, D | 0.99 ± 1.01 (547) | 0.91 ± 0.94 (285) | 0.80 ± 0.86 (342) | 0.92 ± 0.96 (889) | 0.88 ± 0.95 (262) | 0.70 ± 0.82 (23) |
Numbers in parentheses denote the number of subjects providing the data.
Table 3.
Pedigree Disequilibrium Test Analyses of the IGF-1 Gene Tag SNPs
| High-Grade Myopia |
Any Myopia |
|||
|---|---|---|---|---|
| (SPH) | (SE) | (SPH) | (SE) | |
| rs6214 | 0.0022 | 0.003 | 0.0029 | 0.0021 |
| rs12821878 | 0.2994 | 0.3458 | 0.0845 | 0.1448 |
| rs5742714 | 0.7582 | 1 | 0.2457 | 0.3312 |
| rs10860862 | 0.0778 | 0.2087 | 0.2553 | 0.2312 |
| rs1520220 | 0.829 | 0.9379 | 0.5557 | 0.5367 |
| rs5742629 | 0.2786 | 0.2695 | 0.2299 | 0.4207 |
| rs35765 | 0.5408 | 0.3023 | 0.4987 | 0.2433 |
| rs35766 | 0.2013 | 0.0668 | 0.245 | 0.1368 |
| rs2946834 | 0.2803 | 0.1175 | 0.0549 | 0.0326 |
| rs1019731 | 0.4772 | 0.3507 | 0.9383 | 0.7533 |
| rs10860860 | 0.0312 | 0.0167 | 0.024 | 0.0248 |
| rs5742620 | 0.1699 | 0.1495 | 0.3827 | 0.4733 |
Probabilities shown in bold denote significant differences.
Discussion
IGF-1 has been a target candidate gene of genetic association studies for numerous human diseases and characteristics such as diabetes, diabetic retinopathy (DR), cancer susceptibility, osteoarthritis, body weight, growth, menarche, and longevity.19 It has been implicated in ocular diseases such as proliferative DR, retinopathy of prematurity (ROP), and age-related macular degeneration (AMD).20–24 In the present study, IGF-1 polymorphisms were tested for possible association with myopia and the SNP rs6214 showed significant association with the high-grade– and any-myopia phenotypes.
The SNP rs6214 is located in the 3′-untranslated region (UTR) of the IGF-1 gene. The minor allele frequencies in European, Han Chinese, Japanese, and Yoruba populations are 0.421, 0.467, 0.568, and 0.533, respectively, based on HapMap data (www.hapmap.org). 3′-UTR variants may play an important role in disease by regulating gene expression, as 3′ regulatory regions control mRNA 3′ end formation, stability, nuclear export, subcellular localization, and translation.25,26 The association of IGF-1 with high-grade myopia may be due to a similar mechanism involving regulation of gene expression levels. Arends et al.27 hypothesized that minor genetic variation in the IGF-1 gene could influence pre- and postnatal growth. Thus, the speculation that IGF-1 may also influence ocular growth is in line with the findings that body size parameters can predict eye size to a large extent, indicating common genetic influences on both body and eye growth.28 However, it should be noted that the referenced study reports findings from a breeding experiment in chickens, and it is unclear whether such findings would translate to mammals or humans.
It is more than likely that a functional polymorphism that is in strong LD with rs6214 may be involved in myopia. Although we did not observe any stronger association with other IGF-1 SNPs, genes centromeric to IGF-1, which include nucleoporin 37 kDa (NUP37), coiled-coil domain containing 53 (CCDC53), promelanin-concentrating hormone (PMCH), and hypothetical protein LOC55010 (C12orf48), are not obvious candidates and are only in modest LD with the IGF-1 gene (based on HapMap CEU population data). Although the exact role of IGF-1 in myopia development is not known, the finding that insulin and IGF-1 promote ocular axial elongation in the chick model of myopia make IGF-1 an obvious candidate for further investigations in both human and animal studies.
Hypothesis-driven candidate gene association studies have been successfully used to identify causative genes in other diseases. Genetic association studies have recently generated much interest in the field of myopia genetics, and it is important to be cautious regarding potential false-positive findings, although this line of research is still relatively young. Validation and replication of findings is important, the lack of which can be due to several factors, from publication bias to interstudy heterogeneity. Careful study design and analyses play an important role in avoiding this problem and in making cautious yet meaningful interpretations. In the present study, we used an effective SNP selection strategy, definitive ascertainment criteria, and phenotype classification to perform a comprehensive search of markers within the IGF-1 gene that may be associated with myopia. Rigorous multiple-correction criteria were applied that pinpointed the association of rs6214 with myopia.
In summary, we found that the IGF-1 gene polymorphisms were genetically associated with high-grade myopia in a Caucasian family-based dataset. The current findings, along with the evidence that IGF-1 was located within a myopia susceptibility locus and its direct involvement in myopia induction in an animal model, warrant further studies on its role in myopia.
Acknowledgments
The authors thank the members of all myopia families for their participation and Bei Zhao for assistance with data analysis.
Footnotes
Supported by Grants R01 EY014685 from National Institute of Health (NIH) and a Research to Prevent Blindness, Inc. Lew Wasserman Award (TLY).
Disclosure: R. Metlapally, None; C.-S. Ki, None; Y.-J. Li, None; K.-N. Tran-Viet, None; D. Abbott, None; F. Malecaze, None; P. Calvas, None; D.A. Mackey, None; T. Rosenberg, None; S. Paget, None; J.A. Guggenheim, None; T.L. Young, None
References
- 1.Vitale S, Ellwein L, Cotch MF, Ferris FL, III, Sperduto R. Prevalence of refractive error in the United States 1999–2004. Arch Ophthalmol. 2008;126:1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norton TT, Metlapally R, Young TL. Myopia. In: Klintworth GK, Garner A. eds. Garner and Klintworth's Pathobiology of Ocular Disease. 3rd ed.London, UK: Informa Healthcare; 2008:537–556 [Google Scholar]
- 3.Vitale S, Cotch MF, Sperduto R, Ellwein L. Costs of refractive correction of distance vision impairment in the United States 1999–2002. Ophthalmology. 2006;113:2163–2170 [DOI] [PubMed] [Google Scholar]
- 4.Curtin BJ. The Myopias: Basic Science and Clinical Management. Philadelphia: Harper & Row; 1985 [Google Scholar]
- 5.Young TL, Metlapally R, Shay AE. Complex trait genetics of refractive error. Arch Ophthalmol. 2007;125:38–48 [DOI] [PubMed] [Google Scholar]
- 6.Young TL, Ronan SM, Alvear AB, et al. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farbrother JE, Kirov G, Owen MJ, Pong-Wong R, Haley CS, Guggenheim JA. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci. 2004;45:2879–2885 [DOI] [PubMed] [Google Scholar]
- 8.Li YJ, Guggenheim JA, Bulusu A, et al. An international collaborative family-based whole genome linkage scan for high-grade myopia. Invest Ophthalmol Vis Sci. 2009;50(7):3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nurnberg G, Jacobi FK, Broghammer M, et al. Refinement of the MYP3 locus on human chromosome 12 in a German family with Mendelian autosomal dominant high-grade myopia by SNP array mapping. Int J Mol Med. 2008;21:429–438 [PubMed] [Google Scholar]
- 10.Feldkaemper MP, Neacsu I, Schaeffel F. Insulin acts as a powerful stimulator of axial myopia in chicks. Invest Ophthalmol Vis Sci. 2009;50:13–23 [DOI] [PubMed] [Google Scholar]
- 11.Zhu X, Wallman J. Opposite effects of glucagon and insulin on compensation for spectacle lenses in chicks. Invest Ophthalmol Vis Sci. 2009;50:24–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordain L, Eaton SB, Brand MJ, Lindeberg S, Jensen C. An evolutionary analysis of the aetiology and pathogenesis of juvenile-onset myopia. Acta Ophthalmol Scand. 2002;80:125–135 [DOI] [PubMed] [Google Scholar]
- 13.Jacobsen N, Jensen H, Lund-Andersen H, Goldschmidt E. Is poor glycaemic control in diabetic patients a risk factor of myopia? Acta Ophthalmol. 2008;86:510–514 [DOI] [PubMed] [Google Scholar]
- 14.Bourla DH, Laron Z, Snir M, Lilos P, Weinberger D, Axer-Siegel R. Insulinlike growth factor I affects ocular development: a study of untreated and treated patients with Laron syndrome. Ophthalmology. 2006;113:1197–2005 [DOI] [PubMed] [Google Scholar]
- 15.Xu H, Gregory SG, Hauser ER, et al. SNPselector: a web tool for selecting SNPs for genetic association studies. Bioinformatics. 2005;21:4181–4186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaykin D, Zhivotovsky L, Weir BS. Exact tests for association between alleles at arbitrary numbers of loci. Genetica. 1995;96:169–178 [DOI] [PubMed] [Google Scholar]
- 17.Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storey JD. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498 [Google Scholar]
- 19.Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am J Epidemiol. 2006;164:1–4 [DOI] [PubMed] [Google Scholar]
- 20.Simo R, Lecube A, Segura RM, Garcia AJ, Hernandez C. Free insulin growth factor-I and vascular endothelial growth factor in the vitreous fluid of patients with proliferative diabetic retinopathy. Am J Ophthalmol. 2002;134:376–382 [DOI] [PubMed] [Google Scholar]
- 21.Ruberte J, Ayuso E, Navarro M, et al. Increased ocular levels of IGF-1 in transgenic mice lead to diabetes-like eye disease. J Clin Invest. 2004;113:1149–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rietveld I, Ikram MK, Vingerling JR, et al. An igf-I gene polymorphism modifies the risk of diabetic retinopathy. Diabetes. 2006;55:2387–2391 [DOI] [PubMed] [Google Scholar]
- 23.Hellstrom A, Perruzzi C, Ju M, et al. Low IGF-I suppresses VEGF-survival signaling in retinal endothelial cells: direct correlation with clinical retinopathy of prematurity. Proc Natl Acad Sci U S A. 2001;98:5804–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambooij AC, van Wely KH, Lindenbergh-Kortleve DJ, Kuijpers RW, Kliffen M, Mooy CM. Insulin-like growth factor-I and its receptor in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2003;44:2192–2198 [DOI] [PubMed] [Google Scholar]
- 25.Chen JM, Ferec C, Cooper DN. A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet. 2006;120:1–21 [DOI] [PubMed] [Google Scholar]
- 26.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6:637–641 [DOI] [PubMed] [Google Scholar]
- 27.Arends N, Johnston L, Hokken-Koelega A, et al. Polymorphism in the IGF-I gene: clinical relevance for short children born small for gestational age (SGA). J Clin Endocrinol Metab. 2002;87:2720. [DOI] [PubMed] [Google Scholar]
- 28.Prashar A, Hocking PM, Erichsen JT, Fan Q, Saw SM, Guggenheim JA. Common determinants of body size and eye size in chickens from an advanced intercross line. Exp Eye Res. 2009;89:42–48 [DOI] [PubMed] [Google Scholar]