The RIM101 pathway of Candida albicans signals the expression of fungal genes involved in adaptation and morphogenesis. Fungal mutants lacking Rim8p, Rim13p, Rim20p, or Rim101p or the downstream cell-wall protein Phr1p, showed that this pH-responsive pathway contributes to the pathogenicity of C. albicans for the cornea.
Abstract
Purpose.
To examine the role of the fungal RIM101 signal transduction pathway in the pathogenesis of Candida albicans keratitis.
Methods.
C. albicans wild-type strain SC5314, prototrophic mutant control DAY185, and homozygous fungal mutants for the rim8, rim13, rim20, rim101, and phr1 genes were evaluated in vitro using proliferation and filamentation assays. Scarified corneas of BALB/c and C57BL/6J mice were topically inoculated and observed daily for keratitis severity. Corneal adaptation and pathogenicity were assessed ex vivo by maintaining infected porcine corneas for 3 days in an explantation culture system for histologic evaluation of hyphal penetration.
Results.
All C. albicans strains had similar growth kinetics, and SC5314 and DAY185 demonstrated pH-induced filamentation. Fungal mutants had reduced hyphal formation at alkaline and neutral pH, but normal acidic assays ascertained that mutant strains did not have a generalized filamentation defect. SC5314 and DAY185 caused moderate to severe keratitis in mice, whereas fungal strains lacking constituents of the RIM101 pathway had significantly (P < 0.05) attenuated severity in vivo. Three days after inoculation of porcine corneas, SC5314 and DAY185 produced hyphae that penetrated 28% and 25%, respectively, of the corneal thickness, and all five mutant strains showed significantly (P < 0.05) less stromal penetration.
Conclusions.
The RIM101 signal transduction pathway plays an important role in the development of C. albicans keratitis. The fungal pathway intermediates Rim8p, Rim13p, Rim20p, and Rim101p and the downstream cell-wall protein Phr1p are pivotal in the process of corneal invasion by C. albicans.
The fungus Candida albicans causes systemic mycosis, opportunistic mucosal candidiasis, endogenous endophthalmitis, and suppurative keratitis.1–4 Attributes of C. albicans the contribute to adaptation and tissue invasion include the secretion of degradative enzymes and the ability to switch from yeasts to filamentous forms.5 C. albicans keratitis occurs when, in response to conditions at the ocular surface, blastospores transition from saprophytic commensals into invasive microorganisms. After attachment to the injured cornea, pseudohyphae and hyphae invade the corneal stroma.6
Morphologic versatility is a distinctive feature of dimorphic fungi such as Candida species. Specifically, the pleomorphic nature of C. albicans allows a range of growth patterns from yeasts to filaments that develop as apical extensions from blastospores.7,8 Pseudohyphae and hyphae facilitate attachment and invasiveness, properties associated with fungal virulence for host tissues and the pathogenic process.
C. albicans uses a set of conserved pathways to regulate its morphogenic state, including the RIM101 pathway, the mitogen-activated protein (MAP) kinase pathway, a cAMP-dependent protein kinase pathway, the CZF1 matrix pathway, and Tup1-mediated repression.9 When environmental cues are sensed by fungi, the transduction pathway specific for a particular signal is activated, resulting in corresponding transcription factors that induce fungal genes to produce proteins enabling hyphal filamentation.10–13
Ambient pH influences fungal filamentous growth through a conserved pathway first identified in Saccharomyces cerevisiae and C. albicans,14,15 with a homologous cascade found in ascomycetes and basidiomycetes.16–19 Named the RIM101 pathway or PacC pathway, depending on the fungal genus, a key intermediate is the zinc finger–containing transcription factor Rim101p/PacC.19–23 Under acidic conditions Rim101p/PacC is full-length and inactive. However, at neutral to alkaline conditions its C-terminal portion is cleaved, resulting in activation of the protein subunit that alters gene expression.24–27 In the RIM101 signal transduction pathway, Rim13p is responsible for the proteolytic activation of the transcription factor Rim101p.15,20
Rim101p contributes to virulence in models of disseminated candidiasis.15,22,28 Previous studies suggest that the pH responsive RIM101 pathway may also be involved in the pathogenesis of fungal corneal infection. For example, in a model of experimental murine keratitis, an insertional mutation of rim13 results in attenuated virulence of C. albicans.6,29 In addition, the transcription factor Efg1p, a key component of several signal transduction pathways including the RIM101 pathway, is pivotal for filamentous growth of C. albicans during the establishment of keratomycosis. A lack of efg1 diminishes corneal virulence, whereas a reintegrant is invasive and virulent.30,31
Based on this evidence, we hypothesized that Rim101p affects virulence in the experimental keratomycosis model. This current investigation examined the role of the RIM101 signal transduction pathway in the pathogenesis of keratomycosis. Using genetic knockout mutants of C. albicans, we confirmed the need for Rim13p and determined the importance of pathway components Rim8p, Rim20p, and Rim101p. We also studied whether corneal virulence depends on Phr1p, a pH-regulated protein with transcription that is governed by Rim101p and is involved in assembly of the fungal cell wall during filamentous growth at neutral and alkaline conditions.15 Using models of in vivo murine keratomycosis and ex vivo porcine corneal infection we showed that these components of the RIM101 pathway allow C. albicans to infect the cornea.
Materials and Methods
Fungal Strains
A wild-type strain (SC5314) of C. albicans, originally isolated from human infection; a prototrophic mutant control reference strain (DAY185); a homozygous Tn7 transposon mutant (GK088); and four engineered homozygous deletion mutants (DAY25, DAY111, DAY117, and CAS10) were evaluated (Table 1). Strain SC5314 has been used extensively for genetic studies of C. albicans and experimentally causes corneal disease in rabbits and mice.29,32,33 The prototrophic mutant control reference strain DAY185 was created by transformation of strain BWP17 and has a wild-type Ura+Arg+His+ genotype.28 Strain GK088 (Tn7-rim13) is a homozygous mutant with a transposon insertion at position 239 of the rim13 coding sequence20,29 that was transformed into the SC5314-derivative strain BWP1719 as part of a homozygous insertion mutant library.34 The mutant strains DAY25, DAY111, and DAY117 were generated and described by PCR-directed gene knockout by using consecutive transformation of strain BWP17.19,28 The mutant strain CAS10 was generated by homozygous deletion of phr1 from the SC5314 derivative CAF3–1.35
Table 1.
Genotypes, Phenotypes, and the Effects of pH Variation on Growth Rates for C. albicans Strains
| Strain | Genotype | Phenotype | Doubling Time (h)* |
||
|---|---|---|---|---|---|
| pH 6.0 | pH 7.3 | pH 8.0 | |||
| SC531432 | Clinical isolate | Wild-type | 1.17 ± 0.02 | 1.28 ± 0.02 | 1.45 ± 0.01 |
| DAY18528 | ura3Δ::λimm434pHIS1::his1::hisGarg4::URA3::arg4::hisG | Wild-type | 1.54 ± 0.02 | 1.46 ± 0.02 | 1.61 ± 0.06 |
| ura3Δ::λimm434 his1::hisG arg4::hisG | |||||
| DAY2528 | ura3Δ::λimm434pHIS1::his1::hisGarg4::hisG rim101::URA3 | Rim101p-negative | 1.46 ± 0.02 | 1.66 ± 0.01 | 1.71 ± 0.04 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim101::ARG4 | |||||
| DAY11119 | ura3Δ::λimm434his1::hisG arg4::hisG rim20::ARG4 pRIM101 | Rim20p-negative | 1.50 ± 0.03 | 1.45 ± 0.02 | 1.83 ± 0.03 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim20::URA3 RIM101 | |||||
| DAY11728 | ura3Δ::λimm434HIS1::his1::hisGarg4::hisGrim8::ARG4 | Rim8p-negative | 1.48 ± 0.02 | 1.46 ± 0.02 | 1.86 ± 0.02 |
| ura3Δ::λimm434 his1::hisG arg4::hisG rim8::URA3 | |||||
| GK08820 | ura3Δ::limm434his1::hisGarg4::hisGrim13::URA3 | Rim13p-negative | 1.44 ± 0.10 | 1.56 ± 0.06 | 1.97 ± 0.03 |
| ura3Δ::limm434 his1::hisG arg4::hisG rim13::ARG4 | |||||
| CAS1035 | Δura3::imm434 Δphr1::hisG | Phr1p-negative | 1.24 ± 0.02 | 1.59 ± 0.02 | 2.00 ± 0.03 |
| Δura3::imm434 Δphr1 | |||||
Mean ± SD.
Corneal inocula were prepared by culturing yeasts on Sabouraud dextrose agar (Difco, Detroit, MI) for 3 days at 25°C. Colonies were harvested and diluted in sterile phosphate-buffered saline (PBS) to yield 2 × 104 colony-forming units (CFU)/μL. A 5-μL aliquot was applied to the right eyes.
In Vitro Growth Kinetics
Yeast strains were grown in 1% yeast extract, 2% peptone, and 2% dextrose (YPD) liquid medium at 30°C overnight and then harvested and suspended in sterile phosphate-buffered saline (PBS). Optical density (OD) was measured with a spectrophotometer (Ultraspec 2000; Pharmacia Biotech, Princeton, NJ) at a wavelength of 600 nm (OD600). A conversion factor of one OD600 unit equivalent to 3 × 107 colony-forming units (CFU)/mL was used to estimate fungal concentration.36,37 Triplicate samples of 3 × 105 CFU for each strain were inoculated into 25 mL M199 liquid media (Invitrogen, Grand Island, NY) at pH 6.0, 7.3, and 8.0 and incubated at 27°C with a shaking speed of 150 rpm. C. albicans concentrations were determined spectrophotometrically at 1.5, 3, 4.5, 6, 9, 12, 15, and 24 hours postinoculation (PI). The doubling time for each strain at each pH was calculated from the slope of the linear regression equation describing the exponential growth phase. Student's t-test was used for pairwise comparisons, and P < 0.05 was considered statistically significant.
Filamentation Assays
For pH-induced filamentation assays, yeast strains were grown on Sabouraud dextrose agar for 2 days at room temperature then harvested, diluted in sterile PBS, and inoculated onto M199 agar plates containing Earle's salts and glutamine but lacking sodium bicarbonate.6,38 The plates were buffered with 2 M Tris-HCl to yield media of pH 4.0, 7.3, and 8.0. Inoculated plates were incubated at 37°C, and colonies were observed daily by inverted microscopy for 7 days. Filamentation was scored by the pace of onset, according to the postinoculation day that hyphae/pseudohyphae were first observable using a procedure as previously described.6 The degree of filamentation based on the number and length of filaments was also noted.
Animals and Experimental Keratomycosis
Animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research under protocols approved by the Baylor College of Medicine Institutional Animal Care and Use Committee. Female BALB/c and C57BL/6J mice 6 to 8 weeks of age (Harlan Sprague-Dawley, Houston, TX) were anesthetized intraperitoneally with ketamine, xylazine, and acepromazine. The corneas of right eyes were superficially scarified as previously described.37 A 5-μL inoculum of C. albicans (1 × 105 CFU) was topically applied to right eyes (five mice per group). Mice were monitored with a dissecting microscope daily for up to 7 days PI, to determine the severity of keratomycosis by criteria that assigned grades of 0 to 4 for inflammatory area, density, and surface irregularity.6,37 Findings were documented by slit lamp photomicrography.
Ex Vivo Testing
All seven strains of C. albicans were evaluated in porcine corneas by using an explantation model.39 Fresh porcine eyes were obtained from Visiontech, Inc. (Mesquite, TX), after immersion in a saline solution of penicillin, streptomycin, and amphotericin B (Invitrogen, Grand Island, NY). An 18-mm diameter trephine (VisionPak, Lexington, KY) was used to excise corneas that were then fixated onto an artificial chamber (Refractive Technologies, Cleveland, OH). Superficial scarification with a 22-gauge needle was used to produce a 15 × 15 cross-hatch pattern, and 10 μL containing 1 × 105 CFU of the C. albicans strains was topically applied to each cornea (three corneas per fungal strain). Inoculated corneas were placed into a six-well culture dish (Corning, Corning, NY) so that the periphery was immersed in modified supplemented hormonal epithelial medium (SHEM).39 Tissues were incubated at 34°C in 5% CO2 with 95% humidity, and the SHEM was changed daily. After 72 hours, the corneas were embedded in OCT compound (Sakura Finetec, Torrance, CA) and frozen at −80°C for subsequent histopathologic processing.
Hyphal Penetration
The average maximum hyphal penetration of porcine corneas was determined as previously described.39 Ten-micrometer frozen sections were cut and stained with periodic acid-Schiff (PAS) reagent (Sigma-Aldrich, St. Louis, MO). Three sections were examined for each cornea. Images were obtained of an entire limbus-to-limbus region for each corneal section by using a digital camera attached to a microscope (Y-FL; Nikon, Tokyo, Japan). Hyphal penetration depth and total corneal thickness were measured with an image analysis system (NIS-Element 3.0; Nikon). The maximum hyphal penetration was estimated from measurements taken at five regions of each corneal section demonstrating the greatest depth of corneal penetration. Results from the three areas with deepest penetration were averaged for each histologic section. The absolute maximum penetration depth and the percentage of penetration relative to corneal thickness at the points measured were recorded. Results were compared for statistical significance by Student's t-test.
Results
In Vitro Comparison of C. albicans Strains
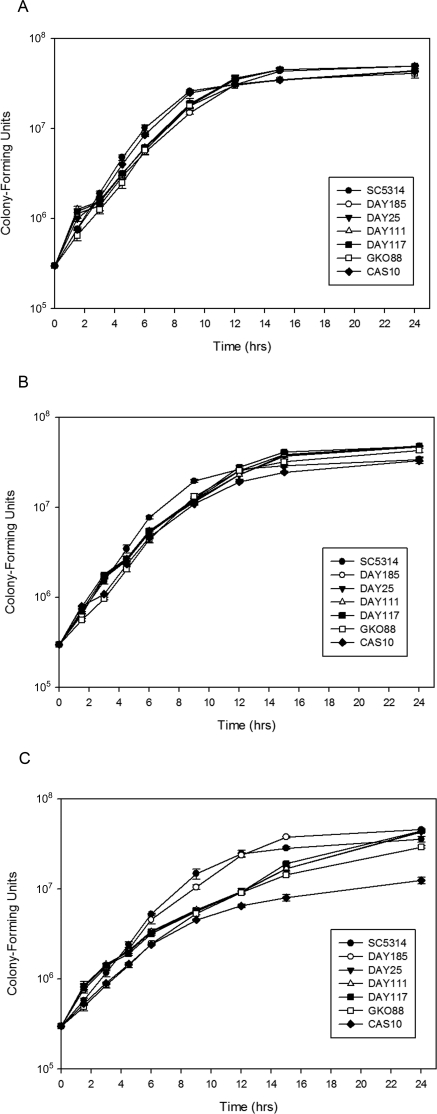
Since an important consideration in studying microbial virulence is the overall growth ability of the organism, all C. albicans strains used in this study were evaluated with acidic, neutral, and alkaline media to determine whether any inherent growth advantages or disadvantages existed. Although there were absolute growth differences among the strains that varied in different pH conditions, strains SC5314, DAY185, DAY25, DAY111, DAY117, GKO88, and CAS10 demonstrated similar lag, log growth, and plateau phases (Fig. 1), with strain SC5314 tending to grow more rapidly than other strains (P < 0.05). All strains demonstrated a trend for reduced growth rates as pH increased (Table 1), resulting in a significant difference between pH 6.0 and 8.0 (P < 0.05). Consistent with the removal of alkaline-response genes, the mutant strains were more affected by increased pH conditions with respect to growth rates than SC5315 or the prototrophic strain DAY185 (Fig. 1C).
Figure 1.
In vitro growth kinetics of Candida albicans strains SC5314, DAY185, DAY25, DAY111, DAY117, GKO88, and CAS10. Triplicate samples of each strain were inoculated into M199 media at pH 6.0 (A), 7.3 (B), and 8.0 (C) and monitored by spectrophotometry over time. Mean OD600 measurements converted to colony-forming unit equivalents (± SD) are plotted.
Further characterization of the fungal strains involved in vitro evaluation of filamentation phenotype. The relative filamentation results are shown in Table 2. C. albicans produced hyphae and pseudohyphae on artificial media, and filamentous growth was enhanced as the culture media pH increased. At neutral and alkaline pH conditions the wild-type strain SC5314 and the prototrophic strain DAY185 demonstrated rapid filamentation: obvious filaments were observed on day 1 PI and progressed throughout 7 days of observation. Strains deficient in genes of the RIM101 alkaline response pathway demonstrated greatly reduced filamentation phenotypes at neutral and alkaline conditions based on the time required for hyphae or pseudohyphae to initially present as well as the overall number and length of the filaments. In contrast to the wild-type and prototrophic controls, the mutant strains demonstrated little to no hyphal production at pH 7.3 and 8.0, even at 7 days PI. Consistent with the pathway mutations being evaluated, filamentation was not affected under acidic conditions (Table 2).
Table 2.
In Vitro Filamentation of C. albicans Strains
| Strain | pH 4.0 | pH 7.3 | pH 8.0 |
|---|---|---|---|
| SC5314 | ++ | +++ | +++ |
| DAY185 | ++ | +++ | +++ |
| DAY25 | ++ | + | + |
| DAY111 | ++ | + | + |
| DAY117 | ++ | + | + |
| GK088 | ++ | 0 | 0 |
| CAS10 | ++ | 0 | 0 |
+++, filamentation initially observed 1 day PI; ++, filamentation initially observed 3 days PI; +, filamentation initially observed 7 days PI; 0, no filamentation observed by 7 days PI.
In Vivo Experimental Fungal Keratitis
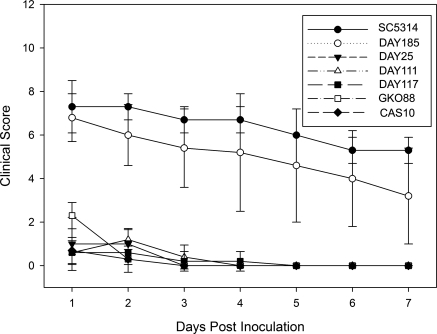
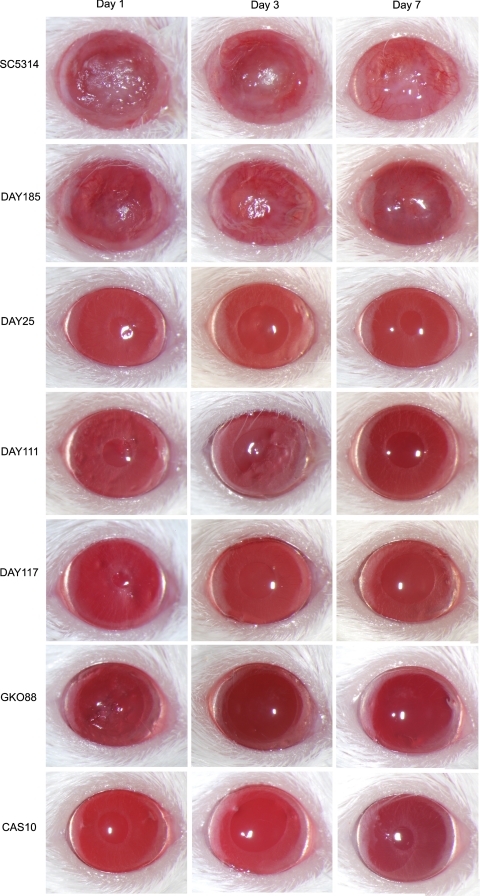
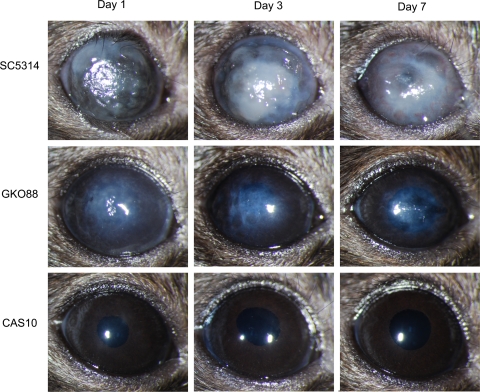
Scarified corneas of BALB/c mice inoculated with different strains of C. albicans were evaluated to determine the relative virulence of the homozygous mutants in vivo. The severity of corneal disease was scored daily for 7 days and assigned a categorical score of 0 to 12 as previously described.37 An inoculation of 1 × 105 CFU of the wild-type strain SC5314 and the prototrophic control strain DAY185 caused moderate corneal disease, whereas the five mutant strains were attenuated in relative virulence (Fig. 2). Representative findings for each strain at 1, 3, and 7 days PI are shown in Figure 3. SC5314 produced more severe disease with an average score of 7.3 ± 1.2 on day 1 PI and a mean score of 5.3 ± 0.6 on day 7 PI. Strain DAY185 produced slightly less corneal disease than SC5314, with mean scores of 6.8 ± 1.1 at 1 day PI and 3.2 ± 2.2 at 7 days PI, but the differences were not significantly different from the scores of SC5314 at any point during the study period (P > 0.05). Eyes infected with the mutant strains had mean disease severity scores ranging from 0.6 ± 0.6 to 2.3 ± 0.6 at day 1 PI depending on the particular strain. By day 7 PI, all indication of infection and corneal disease had resolved in the fungal mutant-infected BALB/c mice, resulting in null scores. Statistical comparison demonstrated no significant difference in disease severity among the mutant strains (P > 0.05) at any day other than GK088 at day 1 PI. Comparison of the mutants to SC53114 and DAY185 revealed a significant difference (P < 0.05) for each day of observation.
Figure 2.
Murine keratomycosis severity induced by C. albicans in BALB/c mice. Immunocompetent BALB/c mice were infected with 105 CFU of C. albicans used in the study and were followed and scored for disease severity for 7 days. Each point represents the mean score (± SD) of five eyes.
Figure 3.
Progression of murine keratomycosis induced by C. albicans in BALB/c mice. Scarified corneas of immunocompetent BALB/c mice were infected with 105 CFU of C. albicans used in the study and photographed on days 1, 3, and 7 PI. Representative images among five eyes per group are shown.
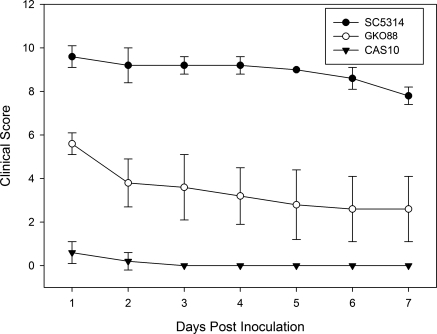
Previous work demonstrated that experimental fungal keratitis can be induced in both inbred strains and outbred stocks of mice.37 To extend these findings and to uncover additional insight into the role of the RIM101 signal transduction pathway in corneal disease, scarified corneas of C57Bl/6J mice were inoculated with 1 × 105 CFU of SC5314, GK088, or CAS10. Eyes were evaluated and scored daily for 7 days (Fig. 4). Representative results for 1, 3, and 7 days PI are shown in Figure 5. As in BALB/c mice, SC5314 induced severe disease that was significantly greater than the disease caused by either GK088 or CAS10 (P < 0.005). SC5314 and GK008 caused greater disease in C57BL/6J mice than in BALB/c mice with mean scores 2 to 3 units higher for each time point evaluated. CAS10 did not produce measurable inflammatory disease in either BALB/c or C57BL/6J mice.
Figure 4.
Murine keratomycosis severity induced by C. albicans in C57BL/6J mice. Immunocompetent C57BL/6J mice were infected with 105 CFU of C. albicans strains SC5314, GKO88, or CAS10 and were scored for disease severity for 7 days. Each point represents the mean score (± SD) of five eyes.
Figure 5.
Progression of murine keratomycosis induced by C. albicans in C57BL/6J mice. Scarified corneas of immunocompetent C57BL/6J mice were infected with 105 CFU of C. albicans strains SC5314, GKO88, or CAS10 and were photographed on days 1, 3, and 7 PI. Representative images among five eyes per group are shown.
Ex Vivo Keratomycosis in Porcine Corneas
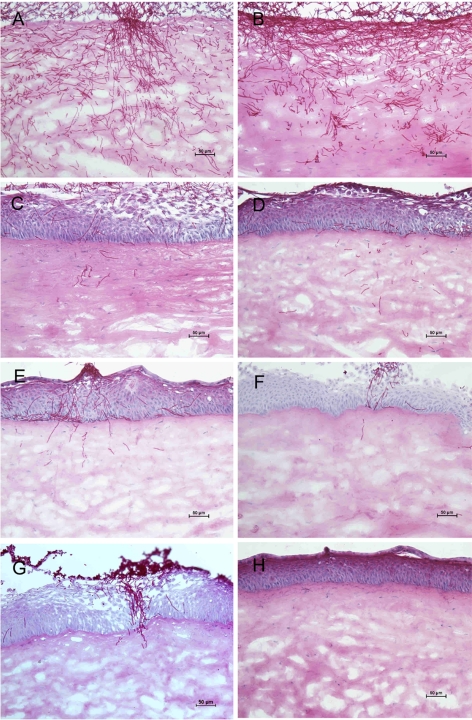
Porcine corneas maintained in an explantation culture system were inoculated with 1 × 105 CFU of SC5314, DAY185, DAY25, DAY111, DAY117, GK088, or CAS10 (three corneas per fungal strain). Three days PI corneas were cryosectioned and histopathologically evaluated. Representative results are shown in Figure 6. Extensive epithelial sloughing and abundant fungi in the corneal stroma were seen in SC5314- and DAY185-infected corneas. Hyphae and pseudohyphae were the predominant fungal forms. Fungal mutant-inoculated corneas showed significantly less fungal burden with fewer hyphae and pseudohyphae within the stroma. Although some epithelial thickening was observed, the epithelial layer was largely intact in mutant-infected corneas.
Figure 6.
Histopathology of porcine keratomycosis induced by C. albicans strains. Porcine corneas inoculated ex vivo with 105 CFU of C. albicans strains SC5314 (A), DAY185 (B), DAY25 (C), DAY111 (D), DAY117 (E), GKO88 (F), or CAS10 (G) or with PBS (H) were cryosectioned at 10-μm thickness 3 days PI and stained with periodic acid-Schiff. Representative sections among three corneas per strain are shown. Original magnification ×200.
The extent of hyphal penetration was quantified by measuring the maximum penetration depth for each experimental group (Table 3). SC5314 and DAY185 showed equivalent maximum penetration as determined by absolute penetration (791.7 ± 235.4 μm and 743.7 ± 209.1 μm, respectively) and by penetration relative to the corneal thickness at the point of penetration (27.8% ± 10.9% and 25.0% ± 7.0%, respectively). No significant difference in penetration ability was seen between these two strains with respect to either absolute (P = 0.93) or relative penetration (P = 0.73). The five mutant strains each demonstrated less than half the average maximum penetration of SC5214 and DAY185, measured by absolute or relative penetration (Table 3). Histologic data demonstrated that the mutant strains were significantly less invasive than the wild-type and prototrophic control strains (P < 0.05).
Table 3.
Maximum Penetration of C. albicans Strains into Porcine Corneal Stroma
| Strain | Penetration Depth* (μm) | P† | Penetration Ratio‡ (%) | P† |
|---|---|---|---|---|
| SC5314 | 761.7 ± 235.4 | 0.93 | 27.8 ± 10.9 | 0.73 |
| DAY185 | 743.7 ± 209.1 | Referent | 25.0 ± 7.0 | Referent |
| DAY25 | 168.5 ± 85.7 | 0.01 | 4.9 ± 1.9 | 0.009 |
| DAY111 | 295.1 ± 79.7 | 0.03 | 9.4 ± 2.2 | 0.02 |
| DAY117 | 340.3 ± 144.8 | 0.03 | 11.1 ± 1.6 | 0.03 |
| GK088 | 207.9 ± 110.6 | 0.02 | 9.7 ± 2.9 | 0.03 |
| CAS10 | 55.4 ± 62.2 | 0.005 | 2.7 ± 3.6 | 0.008 |
C. albicans maximum infiltrating depth, mean ± SD.
Pairwise comparison of each strain to prototrophic strain DAY185.
Mean maximum penetration depth relative to corneal thickness, % ± SD.
Discussion
Sequencing of the C. albicans genome and development of techniques for targeting specific genes have led to a research upsurge into investigating how particular fungal genes contribute to the pathogenesis of candidiasis.13,40,41 Selective mutants have helped to identify key intermediates affecting fungal survival and pathogenicity. Stemming from ophthalmic studies on the morphogenesis and pathogenicity of C. albicans,6,29,32,37 we used a systematic gene-deletion approach to examine how fungal keratitis entails genetically regulated mechanisms enabling the formation of invasive hyphae into the corneal stroma.
Pathways affecting the morphologic plasticity of C. albicans are interactive and interdependent.5,7–13 Environmental cues are detected by the microorganism resulting in intracellular signaling that culminates in responses at the gene level via various transcription factors. Signals known to elicit the transition of C. albicans blastospores into hyphal forms include temperature and ambient pH.42 Conditions at the ocular surface, such as a temperature of 34°C and a pH of 7.5,43–45 favor hyphal growth of C. albicans.46,47
Several gene products are necessary for pH sensing and signaling, and the molecular process has been reviewed in detail.5,7–11,48 In brief, surface receptor proteins Dfg16p and Rim21p/PalH detect environmental pH, and Rim9p/PalI assists in the localization of Rim21p/PalH to the plasma membrane.49–52 Stimulation of the membrane sensor prompts ubiquitination and promotes endocytosis of the Rim8p/PalF protein that is associated with Rim21p/PalH.53 This interplay results in the recruitment of endosomal sorting complexes that include Vps20-Snf7p/Vps32 proteins.54 Snf7p/Vps32 oligomerizes on the endosome and recruits Rim20p/PalA and the calpain-like protease Rim13p/PalB.20,54,55 Rim20p/PalA binds to the inhibitory domain on the carboxyl end of the full-length, inactive transcription factor Rim101p/PacC.56 Rim13p/PalB has proteolytic activity and removes the inhibitory C-terminal domain of Rim101p/PacC. This processed, active form translocates into the nucleus to regulate transcriptional changes promoting pH-dependent responses.38,57,58
The present study confirms our previous observation that a rim13−/− mutant of C. albicans has significantly reduced corneal virulence.29 Compared with wild-type C. albicans, the rim13-null mutant had minimal filamentation ability on explanted corneas and failed to establish progressive corneal infection in vivo. We also examined the role of other components of the conserved RIM101 signal transduction pathway. Although the proteolytic activity of Rim13p or a previously unknown biological activity independent of the RIM101 pathway cannot be excluded as mechanisms influencing fungal invasion, our observations imply that the RIM101 signal transduction pathway regulates the development of C. albicans keratitis.
Rim101p is one of the principal regulators of host–pathogen interactions, as exemplified by a rim101−/− strain demonstrating a severe virulence defect in a murine model of systemic candidiasis.28 Similarly, based on previous studies demonstrating a correlation between clinical appearance and histopathology in a murine model of experimental keratomycosis,29,37,59 our current results show that this regulatory transcription factor is a determinant of virulence in corneal infection. The rim101−/− homozygous mutant strain DAY25 induced only mild corneal inflammation without obvious infection, and mice recovered fully within 1 to 2 days. Furthermore, in explanted corneas the rim101−/− mutant strain only invaded the superficial corneal layer compared with deep stromal penetration by the wild-type and prototrophic strains. These in vivo and ex vivo findings provide evidence implicating Rim101p in mediating C. albicans virulence. Together with our previous studies of a Rim13p-deficient mutant, these results implicate the RIM101 signal transduction pathway as being directly involved in the pathogenesis of C. albicans keratitis.
Other constituents of the RIM101 pathway including Rim20p and Rim8p have been identified in C. albicans,19 and our results confirmed that these molecules are also required for corneal virulence. These findings demonstrate that several components leading to the activation of Rim101p affect C. albicans virulence during corneal infection. In comparison, restoration of virulence to the prototrophic mutant control DAY185 confirms the effects of selective gene disruption and shows that attenuation of the fungal mutants was not due to a nonspecific consequence of the molecular techniques used to generate the genetic deletions. The highly conserved nature of the RIM101/PacC signal transduction pathway across fungal species further suggests that these genes or their respective homologues have corresponding roles in virulence for other fungi.
We also examined a downstream product of Rim101p activation. Activated Rim101p directly and indirectly regulates the expression of alkaline pH-induced genes that are involved in fungal filamentation.15 In particular, the phr1 gene is a target of the RIM101 pathway that is involved in the yeast-to-hypha transition. This gene encodes a glycosylphosphatidylinositol-anchored cell surface glycosidase that cross-links carbohydrate polymers needed for integrity of the fungal cell wall.60 A phr1-null mutant is unable to undergo apical growth at neutral-to-alkaline pH and has reduced virulence for producing systemic infection in BALB/c mice.35,61 Strain CAS10, the phr1−/− mutant that we used in the present study, did not produce filamentous forms in explanted porcine corneas and was avirulent in the immunocompetent murine cornea. Furthermore, in a parallel study, we found that the relative regulation of phr1 under alkaline conditions is significantly lower in VE175, a wild-type strain of C. albicans having attenuated corneal virulence and altered in vitro morphology compared with the virulent SC5314 strain.39 Phr2, a functional homologue of phr1 that is preferentially expressed at acidic pH and repressed at alkaline pH in a Rim101p-dependent fashion,19,62 was significantly upregulated in the hypovirulent VE175 strain.39 Thus, our findings are consistent with other recent studies on the role of the RIM101 pathway and its downstream targets in fungal pathogenicity.
This investigation indicates that the RIM101 pathway is important for corneal virulence by C. albicans, but other virulence mechanisms may also be involved. Mds3p is necessary for growth and hyphal formation at alkaline pH, independent of the RIM101 pathway,27,34 and is involved in virulence in both a mouse systemic model34 and the ocular model.6 Sla2p is an actin-binding protein necessary for alkaline pH–induced hyphal formation.63,64 Sch9p is a serine-threonine protein kinase needed for embedded hyphal growth.27 Suv3p is an ATP-dependent RNA helicase involved in mitochondrial RNA catabolism, embedded filamentation, and growth under oxygen-limited conditions.27 Sap6p is one of 10 known secreted aspartyl proteinases and can influence virulence through several possible mechanisms, but its effect on corneal virulence is likely associated with filamentation.31 Interrelated pathways affect fungal morphogenesis as well as pathogenicity.
In addition to the mycological processes of candidal infection, the murine models used in this study indicate that host factors modulate the severity of keratomycosis. We found that C57BL/6J and BALB/c mice had inherent differences in susceptibility to C. albicans and developed disparate levels of severity of fungal keratitis. Although both mouse strains were vulnerable to corneal infection, C57BL/6J mice tended to develop more severe disease. This interstrain difference is similar to reports for models of bacterial keratitis in which C57BL/6 mice were more susceptible to Pseudomonas aeruginosa infection than BALB/c mice.65 In addition, we previously reported a difference between inbred BALB/c mice and outbred NIH Swiss mice with respect to C. albicans keratitis.37 These findings disclose an opportunity to explore how host resistance contributes to the susceptibility and severity of fungal keratitis.
In summary, model systems and molecular biological techniques provide the means to dissect the mechanisms of infectious eye disease. This study indicates that pathogens respond to local conditions at the ocular surface and provides insight into the molecular machinery directing fungal infection of the cornea. Ongoing research into the pathogenesis of keratomycosis offers prospects for translational studies on improved control and prevention.
Acknowledgments
The authors thank Aaron P. Mitchell (Department of Microbiology and Institute of Cancer Research, Columbia University, New York, NY) for providing the GK088 strain and William A. Fonzi (Department of Microbiology and Immunology, Georgetown University, Washington, DC) for providing the CAS10 strain.
Footnotes
Supported by Core Grant EY02520 from the National Institutes of Health, Research to Prevent Blindness, and Sid W. Richardson Foundation.
Disclosures: X. Yuan, None; B.M. Mitchell, None; X. Hua, None; D.A. Davis, None; K.R. Wilhelmus, None
References
- 1.Ruhnke M. Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Curr Drug Targets. 2006;7:495–504 [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun RL, Jones DB, Wilhelmus KR. Clinical characteristics and outcome of Candida keratitis. Am J Ophthalmol. 2007;143:1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah CP, McKey J, Spirn MJ, Maguire J. Ocular candidiasis: a review. Br J Ophthalmol. 2008;92:466–468 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell AP. Dimorphism and virulence in Candida albicans. Curr Opin Microbiol. 1998;1:687–692 [DOI] [PubMed] [Google Scholar]
- 6.Jackson BE, Wilhelmus KR, Mitchell BM. Genetically regulated filamentation contributes to Candida albicans virulence during corneal infection. Microb Pathog. 2007;42:88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324 [DOI] [PubMed] [Google Scholar]
- 8.Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H. Co-regulation of pathogenesis with dimorphism and phenotypic switching in Candida albicans, a commensal and a pathogen. Int J Med Microbiol. 2002;292:299–311 [DOI] [PubMed] [Google Scholar]
- 10.Ernst JF. Transcription factors in Candida albicans: environmental control of morphogenesis. Microbiology. 2000;1461763–1774 [DOI] [PubMed] [Google Scholar]
- 11.Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4:728–735 [DOI] [PubMed] [Google Scholar]
- 12.Rooney PJ, Klein BS. Linking fungal morphogenesis with virulence. Cell Microbiol. 2002;4:127–137 [DOI] [PubMed] [Google Scholar]
- 13.Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peñalva MA, Arst HN., Jr Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol Mol Biol Rev. 2002;66:426–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet. 2003;44:1–7 [DOI] [PubMed] [Google Scholar]
- 16.Su SS, Mitchell AP. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics. 1993;133:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su SS, Mitchell AP. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res, 1993;21:3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tilburn J, Sarkar S, Widdick DA, et al. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis D, Wilson RB, Mitchell AP. RIM101-dependent and -independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 2000;20:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Martin SJ, Bruno VM, Mitchell AP, Davis DA. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot Cell. 2004;3:741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porta A, Ramon AM, Fonzi WA. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J Bacteriol. 1999;181:7516–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porta A, Wang Z, Ramon A, Mühlschlegel FA, Fonzi WA. Spontaneous second-site suppressors of the filamentation defect of prr1Δ mutants define a critical domain of Rim101p in Candida albicans. Mol Genet Genomics. 2001;266:624–631 [DOI] [PubMed] [Google Scholar]
- 24.Lamb TM, Xu W, Diamond A, Mitchell AP. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J Biol Chem. 2001;276:1850–1856 [DOI] [PubMed] [Google Scholar]
- 25.Diez E, Alvaro J, Espeso EA, et al. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002;21:1350–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb TM, Mitchell AP. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol, 2003;23:677–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nobile CJ, Bruno VM, Richard ML, Davis DA, Mitchell AP. Genetic control of chlamydospore formation in Candida albicans. Microbiology. 2003;149:3629–3637 [DOI] [PubMed] [Google Scholar]
- 28.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell BM, Wu TG, Jackson BE, Wilhelmus KR. Candida albicans strain-dependent virulence and Rim13p-mediated filamentation in experimental keratomycosis. Invest Ophthalmol Vis Sci. 2007;48:774–780 [DOI] [PubMed] [Google Scholar]
- 30.Andrutis KA, Riggle PJ, Kumamoto CA, Tzipori S. Intestinal lesions associated with disseminated candidiasis in an experimental animal model. J Clin Microbiol. 2000;38:2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson BE, Wilhelmus KR, Hube B. The role of secreted aspartyl proteinases in Candida albicans keratitis. Invest Ophthalmol Vis Sci. 2007;48:3559–3565 [DOI] [PubMed] [Google Scholar]
- 32.O'Day DM, Head WS, Csank C, et al. Differences in virulence between two Candida albicans strains in experimental keratitis. Invest Ophthalmol Vis Sci. 2000;41:1116–1121 [PubMed] [Google Scholar]
- 33.Lo HJ, Köhler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949 [DOI] [PubMed] [Google Scholar]
- 34.Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics. 2002;162:1573–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saporito-Irwin SM, Birse CE, Sypherd PS, Fonzi WA. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol Cell Biol. 1995;15:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan X, Mitchell BM, Wilhelmus KR. Gene profiling and signaling pathways of Candida albicans keratitis. Mol Vis. 2008;14:1792–1798 [PMC free article] [PubMed] [Google Scholar]
- 37.Wu TG, Wilhelmus KR, Mitchell BM. Experimental keratomycosis in a mouse model. Invest Ophthalmol Vis Sci. 2003;44:210–216 [DOI] [PubMed] [Google Scholar]
- 38.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351 [DOI] [PubMed] [Google Scholar]
- 39.Hua X, Yuan X, Mitchell BM, Lorenz MC, O'Day DM, Wilhelmus KR. Morphogenic and genetic differences between Candida albicans strains are associated with keratomycosis virulence. Mol Vis. 2009;15:1476–1484 [PMC free article] [PubMed] [Google Scholar]
- 40.Magee PT, Gale C, Berman J, Davis D. Molecular genetic and genomic approaches to the study of medically important fungi. Infect Immun. 2003;71:2299–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruno VM, Mitchell AP. Large-scale gene function analysis in Candida albicans. Trends Microbiol. 2004;12:157–161 [DOI] [PubMed] [Google Scholar]
- 42.Buffo J, Herman MA, Soll DR. A characterization of pH-regulated dimorphism in Candida albicans. Mycopathologia. 1984;85:21–30 [DOI] [PubMed] [Google Scholar]
- 43.Bonanno JA, Polse KA. Measurement of in vivo human corneal stromal pH: open and closed eyes. Invest Ophthalmol Vis Sci. 1987;28:522–530 [PubMed] [Google Scholar]
- 44.Yamada M, Mochizuki H, Kawai M, Yoshino M, Mashima Y. Fluorophotometric measurement of pH of human tears in vivo. Curr Eye Res. 1997;16:482–486 [DOI] [PubMed] [Google Scholar]
- 45.Girardin F, Orgul S, Erb C, Flammer J. Relationship between corneal temperature and finger temperature. Arch Ophthalmol. 1999;117:166–169 [DOI] [PubMed] [Google Scholar]
- 46.De Bernardis F, Mühlschlegel FA, Cassone A, Fonzi WA. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nantel A, Dignard D, Bachewich C, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009;12:365–370 [DOI] [PubMed] [Google Scholar]
- 49.Castrejon F, Gomez A, Sanz M, Duran A, Roncero C. The RIM101 pathway contributes to yeast cell wall assembly and its function becomes essential in the absence of mitogen-activated protein kinase Slt2p. Eukaryot Cell. 2006;5:507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, et al. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot Cell. 2007;6:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rothfels K, Tanny JC, Molnar E, Friesen H, Commisso C, Segall J. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:6772–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barwell KJ, Boysen JH, Xu W, Mitchell AP. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot Cell. 2005;4:890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herranz S, Rodriguez JM, Bussink HJ, et al. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci U S A. 2005;102:12141–12146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu W, Smith FJ, Jr, Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baek YU, Martin SJ, Davis DA. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall β-glycosidase Phr2. Eukaryot Cell. 2006;5:1550–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baek YU, Li M, Davis DA. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot Cell. 2008;7:1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jackson BE, Mitchell BM, Wilhelmus KR. Corneal virulence of Candida albicans strains deficient in Tup1-regulated genes. Invest Ophthalmol Vis Sci. 2007;48:2535–2539 [DOI] [PubMed] [Google Scholar]
- 60.Chauhan N, Li D, Singh P, Calderone R, Kruppa M. The cell wall of Candida spp. In: Calderone RA. ed. Candida and Candidiasis. Washington, DC: ASM Press; 2002:159–175 [Google Scholar]
- 61.Ghannoum MA, Spellberg B, Saporito-Irwin SM, Fonzi WA. Reduced virulence of Candida albicans PHR1 mutants. Infect Immun. 1995;63:4528–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mühlschlegel FA, Fonzi WA. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol Cell Biol. 1997;17:5960–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Asleson CM, Bensen ES, Gale CA, Melms AS, Kurischko C, Berman J. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol Cell Biol. 2001;21:1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Melms AS, Gausmann U, Swoboda RK, Dominguez A, Kurischko C. Sequence analysis of SLA2 of the dimorphic yeasts Candida albicans and Yarrowia lipolytica. Yeast. 1999;15:1519–1528 [DOI] [PubMed] [Google Scholar]
- 65.Hazlett LD. Bacterial infections of the cornea (Pseudomonas aeruginosa). Chem Immunol Allergy. 2007;92:185–194 [DOI] [PubMed] [Google Scholar]