Expression of the keratin 12 (Krt12) gene is subjected to monoallelic regulation, as demonstrated by the use of bitransgenic Krt12cre/+/ROSAEGFP mice. This allelic selection of Krt12 gene expression is advantageous for retaining epithelial cells expressing the Krt12+ allele and allows tolerance to structural mutations of Krt12 in persons carrying heterozygous mutations.
Abstract
Purpose.
The purpose of this study was to characterize a Krt12-Cre knock-in mouse line for corneal epithelium-specific gene ablation and to analyze the allelic selection of the keratin 12 (Krt12) gene during corneal type-epithelium differentiation.
Methods.
Knock-in mice were generated by gene targeting. The authors examined the expression patterns of several reporter genes in the corneas of bitransgenic Krt12cre/+/ROSAEGFP, Krt12Cre/+/ZEG, and Krt12Cre/+/ZAP mouse lines. Krt12 and cre recombinase (Cre) immunostaining was performed. Corneal epithelial cells from bitransgenic Krt12Cre/+/ROSAEGFP mice were examined by fluorescence-activated cell sorter.
Results.
Mosaic and spiral expression patterns of EGFP were observed in young and adult bitransgenic Krt12Cre/+/ZEG mice, respectively. Immunostaining revealed that Cre− cells were also Krt12 negative in the corneal epithelia of Krt12Cre/−/ZAP mice. Using FACS analysis, 60% to 70% of the corneal epithelial cells from Krt12Cre/+/ROSAEGFP mice were EGFP positive, whereas 20% to 30% were negative. RT-PCR revealed that EGFP+ cells express both Krt12Cre and Krt12+ alleles, whereas EGFP− cells express only Krt12+. In the Krt12Cre/− cornea, the number of epithelial cells expressing Cre is the same as that found in Krt12Cre/Cre, which can be explained by the fragility of corneal epithelial cells that did not produce Krt12 because the Krt12Cre allele was not transcribed.
Conclusions.
These observations are consistent with the notion that clonal limbal stem cells randomly activate Krt12 alleles in the process of terminal differentiation. The authors suggest that this selection is advantageous for retaining epithelial cells expressing the Krt12+ allele and that it allows tolerance to structural mutations of Krt12.
Mammals are mosaic organisms because both alleles of many genes are not ubiquitously or equally expressed by individual cells of the same cell type in a given differentiated tissue. Mosaicism results from monoallelic expression, an unusual form of gene expression in which one allele of a gene is expressed.
When a postzygotic mutation occurs in either the germ line or somatic cells, it leads to an organism with two or more genetically different populations of cells originating from a genetically homozygous zygote. Random X-chromosome inactivation in females was first described by Lyon1 and is one of the best examples of monoallelic selection. Inheritance of hair color in mammals is another example of mosaic expression patterns of clonal allele activation of melanocytes. For example, many single-gene mutations at a wide variety of mouse pigmentary loci lead to multicolored coats.2 Those color patterns that are caused by genes expressed directly in melanocytes are clearly clonal patterns reflecting the proliferation and migration of pigment cells3,4 as they originate from the neural crest.5 Mintz2 proposed the phenoclone hypothesis to explain such observations, according to which virtually all mammalian cell types comprise phenotypically different developmental clones, or phenoclones, and virtually all loci have some alleles subjected to such ambiguous cis-acting controls. However, there was no other experimental evidence than hair color inheritance derived from melanocytes to substantiate the phenoclone hypothesis.2 Apart from single-allele activity of genes in cells of X chromosome-linked heterozygotes,6 the basis for this clonal phenotypic heterogeneity is unknown.
Mosaicism has been used as one mechanism that accounts for segmental skin disease in which the diseased cells are virtually derived from a mutated cell clone at postzygotic stages. In skin disorders, type 1 mosaicism is characterized by segmental lesions according to Blaschko's line, with mutations found in affected tissues but not in healthy tissues. Blaschko's line, named after the German dermatologist Alfred Blaschko, is an invisible skin line thought to trace the migration of embryonic cells.7 Type 2 mosaicism of autosomal dominant mutation exhibits mild diffused manifestations in most of the body, but linear patterns of exacerbation occur along the Blaschko's line. Recent studies of a patient with Hailey-Hailey disease, also known as familial benign chronic pemphigus, a genetic disorder that causes blisters to form on the skin, demonstrated the loss of heterozygosity of the ATP2C1 gene in the affected regions of skin.8,9 However, it remains unclear why those regions of skin that maintain heterozygosity have mild clinical manifestations. One possible explanation is that the mRNA of the mutant allele is unstable and does not lead to the synthesis of a significant amount of mutant protein, causing perturbation of cellular function. Alternatively, the phenomenon can be attributed to clonal activation of an individual allele of the affected gene, which may also lead to the mosaic phenotype in tissue.
In the present study, we examined the possibility of clonal allelic activation taking place within keratinocytes (e.g., corneal epithelial cells). Conventional gene targeting was used to create Krt12-Cre knock-in mice in which an internal ribosome entry site (IRES)-Cre minigene was inserted immediately after the stop codon in exon 8 of the mouse Krt12 gene. As a result, transcription of the modified Krt12Cre allele leads to simultaneous synthesis of K12 keratin and Cre recombinase. The expression pattern of Cre was assessed by the expression of reporter genes EGFP and AP during corneal-type epithelial differentiation of limbal stem cells in the corneal epithelium of bitransgenic Krt12-Cre/ROSA-EGFP, Krt12-Cre/ZEG, and Krt12-Cre/ZAP mice, respectively. Our results suggest that clonal activation of individual Krt12 alleles occurred in limbal stem cells undergoing corneal type-epithelium differentiation.
Methods
Generation of Krt12-IRES-Cre Knock-in Mice by Gene Targeting
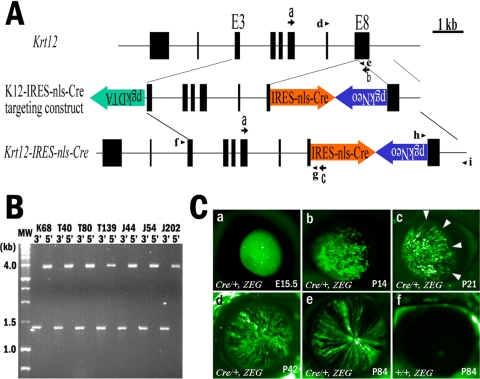
Conventional gene targeting was used to create Krt12-Cre knock-in mice. A 3.7-kb BamHI-StuI genomic DNA fragment of Krt12, constituting a region extending from intron 2 to the 5′ part of exon 8, was cloned into the BamHI/SmaI site of pBluescriptSKII+ (Stratagene, La Jolla, CA). A 1.1-kb 3′ Krt12 gene fragment, 3′ part of exon 8 to the 3′ untranslated region, including the poly adenylation signal, was prepared by PCR with the upstream sense primer K12/7152KpnI+, 5′-CGGGGTACCCCGGGCCTCACACGGGCTCCTCTGG, and downstream antisense primer K12/8213AscIKpnI-, 5′-CGGGGTACCCGGTCCGGGCGCGCCTCAGCTGCTGCCAGGTAGGAGAAAG, and was cloned into the KpnI site of pBluescriptSKII+. A minigene, nls-Cre containing nuclear localization signals and the Cre recombinase coding sequence was cloned into the pIRES vector (Clontech), and the IRES-Cre EcoRI-DraI fragment cloned into EcoRI/EcoRV of pBluescriptSKII+. Neomycin resistance cassette (NeoR) was ligated to the XhoI site between IRES-Cre and the 3′ Krt12 genomic DNA fragment. Then the phosphoglucokinase diphtheria toxin A (pgkpr-DTa) minigene cassette was inserted into the NotI site in front of the 3.7-kb BamHI-StuI genomic fragment. The orientation and fidelity of these inserts were verified by enzyme digestion and nucleotide sequence. Fifty micrograms of AscI linearized targeting vector was used for electroporation of R1 embryonic stem (ES) cells. Homologous recombinants were identified by PCR of genomic DNA isolated from neomycin-resistant ES clones for 3′-end recombination with the upstream primer pgk472-, 5′-AAAGCGCATGCTCCAGACTGCC, and the downstream primer K12/8431-, 5′-GCAACAGAGTTAGGACTTGAACCC. They were verified by PCR of genomic DNA for 5′-end recombination with upstream primer K12/3404+, 5′-GTACTAGGATTACAGACATGGGCCACATAGCCC, and downstream primer IRES37-, 5′-GTAACGTTAGGGGGGGGGGAGGGAGAGGGGCGG, as shown in Figure 1. Recombinant ES cells were used for blastocyst injection with C57BL/6 embryos to obtain germ line chimeras by the transgenic core in the Department of Molecular Genetics, University of Cincinnati. Sibling breeding was used to obtain homozygous Krt12Cre/Cre mice. Genotypes of knock-in mice were identified by PCR with genomic DNA, with primers shown (Fig. 1A) for Krt12Cre and Krt12 wild-type (+) alleles.
Figure 1.
Generation of Krt12Cre/+ knock-in mice by gene targeting. (A) To generate the Krt12-IRES-nls-Cre targeting construct, an IRES-nls-Cre cassette containing IRES, nls, Cre, and SV40-polyA was cloned between the stop codon and the polyadenylation signal in exon 8 of the Krt12 gene. The pgkNeo cassette (positive selection marker) in a reverse orientation, followed by IRES-Cre and diphtheria toxin A fragment (pgkDTA) cassette (negative selection marker), were placed on the 5′ end of the targeting vector. The Krt12-IRES-nls-Cre gene shows the predicted structure of a targeted knock-in allele after homologous recombination. Arrowheads: genotyping primer pairs for wild-type (d, e) and for the 5′ (f, g) and 3′ (h, i) ends of K12-IRES-nls-Cre gene. Arrows: RT-PCR primer pairs for Krt12 wild-type (a, b) and K12-IRES-nls-Cre (a, c). (B) Homologous recombinant ES cell clones were identified by PCR, by which the 3′ recombination yields a 1.4-kb product and the 5′ recombination yields a 4-kb product. Locations of primer pairs are indicated in (A). (C) EGFP expression pattern of Krt12Cre/+/ZEG bitransgenic mice (Ca–e) during development and maturation. EGFP expression starts from embryonic day (E) 15.5 as a sporadic pattern. After birth, mosaic expression patterns of EGFP are observed throughout the entire corneal epithelium until 2 weeks of age (Cb). After 2 weeks, a spiral pattern can be seen invading from the limbus area (arrowheads, Cc) to the center of the cornea. Mosaic expression patterns gradually disappeared from the center of cornea (Cd). After 12 weeks, only spiral expression patterns were observed throughout the entire corneal epithelium (Ce). Twelve-week-old Krt12+/+/ZEG mouse cornea is shown as negative control (Cf). Krt12 knock-in allele (cre); Krt12 wild-type allele (+).
Animal Protocol
Krt12-IRES-Cre mice (Krt12-Cre/+) and Krt12 knockout10 (Krt12−/−) mice were crossed with ZEG (Tg(ACTB-Bgeo/GFP)21Lbe/J), ROSA-EGFP (B6;129-Gt(ROSA)26Sortm2Sho/J), and ZAP (Tg(ACTB-Bgeo/ALPP)1Lbe/J) reporter mice (Jackson Laboratory, Bar Harbor, ME).11–13 All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of Cincinnati and were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
LacZ and AP Staining
LacZ Staining.
Cryosections prepared from enucleated eyes of 12-week-old mice were fixed in 0.2% glutaraldehyde, 50 mM ethylene glycol-bis (2-aminoethylether)-N,N,N′,N′-tetraacetic acid, and 100 mM MgCl2 in phosphate-buffered saline (PBS), pH 7.4, for 30 minutes at 4°C. Sections were washed three times for 15 minutes each with LacZ wash buffer (2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% Nonidet-P40 in PBS) and then incubated with staining solution containing 0.5 mg/mL X-gal, 4.5 mM potassium ferrocyanide and 4.5 mM potassium ferricyanide in LacZ wash buffer at 25°C for 12 hours.
Alkaline Phosphatase Staining.
Cryosections were rinsed in PBS and heated at 70°C for 30 minutes to inactivate endogenous alkaline phosphatases. Alkaline phosphatase was visualized with a staining kit according to the manufacturer's protocol (Fast Red; Sigma-Aldrich, St. Louis, MO).
Histology and Immunostaining
Fifteen-micrometer cryosections were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, at 4°C for 30 minutes, washed three times for 10 minutes each with PBS, blocked with 5% dry milk in PBS at 4°C overnight. The sections were stained with rabbit polyclonal anti–K12 antibody14 and anti–Cre antibody (Novagen, San Diego, CA) and were visualized by indirect immunofluorescence with appropriate secondary antibody conjugates (Jackson Laboratory; Molecular Probes, Eugene, OR). Specimens were counterstained with reagent containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Shield; Vector Laboratories, Burlingame, CA). Sections were observed with a confocal laser scanning microscope (LSM 510; Carl Zeiss MicroImaging, Thornwood, NY).
Fluorescence-Activated Cell Sorting and RT-PCR
Enucleated eyes from bitransgenic Krt12+/+/ROSAEGFP, Krt12Cre/+/ROSAEGFP, Krt12Cre/Cre/ROSAEGFP, Krt12Cre/−/ROSAEGFP and Krt12−/−/ROSAEGFP mice were incubated in 1% dispase II (Roche, Indianapolis, IN) in PBS (pH 7.4) at 4°C for 12 hours. The corneal epithelium was peeled from the eye and incubated in 0.25% trypsin-EDTA (Gibco, Carlsbad, CA) at 25°C for 40 minutes and was washed twice in PBS. Fluorescence-activated cell sorting (FACS) was performed (FACSVantage; BD Biosciences, San Jose, CA) with a solid-state laser (Lyt 200 S 488; iCyte, Champaign, IL). Sort gates were set, and the data were analyzed (DIVA software; BD Biosciences). Total RNA was isolated from EGFP+ and EGFP− cells using reagent (Trizol; Gibco). cDNA was synthesized (AMV Reverse Transcriptase; Promega, Madison WI) and subjected to RT-PCR for detection of Krt12 wild-type and Krt12 IRES-Cre mRNA with the following primer pairs: 5′ common primer Krt12/5202+, 5′-GCTGGGCGTCAAGGCTCGCCTGGAG, and 3′ primer Krt12/7282-, 5′-CAAGACCCAACCTGCATAGAGAATCC, for wild-type Krt12 mRNA; 3′ primer IRES263-, 5′-CGCTACAGACGTTGTTTGTCTTC, for K12-IRES-Cre mRNA; 5′ primer mGAPDH333+, 5′-GGGTGGAGCCAAACGGGTCATC, and 3′ primer mGAPDH864-, 5′-GGAGTTGCTGTTGAAGTCGCAGG, for GAPDH mRNA as control. PCR was performed as follows: 94°C for 5 minutes, followed by 30 cycles of 94°C for 30 seconds, 65°C for 30 seconds, and 72°C for 30 seconds, and 72°C for 5 minutes. PCR products were analyzed by electrophoresis in 2% agarose gels.
Results
Preparation of Krt12-Cre Knock-in Mouse Line
The knock-in strategy of gene targeting was used to prepare a mouse line, which expresses Cre recombinase under the control of the Krt12 promoter. The targeting construct contained IRES-Cre and NeoR minigenes and was used for electroporation of ES cells as shown in Figure 1A. Homologous recombination resulted in the insertion of the minigenes between the stop codon of exon 8 and the polyadenylation signal of the Krt12 gene. Seven homologous recombinant ES cell clones were obtained (Fig. 1B). Conventional blastocyst injection was used to obtain germ line chimeras. Three chimeras were bred with Swiss black mice to generate heterozygous Krt12-IRES-nls-Cre (Krt12Cre/+) knock-in mice. The modified Krt12Cre allele leads to the synthesis of a bicistronic mRNA that allows simultaneous synthesis of keratin 12 and Cre recombinase when the modified Krt12Cre alleles are transcribed by the corneal epithelial cells of knock-in mice.
K12 Keratin and Reporter Gene Expression Pattern in Corneas of Bitransgenic Mice
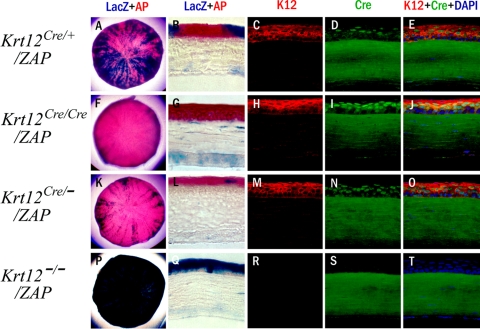
To examine the functionality of the knock-in mouse line, Krt12Cre/+ mice were crossed with the ZEG reporter mouse line (Jackson Laboratory) to obtain Krt12Cre/+/ZEG bitransgenic mice. ZEG mice contain the LacZ gene and a stop codon that are flanked by lox P sites followed by the EGFP gene. In the absence of Cre, mice under the control of the chicken β-actin promoter will express β-galactosidase but not EGFP. In the presence of Cre, recombination occurs, removing LacZ and turning on the expression of EGFP. These mice allow us to identify K12-expressing cells based on the expression of EGFP. Interestingly, mosaic and spiral expression patterns of EGFP were observed in the corneal epithelium of young and adult Krt12Cre/+/ZEG bitransgenic mice, respectively (Fig. 1C). One possible explanation for such expression patterns might be derived from the selected activation of either the Krt12+ wild-type allele or the Krt12Cre knock-in allele by individual limbal stem cell clones undergoing corneal-type epithelial differentiation. Interestingly, sibling breeding of Krt12Cre/+/ZEG revealed that KrtCre and ZEG alleles were always cosegregated and did not yield Krt12Cre/Cre/ZEG offspring, suggesting that, like Krt12, ZEG is localized on mouse chromosome 11. Thus, Krt12Cre/+/ZEG bitransgenic mice are not suitable for further analysis into the clonal activation of Krt12 alleles during corneal type-epithelial differentiation. To circumvent this difficulty, knock-in Krt12Cre/+ and knockout Krt12−/− mice were crossed with ZAP reporter mice (Jackson Laboratory). The ZAP reporter mouse line works similarly to the ZEG reporter mouse except that EGFP is replaced by alkaline phosphatase. One benefit of using this mouse line is that β-galactosidase and alkaline phosphatase staining can be performed and visualized on a single section. Thus, those corneal epithelial cells expressing Cre were stained red (Fast Red; Sigma-Aldrich), whereas those that were negative for Cre were stained blue by X-gal stain because of the synthesis of β-galactosidase. In addition, immunofluorescence staining for keratin K12 and Cre recombinase and histochemistry of AP and β-galactosidase activities were performed with corneas of 12-week-old Krt12Cre/+/ZAP, Krt12Cre/Cre/ZAP, Krt12Cre/−/ZAP, and Krt12−/−/ZAP mice. As shown in Figure 2 in Krt12Cre/+/ZAP mouse corneas, a spiral pattern was observed for both AP and β-galactosidase (Fig. 2A). Immunostaining with an anti–K12 antibody showed ubiquitous expression of keratin 12 by almost all corneal epithelial cells except for a few basal corneal epithelial cells (progenitor cells) that are still undifferentiated, as had been shown previously.15 Despite the fact that nearly all cells in the corneal epithelium expressed K12, many corneal epithelial cells were negative for Cre (Figs. 2C-E). These observations are consistent with the notion of clonal activation of one and both of the two Krt12 alleles in the process of corneal-type epithelial differentiation. This suggestion is further supported by the expression pattern of the AP reporter gene in corneas of Krt12Cre/Cre/ZAP mice in which the expressions of K12 keratin and Cre genes were ubiquitous by all differentiated corneal epithelial cells except basal cells at the limbus and a few undifferentiated cells in the mid-peripheral regions of the cornea (Figs. 2F-J). To further elucidate the hypothesis of clonal activation of Krt12 alleles, we investigated the expression pattern of the AP reporter gene, Cre, and Krt12 in the corneal epithelium of Krt12Cre/−/ZAP mice in which one Krt12 allele was ablated.10 Interestingly, cells expressing K12 also expressed Cre (Figs. 2K-O). The corneal epithelium of Krt12−/−/ZAP mice lacked AP, K12, and Cre, but β-galactosidase was detected in all corneal epithelial cells.
Figure 2.
Reporter gene expression. Cre recombinase expression in K12Cre/+/ZAP, K12Cre/Cre/ZAP, and K12Cre/−/ZAP mouse corneas. Corneas from 12-week-old mice were used for whole mount staining for β-galactosidase (Cre− cells, blue) and AP (Cre+ cells, red) (A, F, K, P). K12Cre/+/ZAP cornea is stained by a red and blue spiral pattern. In K12Cre/Cre/ZAP and K12Cre/−/ZAP cornea, most of the epithelium is stained red with small slivers of blue. Cryosections (B, G, L, Q) are stained by β-galactosidase (blue) and AP (red), and adjacent sections of each genotype are stained by K12 (C, H, M, R, red) and Cre (D, I, N, S, green) and then merged with DAPI (E, J, O, T). In the K12Cre/+/ZAP cornea, LacZ+ and AP+ cell populations appear side by side, and a serial section shows that most of the epithelium is positive for Krt12. K12−/−/ZAP serves as the negative control, and nonspecific binding of anti–Cre antibody can be observed in the corneal stroma. Krt12Cre, Cre knock-in allele; Krt12+, wild-type allele.
Clonal Activation (Allelic Selection) of Krt12Cre Knock-in and Krt12+ Alleles by Corneal Epithelial Cells of Krt12Cre/+/ROSAEGFP Bitransgenic Mice
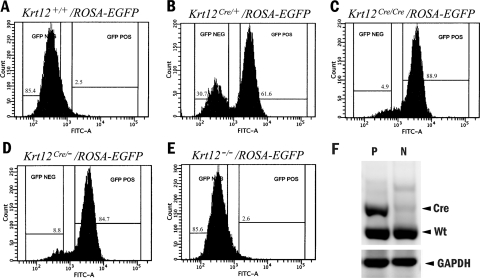
To further examine the possibility of allelic selection of the Krt12 allele during corneal-type epithelial differentiation, Krt12Cre/+ mice were mated with ROSA-EGFP reporter mice to obtain bitransgenic Krt12Cre/+/ROSAEGFP mice in which Cre expression from the activated Krt12Cre allele leads to EGFP expression after the excision of stop elements within the ROSAEGFP allele of corneal epithelial cells.12 Krt12Cre/+/ROSAEGFP bitransgenic corneal epithelial cells were sorted by FACS (Fig. 3B). In Krt12Cre/+/ROSAEGFP bitransgenic mouse corneas, 60% of the cells were EGFP positive, whereas 30% were negative. On the other hand, approximately 85% of the corneal epithelial cells from Krt12Cre/−/ROSAEGFP mice were EGFP positive. In Krt12Cre/Cre/ROSAEGFP bitransgenic mouse corneas, 85% to 90% of the corneal epithelial cells were EGFP positive, and 5% were negative (Figs. 3A-E). At 12 weeks of age, not all basal epithelial cells expressed K12, making the percentage of EGFP+ cells <100%. Krt12Cre/−/ROSAEGFP bitransgenic corneas had nearly the same percentage of cells expressing EGFP as did Krt12Cre/Cre/ROSAEGFPcorneas. To determine the mRNA expression of each Krt12 allele (wild-type or knock-in), RT-PCR was performed using total RNA isolated from EGFP+ and EGFP− cells. Interestingly, EGFP+ cells expressed both the Krt12Cre knock-in allele and the Krt12+ allele, whereas EGFP− cells expressed only the Krt12+ allele (Fig. 3F).
Figure 3.
FACS of EGFP+ and EGFP− cells. FACS of Krt12+/+/ROSAEGFP, Krt12Cre/+/ROSAEGFP, Krt12Cre/Cre/ROSAEGFP, Krt12Cre/−/ROSAEGFP, and Krt12−/−/ROSAEGFP corneal epithelial cells according to the expression of ROSAEGFP, and isolated RNA was subjected to RT-PCR. Corneas from 12-week-old mice of each genotype were sorted based on EGFP fluorescence intensity (A–E). Sort gates were set to include the GFP+ (percentages are shown on the horizontal gate lines) cells. (F) RT-PCR from a Krt12Cre/+/ROSAEGFP cornea. GFP− cells only express mRNA of the Krt12+ allele, whereas GFP+ cells express mRNA of both the Krt12Cre allele and the Krt12+ allele. Krt12Cre, Cre knock-in allele; Krt12+, wild-type allele.
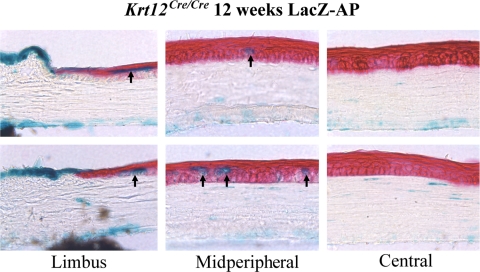
Based on FACS analysis, it is apparent that there is a small population of cells that do not express EGFP. Several possibilities may explain this EGFP− cell population. As mentioned earlier, not all basal epithelial cells express K12 at 12 weeks of age because they are not fully differentiated. In addition, it may be that in this population of cells, both Krt12Cre alleles were not selected and silenced. To gain some insight into this cell population, 12-week-old Krt12cre/cre/ZAP mice were stained for β-galactosidase and alkaline phosphatase. β-Galactosidase–positive cells, which are those cells not expressing K12, can be found within the limbus and within the suprabasal and superficial layers of the corneal epithelium in the midperiphery. Interestingly, β-galactosidase–positive cells are not present in the central epithelium (Fig. 4). The suprabasal and superficial location of these β-galactosidase–positive cells suggest, though not conclusively, that these cells may be the result of selecting both silenced alleles. Typically, cells that are not fully differentiated and, therefore, that do not express K12 are restricted to the basal epithelium.
Figure 4.
Monoallelic selection of Krt12 alleles. Twelve-week-old Krt12cre/cre/ZAP mice were stained for β-galactosidase and alkaline phosphatase. The presence of Cre− cells (blue) can be seen in the suprabasal and superficial epithelial layers of limbus and in the midperipheral region of the cornea. No blue cells are present in the central cornea.
Discussion
Monoallelic expression has been shown to occur in several genes, including T-cell receptor, Toll-like receptor 4, and interleukins, to name a few.16–20 In this article, we provide evidence to show allelic selection of the Krt12 gene occurs within the corneal epithelium. Previous studies have shown that corneal epithelial stem cells are located in the basal layer of the limbal epithelium.21–23 Limbal stem cells divide and centripetally migrate into the cornea to maintain the stratified corneal epithelium. Our previous studies showed that the corneal epithelium-specific Krt12 gene starts expressing when the limbal epithelium migrates onto the cornea.24,25 Although our knowledge of gene silencing and activation has increased in the past few years, there remains a large black box in the genetic and epigenetic regulatory mechanisms dictating gene expression. For example, X-chromosome inactivation has been well characterized, but the mechanism behind autosomal gene silencing is still poorly understood except for imprinting of maternal and paternal inheritance.
In this study, our original purpose was to make a corneal epithelium-specific knockout mouse using the Cre-loxP system. Because of the lack of a functional cornea-specific promoter, we chose to use knock-in gene targeting to modify the Krt12 allele so that it contains an IRES-nls-Cre minigene that allows the synthesis of a bicistronic mRNA for the concurrent synthesis of both Krt12 and Cre recombinase. Krt12Cre/+ showed cornea-specific Cre expression, but 20% to 30% of the epithelial cells did not express Cre (Fig. 2). In the Krt12Cre/Cre cornea, more than 85% of the epithelial cells expressed Cre. These results indicate that to successfully knock out genes within the cornea epithelium in a Krt12-dependent fashion, Krt12Cre/Cre mice would have to be used. Interestingly, Krt12Cre/− corneas had a population of Cre+ epithelial cells similar to that of Krt12Cre/Cre mouse corneas. Our previous studies have shown that in Krt12± mice, most of the corneal epithelial cells express the K12 protein.24,25 Our interpretation of these results was simply that the Krt12+ allele was expressed in all the corneal epithelial cells. However, given the data presented here, it is likely that this occurred because some limbal stem cell clones do not express Krt12+ alleles, resulting in Krt12− cells. The absence of K12 is likely to make these cells fragile and unable to complete their centripetal migration into the central corneal epithelium; however, more research is needed to confirm this hypothesis. In contrast, the cells that express the K12+ allele gain the advantage and become the dominant cell type present in K12± corneas. This hypothesis is partially substantiated by the observation that a small number of K12− suprabasal corneal epithelial cells were found in the limbus and midperipheral cornea of K12Cre/Cre/ZAP mice, which did not complete their centripetal migration into the central corneal epithelium (Fig. 4).
Based on the data presented, we propose that allelic selection of the Krt12 gene occurs within each limbal stem cell clone leading to the generation of four possible outcomes. Figure 5 represents these possibilities and the resultant reporter gene expression within the corneal epithelium for Krt12Cre/Cre, Krt12Cre/+, and Krt12Cre/− mice. It is clear to see that even with a stem cell containing the Krt12Cre allele, there is a chance that this allele will be untranscribed or silenced. If both alleles are silenced, these cells will fail to express EGFP and will eventually be lost before reaching the central cornea possibly cell because of fragility in the absence of K12 keratin that is essential for the maintenance of corneal epithelial cell integrity.10 Naturally there exist progenitor cells in the basal layer of the corneal epithelium that do not express Krt12; however, there remains a population of cells that have silenced Krt12Cre alleles and that are not progenitor cells. This suggestion is substantiated by results shown in Figure 4, in which the cells containing both silenced Krt12Cre alleles can be seen in the suprabasal and superficial layers of the midperipheral cornea but are absent from the central cornea because of cell fragility similar to that seen in Krt12Cre/−/ROSAEGFP mice (Fig. 3). Alternatively, these undifferentiated Krt12 negative cells in the superficial-midperipheral cornea may finally undergo differentiation and express one or both Krt12Cre alleles toward the end of their journey of centripetal migration.
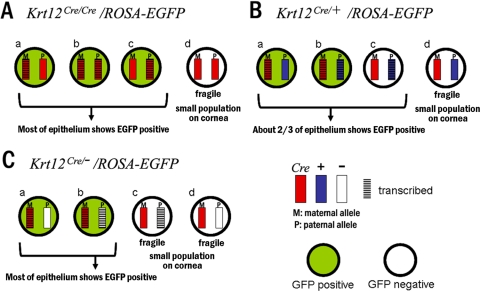
Figure 5.
Model of allelic selection of the Krt12 gene. There are four possible patterns of Krt12 expression based on the random event of allelic selection: (a) maternal allele only, (c) paternal allele only, (b) both alleles, (d) neither allele. Each limbal stem cell selects one of them. (A) In the Krt12Cre/Cre/ROSAEGFP bitransgenic mouse cornea, if the stem cell selects pattern d, this cell lineage will not express Krt12 protein and will become fragile and eventually lost. Therefore, if pattern d is chosen, that cell lineage will have a competitive disadvantage in terms of survival compared with patterns a, b, and c. These three selections will express EGFP throughout nearly all the corneal epithelium. (B) In the Krt12Cre/+/ROSAEGFP bitransgenic mouse cornea, only cell lineage d fails to express Krt12. Patterns a and b express EGFP, but pattern c does not; therefore, approximately two-thirds of the epithelial cells express EGFP. (C) In the Krt12Cre/−/ROSAEGFP bitransgenic mouse cornea, patterns a and b express both Krt12 and EGFP, but patterns c and d do not have Krt12 protein. Therefore, most of the corneal epithelium will become EGFP positive. It should be noted that the ratio of transcriptional activation for each allele is not necessarily 50%. Krt12Cre, Cre knock-in allele; Krt12+, wild-type allele.
It should be noted that allelic expression of the Krt12 gene provides an evolutionary advantage to allow the maintenance of corneal epithelial integrity in those who inherit a heterozygous Krt12 mutation. Krt12 protein is a member of the building blocks of polymeric corneal epithelial intermediate filaments. As such, if both alleles were equally expressed, a heterozygous mutation would lead to interruption in the formation of functional intermediate filaments in all corneal epithelial cells, rendering severe defects to the corneal epithelium. On the contrary, allelic selection would allow cells that do not express the mutant Krt12 allele but express the wild-type allele to gain the advantage in their centripetal movement during corneal epithelial cell differentiation. We have also shown that because of the allelic selection of Krt12, to successfully ablate genes in the corneal epithelium Krt12cre/cre mice have to be used. Although we show the allelic selection of Krt12, the genetic or epigenetic mechanism responsible for such selection remains unclear and will be pursued in the future.
Acknowledgments
The authors thank Sandy Schwemberger for assistance with fluorescence-activated cell sorting of the corneal epithelium.
Footnotes
Supported in part by National Institutes of Health/National Eye Institute Grant EY 10556, Research to Prevent Blindness, and Ohio Lions Eye Research Foundation.
Disclosure: Y. Hayashi, None; M.K. Call, None; C.-Y. Liu, None; M. Hayashi, None; G. Babcock, None; Y. Ohashi, None; W.W.-Y. Kao, None
References
- 1.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature. 1961;190:372–373 [DOI] [PubMed] [Google Scholar]
- 2.Mintz B, Bradl M. Mosaic expression of a tyrosinase fusion gene in albino mice yields a heritable striped coat color pattern in transgenic homozygotes. Proc Natl Acad Sci U S A. 1991;88:9643–9647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradl M, Larue L, Mintz B. Clonal coat color variation due to a transforming gene expressed in melanocytes of transgenic mice. Proc Natl Acad Sci U S A. 1991;88:6447–6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mintz B. Gene control of mammalian pigmentary differentiation, I: clonal origin of melanocytes. Proc Natl Acad Sci U S A. 1967;58:344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawles ME. Some observations on the developmental properties of the presumptive hind-limb area of the chick. Anat Rec. 1947;99:648. [PubMed] [Google Scholar]
- 6.Sano Y, Shimada T, Nakashima H, et al. Random monoallelic expression of three genes clustered within 60 kb of mouse t complex genomic DNA. Genome Res. 2001;11:1833–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Happle R. Monoallelic expression on autosomes may explain an unusual heritable form of pigmentary mosaicism: a historical case revisited. Clin Exp Dermatol. 2009;34:834–837 [DOI] [PubMed] [Google Scholar]
- 8.Hwang LY, Lee JB, Richard G, Uitto JJ, Hsu S. Type 1 segmental manifestation of Hailey-Hailey disease. J Am Acad Dermatol. 2003;49:712–714 [DOI] [PubMed] [Google Scholar]
- 9.Sudbrak R, Brown J, Dobson-Stone C, et al. Hailey-Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca(2+) pump. Hum Mol Genet. 2000;9:1131–1140 [DOI] [PubMed] [Google Scholar]
- 10.Kao WW, Liu CY, Converse RL, et al. Keratin 12-deficient mice have fragile corneal epithelia. Invest Ophthalmol Vis Sci. 1996;37:2572–2584 [PubMed] [Google Scholar]
- 11.Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292 [DOI] [PubMed] [Google Scholar]
- 12.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326 [DOI] [PubMed] [Google Scholar]
- 13.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155 [PubMed] [Google Scholar]
- 14.Moyer PD, Kaufman AH, Zhang Z, Kao CW, Spaulding AG, Kao WW. Conjunctival epithelial cells can resurface denuded cornea, but do not transdifferentiate to express cornea-specific keratin 12 following removal of limbal epithelium in mouse. Differentiation. 1996;60:31–38 [DOI] [PubMed] [Google Scholar]
- 15.Tanifuji-Terai N, Terai K, Hayashi Y, Chikama T, Kao WW. Expression of keratin 12 and maturation of corneal epithelium during development and postnatal growth. Invest Ophthalmol Vis Sci. 2006;47:545–551 [DOI] [PubMed] [Google Scholar]
- 16.Hu-Li J, Pannetier C, Guo L, et al. Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity. 2001;14:1–11 [DOI] [PubMed] [Google Scholar]
- 17.Pereira JP, Girard R, Chaby R, Cumano A, Vieira P. Monoallelic expression of the murine gene encoding Toll-like receptor 4. Nat Immunol. 2003;4:464–470 [DOI] [PubMed] [Google Scholar]
- 18.Riviere I, Sunshine MJ, Littman DR. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228 [DOI] [PubMed] [Google Scholar]
- 19.Bayley JP, van Rietschoten JG, Bakker AM, et al. Allele-specific expression of the IL-1 alpha gene in human CD4+ T cell clones. J Immunol. 2003;171:2349–2353 [DOI] [PubMed] [Google Scholar]
- 20.Grigoriadou K, Boucontet L, Pereira P. T cell receptor-gamma allele-specific selection of V gamma 1/V delta 4 cells in the intestinal epithelium. J Immunol. 2002;169:3736–3743 [DOI] [PubMed] [Google Scholar]
- 21.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209 [DOI] [PubMed] [Google Scholar]
- 22.Davanger M, Evensen A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature. 1971;229:560–561 [DOI] [PubMed] [Google Scholar]
- 23.Schermer A, Galvin S, Sun TT. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu CY, Zhu G, Westerhausen-Larson A, et al. Cornea-specific expression of K12 keratin during mouse development. Curr Eye Res. 1993;12:963–974 [DOI] [PubMed] [Google Scholar]
- 25.Liu CY, Zhu G, Converse RL, et al. Characterization and chromosomal localization of the cornea- specific murine keratin gene Krt1.12. J Biol Chem. 1994;269:24627–24636 [PubMed] [Google Scholar]