Abstract
Zyxin is a dual-function LIM domain protein that regulates actin dynamics in response to mechanical stress and shuttles between focal adhesions and the cell nucleus. Here we show that zyxin contributes to UV-induced apoptosis. Exposure of wild-type fibroblasts to UV-C irradiation results in apoptotic cell death, whereas cells harboring a homozygous disruption of the zyxin gene display a statistically significant survival advantage. To gain insight into the molecular mechanism by which zyxin promotes apoptotic signaling, we expressed an affinity-tagged zyxin variant in zyxin-null cells and isolated zyxin-associated proteins from cell lysates under physiological conditions. A 130-kDa protein that was co-isolated with zyxin was identified by microsequence analysis as the Cell Cycle and Apoptosis Regulator Protein-1 (CARP-1). CARP-1 associates with the LIM region of zyxin. Zyxin lacking the CARP-1 binding region shows reduced proapoptotic activity in response to UV-C irradiation. We demonstrate that CARP-1 is a nuclear protein. Zyxin is modified by phosphorylation in cells exposed to UV-C irradiation, and nuclear accumulation of zyxin is induced by UV-C exposure. These findings highlight a novel mechanism for modulating the apoptotic response to UV irradiation.
Keywords: zyxin, CARP-1, apoptosis
Introduction
Cancer is the second leading cause of death in the United States. Tumors arise when control points regulating cell growth or death are compromised. Chemotherapeutic agents such as taxol interfere with cell division.1 An alternative strategy to eliminate tumor cells is to stimulate apoptosis, or programmed cell death2; thus, understanding of pathways that control cell death has important implications for therapeutic development.3
Cellular programs that control apoptotic machinery can be influenced in multiple ways. For example, genomic damage promotes cell death via the p53 tumor suppressor,4 and the Fas ligand, a member of the tumor necrosis factor family, triggers apoptosis when it binds and induces signaling via the cell surface Fas receptor.5 Integrin-dependent adhesion can also influence apoptosis. For example, the fibronectin receptor (α5β1 integrin) protects against apoptosis by stimulating the PI-3kinase/AKT survival pathway.6
Evidence that loss of integrin-dependent adhesion can trigger apoptosis has fueled an interest in the possible roles of focal adhesion proteins in apoptotic control. The focal adhesion kinase, which is activated by integrin engagement, suppresses apoptosis by binding to the Fas receptor complex,7 integrin-linked kinase promotes cell survival by activating AKT,8 and zyxin promotes apoptosis in Ewing’s sarcoma cells exposed to antibodies that crosslink the cell surface receptor, CD99.9
Zyxin regulates actin assembly10-12 and shuttles between focal adhesions and cell nuclei.13-17 Zyxin displays 3 tandemly arrayed LIM domains, double zinc finger structures that dock proteins and are often found in transcriptional regulators.18,19 Here, we demonstrate that targeted deletion of the zyxin gene confers a survival advantage to fibroblasts subjected to UV-C irradiation, an apoptotic stimulator. We identified Cell Cycle and Apoptosis Regulator Protein-1 (CARP-1) as a zyxin-binding partner. CARP-1 was originally described as a protein required for tumor cell death in response to retinoids and adriamycin.20 CARP-1 is closely related to Deleted in Breast Cancer-1 (DBC-1), cloned from a genomic region that is frequently deleted in human breast tumors.21 DBC-1 interacts with BRCA1, a gene mutated in inherited breast and ovarian cancer, and promotes apoptosis in response to caspase-dependent cleavage.22,23 We report that the LIM region of zyxin is critical for CARP-1 docking as well as zyxin’s proapoptotic effect and suggest that CARP-1 and zyxin cooperate to promote apoptosis.
Results
Loss of zyxin promotes cell survival after UV-C treatment
Apoptosis is stimulated when cells acquire DNA damage as the result of genotoxic stress. Exposure of cultured cells to UV-C irradiation has allowed exploration of the apoptotic machinery and has revealed involvement of p53-dependent and -independent pathways in the cell death response.24 Recent reports using RNA interference suggested a role for zyxin in apoptotic signalling9 and prompted us to explore the involvement of zyxin in apoptosis using cells that harbor a targeted disruption of the zyxin gene and a complete elimination of protein function.
Exposure of wild-type mouse embryonic fibroblasts (MEFs) to UV-C irradiation for 1 minute results in the death of 84% of the cells by 16 hours postexposure, as assayed by an MTT assay of cell viability (Fig. 1a). To distinguish apoptotic from necrotic cell death, we performed TUNEL assays to determine whether the DNA fragmentation that characterizes apoptosis occurs in MEFs exposed to UV-C irradiation. As seen in Figure 1b, UV-C treatment causes a profound increase in the percentage of TUNEL-positive cells, from a baseline of <1% in untreated cells to >65% in UV-C–exposed cells. Activation of the proteolytic capacity of caspase3 is a hallmark of apoptosis.25 Supporting the conclusion that UV-C exposed MEFs die due to apoptosis, caspase3 levels were significantly increased in UV-C– irradiated cells as compared to controls (Fig. 1c).
Figure 1.
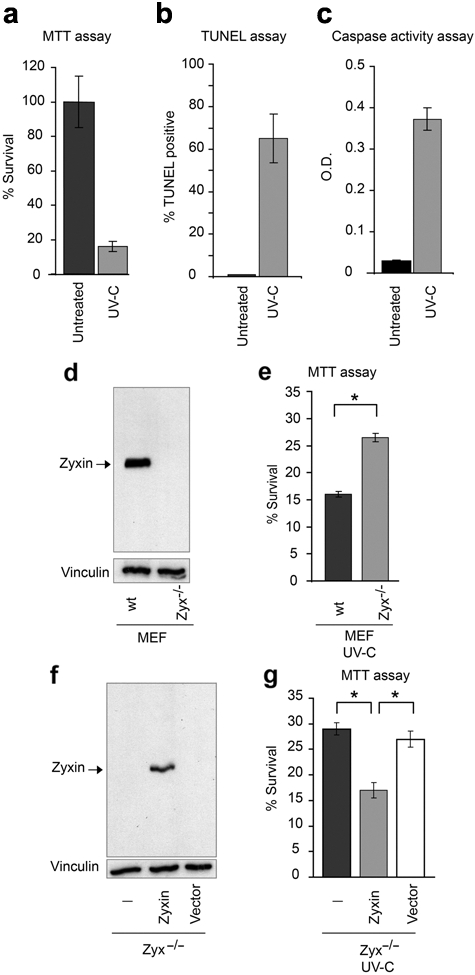
Loss of zyxin promotes cell survival after UV irradiation. Response of mouse embryonic fibroblasts (MEFs) to 1 minute UV-C irradiation after 16 hours using (a) MTT viability assay, (b) TUNEL assay to detect DNA fragmentation, and (c) measurement of caspase3 activation. (d) Immunoblot analysis of zyxin in wild-type (wt) and zyxin-null (Zyx-/-) MEFs, with vinculin detection as a loading control. (e) Survival rate of wild-type and zyxin-null cells after UV-C irradiation as measured by MTT assay. (f) Immunoblot analysis of zyxin protein levels. (g) Survival of cells after UV-C irradiation as measured by MTT assay. Error bars represent SEM; *P < 0.01.
To assess the effect of zyxin expression on the apoptotic response, we compared the sensitivity of wild-type and zyxin (-/-) MEFs (Fig. 1d) to UV-C treatment. As noted above, only 16% of wild-type cells survived UV-C irradiation under our experimental conditions (Fig. 1e). In contrast, 27% of the zyxin (-/-) cells survived (Fig. 1e). To confirm the contribution of zyxin to the apoptotic response, we performed rescue experiments. We established a cell line derived from immortalized zyxin (-/-) cells,26 in which a transgene encoding wild-type zyxin was constitutively expressed following retroviral gene transfer (Fig. 1f). With this approach, the cell lines are genetically identical and vary only in whether or not they express zyxin. After UV-C irradiation, we observed a statistically significant decrease in the viability of cells in which zyxin expression was reconstituted compared to the parental zyxin (-/-) cells or cells infected with control virus (Fig. 1g). Collectively, these results reveal that elimination of zyxin expression promotes the survival of cells exposed to UV-C irradiation.
Zyxin is phosphorylated in response to UV-C irradiation
Western immunoblot analysis revealed a prominent slower mobility zyxin isoform in cells exposed to UV-C irradiation compared to unstimulated cells (Fig. 2a, lanes 1-2). We reasoned that the reduction in zyxin’s electrophoretic mobility might reflect a posttranslational modification that occurs in response to UV-C exposure. Phosphatase treatment of the cell lysates prior to electrophoresis eliminates the occurrence of the slow mobility isoform of zyxin (Fig. 2a, lanes 3-4), illustrating that phosphorylation of zyxin occurs in response to UV-C exposure.
Figure 2.
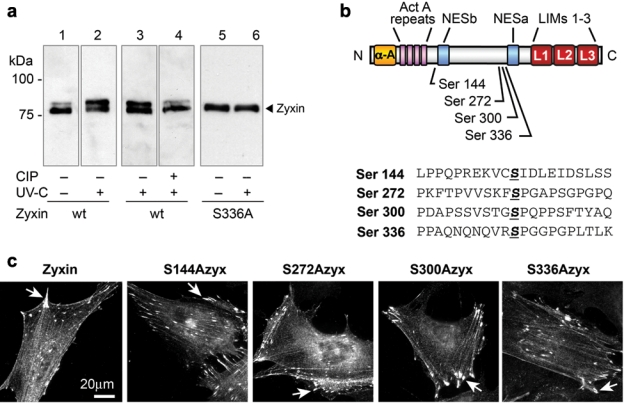
Zyxin is phosphorylated upon UV irradiation. (a) Western immunoblot reveals that zyxin migrates more slowly in SDS-PAGE when isolated from UV-C–irradiated cells (lanes 1-2), and this is reversed if the sample is treated with Calf Intestinal Phosphatase (CIP) prior to electrophoresis (lanes 3-4). Zyxin with Ser336 mutated to alanine to block phosphorylation does not shift to a slower mobility form upon UV-C exposure (lanes 5-6). (b) Diagram of zyxin residues that were shown to be phosphorylated by mass spectroscopy analysis of phosphopeptides generated from purified zyxin. (c) Site directed mutagenesis to convert the phosphorylatable Serine residues to alanine did not adversely impact zyxin’s ability to localize at focal adhesions (arrowheads).
Phosphoproteomic analysis was conducted with technical support from the Cell Migration Consortium (http://www.cellmigration.org/index.shtml). Analysis of TAP-tagged murine zyxin revealed that zyxin is phosphorylated on Serines 144, 272, 300, and 336 (Fig. 2b; http://www.cellmigration.org/resource/proteomics/data/zyxin_total.shtml). By site-directed mutagenesis, we created zyxin variants with alanine substitutions at each of these Serines. When these proteins were expressed in zyxin (-/-) cells, they retained the capacity to localize at focal adhesions (Fig. 2c). Zyxin-S336A did not display slowed electrophoretic mobility after UV exposure, identifying Ser336 as the most likely site for modification in response to UV-C exposure (Fig. 2a, lanes 5-6).
Identification of the apoptotic regulator, CARP-1, as a zyxin binding partner
We performed gene expression profiling to determine whether components of established apoptotic pathways displayed zyxin-dependent changes. To generate genetically identical cells that varied with respect to zyxin expression, the zyxin coding sequence was introduced into zyxin (-/-) cells under the control of the tetracycline promoter (Suppl. Figure S1). A survey of 50 apoptotic regulators failed to reveal altered expression levels (>1.8-fold) in the zyxin (-/-) cells (Suppl. Table S1). These findings suggest that the impact of zyxin on cell survival after UV-C exposure is not explained by altered expression of components of known apoptotic signaling cascades.
In an effort to elucidate zyxin’s role in apoptotic control, we conducted a biochemical screen to identify proteins that associate with zyxin under native conditions. We introduced a modified version of zyxin into a zyxin-null parental cell line to ensure that the engineered zyxin is the only zyxin present in the cells. This cell line, referred to as GZT (GFP-Zyxin-Tag), expresses a zyxin variant with an N-terminal GFP tag to aid in FACS isolation of transgene-expressing cells and a C-terminal ProteinA tag to support purification of protein complexes by affinity to IgG (Fig. 3a). GZT protein is detected by immunoblot at the expected molecular mass (Fig. 3b) and localizes properly to focal adhesions (Fig. 3c).
Figure 3.
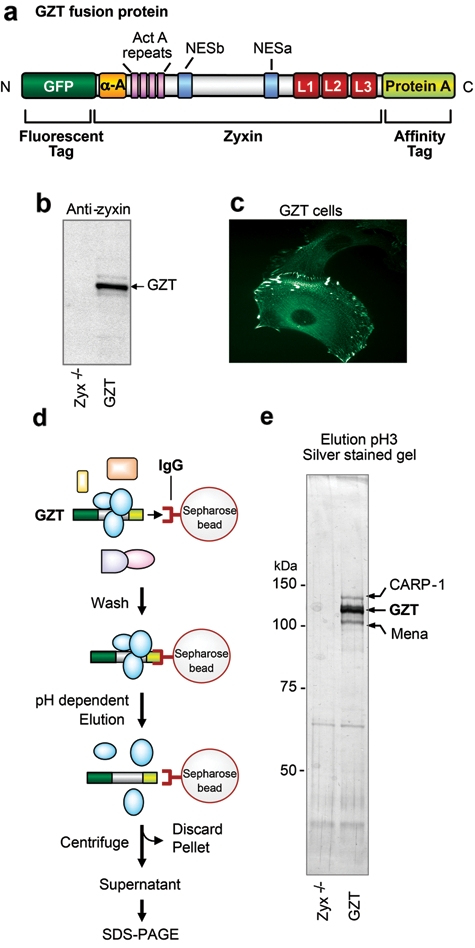
Identification of an apoptotic regulator as a zyxin-binding partner. (a) Schematic of the GZT fusion protein: GFP, green fluorescent protein; α-A, α-actinin binding site; ActA proline rich repeats; NES, nuclear export sequences; L1-2-3, LIM domains; ProteinA tag. (b) Immunoblot analysis of GZT protein expression using anti-zyxin serum, in Zyx-/- and GZT cells. (c) Subcellular localization of the GZT protein mirrors that of wild-type zyxin. (d) Schematic representation of the strategy used to isolate zyxin protein complexes. (e) Silver staining of affinity-purified products from Zyx-/- and GZT cells after SDS-PAGE. Bands specific to GZT cells were identified by mass spectrometric analysis to be CARP-1 and Mena.
The GZT fusion protein was purified in complex with its partners following the strategy presented in Figure 3d. Briefly, a cellular extract from GZT cells was incubated with IgG-Sepharose beads; GZT-containing complexes associate with the beads through the ProteinA-tagged zyxin. After extensive washing of the beads, a low-pH elution step releases the GZT protein and its binding partners. Analysis of the eluted proteins by SDS-PAGE revealed the presence of 3 prominent polypeptides that migrated at apparent molecular masses of 130 kDa, 110 kDa, and 95 kDa (Fig. 3e) and were subsequently identified by mass spectrometric analysis. The central 110-kDa band was identified as the GZT fusion protein. The 95-kDa protein was identified as Mena (NCBI acc no. AAC52864), a member of the Ena/VASP family and a previously established zyxin-binding partner.10 The 130-kDa band was identified as a novel zyxin-binding partner, CARP-1 (NCBI acc no. Q8CH18). CARP-1 was first described in human breast cancer cells as an effector of apoptosis induced by the retinoid compound CD437.20
CARP-1 is concentrated in the nucleus
Analysis of the CARP-1 sequence using Prosite reveals several features (Fig. 4a), including a SAP (SAF, Acinus, PIAS) domain that is implicated in the control of chromatin organization.27 Of interest, a founding member of the SAP domain family, Acinus, has been reported to act downstream of caspases to promote apoptotic chromosomal condensation.28 A NUDIX domain implicated in nucleoside diphosphate binding is found adjacent to the SAP domain.29 Other motifs include a potential calcium-binding EF-hand motif,30 a nucleic acid binding OB-fold domain,31 and several potential bipartite nuclear localization signals.
Figure 4.
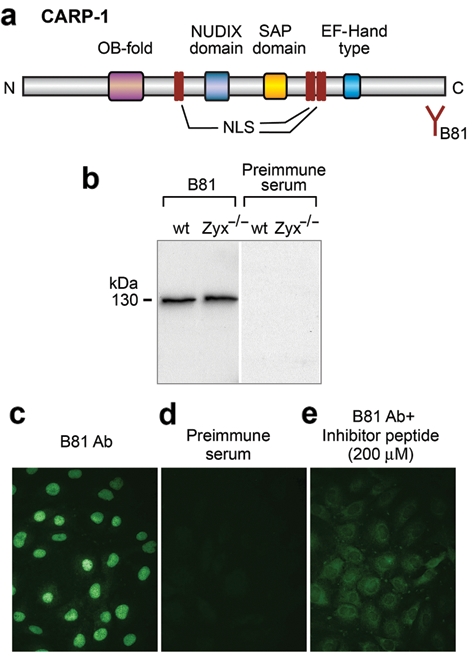
CARP-1 resides in the nuclear compartment. (a) Schematic representation of CARP-1 showing the OB-fold, NUDIX domain, SAP domain, nuclear localization sequences (NLS), and EF–hand-type domain. The epitope for the B81 antibody is at the C-terminus of CARP-1. (b) Immunoblot analysis of CARP-1 in Zyx-/- and wild-type (wt) fibroblasts using B81 anti-CARP-1 antibody and preimmune serum. (c-e) Indirect immunofluorescence of (c) CARP-1 nuclear localization in mouse fibroblasts using B81 anti–CARP-1 antibody. Specificity controls: (d) preimmune serum and (e) B81 anti–CARP-1 antibody incubated with the epitope-containing peptide.
To characterize the subcellular distribution of CARP-1 in mouse fibroblasts, we designed an antibody directed against the C-terminal tail of the protein (amino acids 1130 to 1146). As seen in Figure 4b, anti–CARP-1 antibody detects a single polypeptide of 130 kDa that is present in both wild-type and zyxin (-/-) cells; no immunoreactive band is detected with preimmune serum. By indirect immunofluorescence, anti–CARP-1 labels the nuclei of fibroblasts (Fig. 4c). Similar results were obtained with human Hela cells (not shown). No staining is observed with preimmune serum (Fig. 4d), and the labeling is eliminated when anti–CARP-1 antiserum is preincubated with the immunogenic C-terminal CARP-1 peptide (Fig. 4e).
Zyxin transits to the nucleus in response to UV-C irradiation
In contrast with CARP-1, zyxin is commonly observed in the cytoplasm, most notably at focal adhesions (Fig. 5a). However, zyxin shuttles between focal adhesions and cell nuclei and displays NES sequences that are evolutionarily conserved.13,17 Therefore, we explored whether exposure to UV-C might influence the subcellular distribution of zyxin, facilitating its access to nuclear CARP-1. By blinded immunocytochemistry studies, we detected an increase in nuclear zyxin upon exposure of cells to UV-C irradiation (Fig. 5a). Quantitative analysis revealed that both the percentage of cells with nuclear zyxin (Fig. 5b) as well as the extent of zyxin accumulation in the nucleus (Fig. 5c) increased with UV-C treatment.
Figure 5.
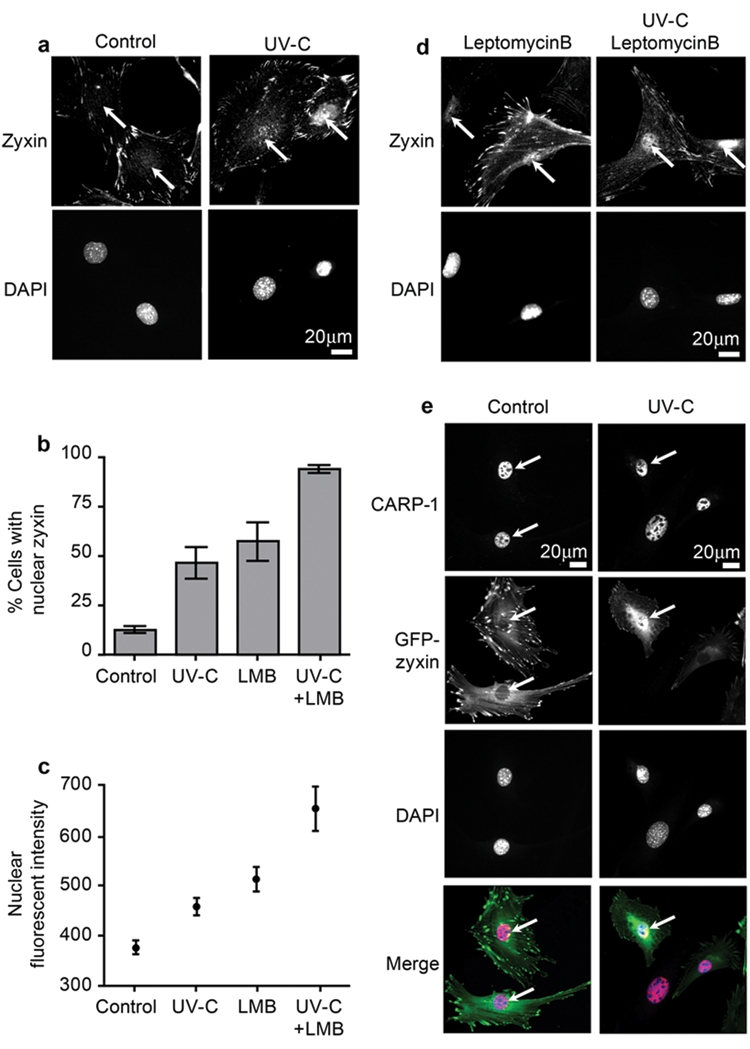
Zyxin transits to the nucleus in response to UV irradiation. (a) Indirect immunofluorescence microscopy of zyxin localization in untreated or UV-C–exposed cells. (b) Quantitative analysis of the percentage of cells with detectable nuclear zyxin in each treament condition. (c) Quantitative analysis of nuclear-localized zyxin by indirect immunofluorescence. (d) Indirect immunofluorescence microscopy of zyxin localization in untreated or UV-C exposed cells in the absence or presence of Leptomycin B (20 nM, 6 hours). (e) CARP-1 is concentrated in cell nuclei both before and after UV exposure. In cells programmed to express GFP-zyxin, which localizes normally to focal adhesions, zyxin is observed to accumulate in the nucleus and co-localize with CARP-1 upon UV-C exposure. For Merge images, zyxin (green), CARP-1 (red), and DAPI (blue) merges to white when all 3 signals overlap. Arrows indicate the position of the nucleus.
Although the accumulation of zyxin in cell nuclei after UV irradiation was statistically significant, we reasoned that an endpoint analysis might underestimate the extent of zyxin nuclear import if the protein displayed a short nuclear dwell time due to rapid export kinetics. Indeed, we showed previously that Leptomycin B, a CRM-1 inhibitor that blocks nuclear export, promotes accumulation of zyxin within cell nuclei.14 Therefore, we measured nuclear zyxin levels in Leptomycin B–treated control cells and cells that were exposed to UV-C. As can be seen in Figures 5b-d, treatment with Leptomycin B augments the ability to detect zyxin in cell nuclei, particularly after UV-C exposure.
DBC-1, a protein closely related to CARP-1, is a substrate for caspases and has been reported to translocate from the nucleus to the cytoplasm during the apoptotic response.23 Therefore, we examined the subcellular distribution of CARP-1 in cells exposed to UV-C that were triggered to undergo apoptosis. As can be seen in Figure 5e, even UV-C–treated cells with significant nuclear accumulation of zyxin fail to show a redistribution of CARP-1 to the cytoplasm.
The LIM domains of zyxin are required for interaction with CARP-1
Further support for the relevance of the zyxin–CARP-1 interaction in vivo comes from immunoprecipitation experiments with endogenous proteins. CARP-1 is co-immunoprecipitated with anti-zyxin antibody from wild-type cell lysates but not from zyxin-null cell lysates, confirming the requirement of zyxin protein for co-isolation (Fig. 6a). To identify the region of zyxin involved in the CARP-1–zyxin interaction, we constructed expression plasmids that encoded either full-length zyxin or a mutant form of the protein lacking the 3 LIM domains (zyxinΔLIM; Fig. 6b). These plasmids were transiently transfected into zyxin-null fibroblasts, and both proteins were expressed as shown by Western immunoblots (Fig. 6c). Both full-length zyxin and zyxinΔLIM are immunoprecipitated with anti-zyxin antibody (data not shown). As can be seen in Figure 6d, CARP-1 is efficiently co-immunoprecipitated when the full-length zyxin is expressed but not when the cells are expressing the zyxinΔLIM protein or are transfected with an empty plasmid. Thus, the interaction of CARP-1 with zyxin requires the LIM domains. To test whether the LIM region of zyxin is sufficient to bind CARP-1, we performed a yeast 2 hybrid-based domain analysis. We generated a panel of deletion variants for the zyxin LIM region, as represented in Figure 6e, and tested their ability to interact with full-length CARP-1. By this approach, we determined that Zyxin-LIM1-3 supports binding of CARP-1, whereas individual or paired LIM domains do not.
Figure 6.
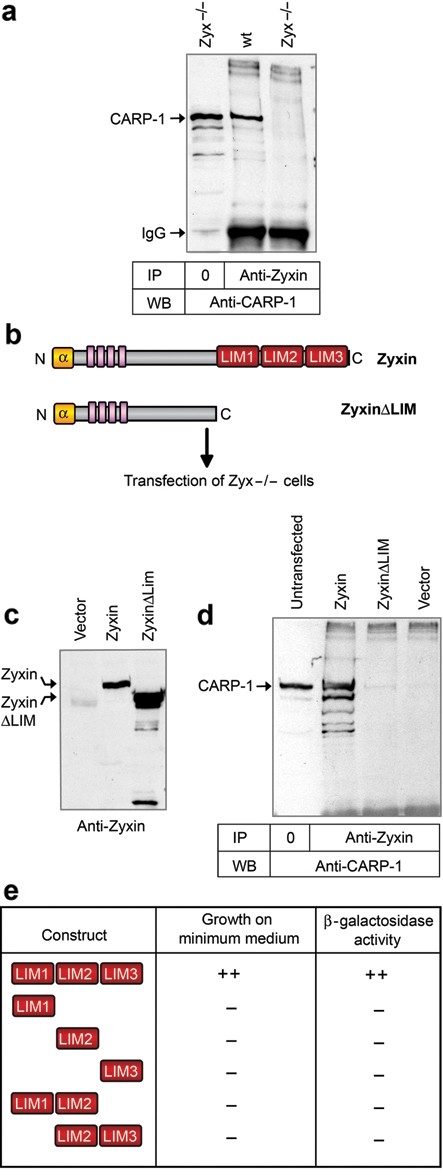
Zyxin’s interaction with CARP-1 is mediated by the LIM domains. (a) Immunoprecipitation of CARP-1 in complex with zyxin; IgG heavy chains are evident in the immunoprecipitation lanes. (b) Schematic representation of Zyxin and ZyxinΔLIM proteins. (c) Expression of zyxin and zyxinΔLIM proteins in transfected Zyxin (-/-) cells. (d) Co-precipitation of CARP-1 with zyxin depends on zyxin’s LIM region. (e) Yeast 2 hybrid analysis of full-length CARP-1 interaction with the indicated zyxin LIM domains. Growth of transformed yeast on minimum medium and β-galactosidase activity were evaluated.
The CARP-1 binding domain of zyxin is required for zyxin’s pro-apoptotic effect
To investigate whether the CARP-1–zyxin interaction is required for zyxin’s proapoptotic effect, we compared the effect of UV-C irradiation on zyxin-null cells stably reconstituted with either full-length zyxin or zyxinΔLIM, a zyxin variant that lacks the CARP-1 binding domain. Full-length zyxin and the deletion variant are expressed at comparable levels and migrate at the expected sizes based on immunoblot analysis (Fig. 7a). Cell lines with different zyxin compositions were UV-C irradiated for 1 minute, and an MTT assay was performed after 16 hours. Cells expressing the zyxinΔLIM mutant display a significant survival advantage relative to cells expressing wild-type zyxin (Fig. 7b). Thus, the LIM region containing the CARP-1 binding site contributes to zyxin’s proapoptotic activity. Interestingly, we also detected a survival advantage of zyxin-null cells relative to cells expressing the zyxinΔLIM variant, suggesting the existence of an additional CARP-1–independent proapoptotic function in the N-terminal region of zyxin.
Figure 7.
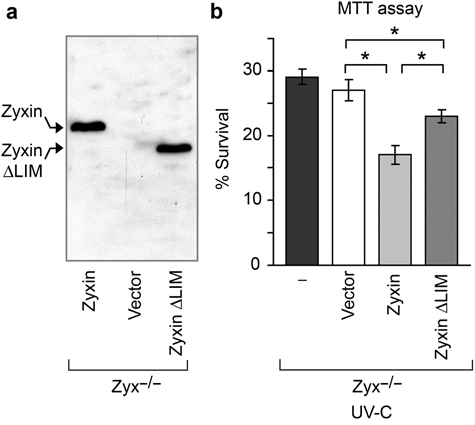
The CARP-1 binding domain of zyxin is required for its full proapoptotic effect. (a) Immunoblot analysis of zyxin variants. (b) Cell survival 16 hours after UV-C irradiation as measured by MTT assay. Error bars represent SEM; *P < 0.01.
Discussion
The results presented in this article demonstrate a role for zyxin in the control of programmed cell death in response to UV-C treatment. Cells in which the zyxin gene has been disrupted by homologous recombination display a survival advantage after UV-C irradiation stress. Compared to wild-type cells that displayed 16% overall survival after UV, 27% of the cells lacking zyxin survived. If one were to express this result in terms of absolute numbers of cells surviving UV irradiation, for every 100 wild-type cells that survive UV irradiation, 169 zyxin-null cells would survive, a significant survival advantage of nearly 1.7-fold. These findings have interesting implications for cancer biology, where suppression of zyxin expression is being identified in a variety of human tumors. For example, zyxin levels are reduced in invasive bladder carcinoma,32 and methylation-sensitive silencing of zyxin expression is a characteristic of several breast cancer cell lines.33 Reduced zyxin expression is also associated with poor prognosis of pediatric acute myeloid leukemia.34
We have demonstrated a direct interaction between zyxin and the apoptotic regulator CARP-1. CARP-1 was first identified in a screen for mediators of apoptotic signaling in response to CD437, a retinoid compound inducing apoptosis of various malignant cell types.20 Knockdown of CARP-1 abrogates apoptotic progression in response to several stimuli, including adriamycin and growth factor receptor inhibitors.20,35 Here we have shown that CARP-1 interacts with the LIM region of zyxin, and zyxin lacking the CARP-1 binding region displays diminished capacity to promote apoptosis in response to UV-C exposure.
Our immunocytochemical studies demonstrate that CARP-1 is prominently concentrated in cell nuclei. It is likely that zyxin and CARP-1 act together in cell nuclei. We showed here that antibodies directed against the C-terminus of CARP-1 reveal CARP-1 concentration in cell nuclei both before and after UV irradiation and UV-C irradiation promotes transit of zyxin to the nucleus. Further support for a role for zyxin in the nucleus comes from the identification of zyxin as a partner of the apoptosis regulator and chromatin condensation factor, acinus, another nuclear SAP domain protein.36
Additional work is required to elucidate how UV irradiation triggers the translocation of zyxin to the nucleus; however, it is interesting that the nuclear:cytoplasmic ratio of a number of proteins is influenced by phosphorylation. We demonstrated here that zyxin undergoes a shift in its electrophoretic mobility upon UV irradiation and that the change in mobility can be attributed to phosphorylation of Ser336. Interestingly, a large-scale nuclear phosphoproteome analysis identified a human zyxin peptide phosphorylated on the residue equivalent to murine zyxin Ser336 enriched in HeLa cell nuclei.37 Thus, phosphorylation of zyxin on Ser336 is both correlated with UV exposure and compatible with nuclear residence of zyxin. Future experiments will be required to determine whether phosphorylation is necessary and/or sufficient for translocation of zyxin to the nucleus and to elucidate the signaling cascades downstream of UV exposure that lead to posttranslational modification of zyxin.
In addition to the proapoptotic role of zyxin in UV-treated cells reported here, zyxin was shown to be important for the development of an apoptotic response by Ewing’s sarcoma cells exposed to antibodies directed against the cell surface protein, CD99.9 CD99 is strongly expressed on the surface of Ewing’s sarcoma cells and anti-CD99 is being explored as a potential therapy for this disease.38 Anti-CD99 promotes apoptosis by a mechanism that appears to depend on actin filament assembly and stabilization of cell-cell adhesion.9 Zyxin promotes actin assembly and also enhances the stability of cell-cell junctions.26,39 Thus, zyxin’s impact on cell adhesion and the cytoskeleton may also be important to modulate apoptotic signaling. In yeast, there is a clear link between regulation of actin dynamics, generation of reactive oxygen species, and cell death,40 and this control may apply in mammals as well.
Zyxin’s contribution to apoptotic signaling appears to be context dependent. As discussed above, zyxin promotes cell death downstream of apoptotic stimuli such as UV-C irradiation and exposure to anti-CD99 antibody (this report and Cerisano et al.9). However, zyxin has also been implicated in Akt-dependent cardiomyocyte survival pathways downstream of growth factor receptor engagement and exposure to the antiapoptotic factor atrial natriuretic peptide.16 Adding further complexity, in this case zyxin was shown to have capacity for either antiapoptotic or proapoptotic function depending on its subcellular location.16 More work is required to dissect the detailed pathways that influence zyxin’s interface with, and influence on, the apoptotic machinery. However, a growing body of evidence highlights an important role for zyxin and its partners in the central regulatory networks that influence programmed cell death.
Materials and Methods
DNA constructs
The FLAG-tag/TEV/proteinA sequence described41 was inserted in the 3′ end of GFP-mouse-zyxin DNA.26 The resulting construct was inserted in the pLINX plasmid42 to generate pLINX-GZT (GFP-zyxin-Tag). Retroviral constructs used for the expression of Zyxin and ZyxinΔLIM proteins derive from the pBABE-puro plasmid.43 PCR products corresponding to full-length zyxin or zyxin amino acids 1-376 were sequenced and inserted in pBABE-puro. Retroviruses were generated using Phoenix-Eco packaging cells (American Type Culture Collection).44
Cell lines and gene expression profiling
Wild-type and Zyxin-null fibroblast preparation and culture were as previously described.26 GZT cells were obtained after retroviral gene transfer from pLINX-GZT in Zyx-/- cells and sorted for GFP to select GZT-expressing cells. Tetracycline suppression of zyxin expression from the pLINX retroviral vector was as described.42 Microarray analysis was performed to compare cells with and without tetracycline treatment to suppress zyxin expression.
Antibodies and protein analysis
The B71 antizyxin polyclonal rabbit serum was previously described.45 B81 anti–CARP-1 antibody was generated by immunization of rabbits with the peptide CVKSEDRDEKSKENGSGV (amino acids 1129-1146) and was diluted 1:500 for indirect immunofluorescence microscopy and 1:5,000 for Western immunoblot detection. Co-immunoprecipitations were performed as previously described.46 Vinculin monoclonal antibody was purchased (Sigma-Aldrich). Zyxin (GZT) complex purification and analysis of binding partners was performed as previously described.41 For phosphorylation studies, GZT protein was purified with IgG-Sepharose then digested with trypsin, followed by HPLC peptide separation, and mass spectrometry analysis for phosphopeptide identification (http://www.cellmigration.org/resource/proteomics/data/zyxin_total.shtml).47-49
Viability/apoptosis tests
For UV-C exposure, cells were washed with PBS and irradiated for 1 minute in PBS using a UV Stratalinker 1800 (Stratagene), fresh medium was added, and cells were grown for 16 hours. Cell viability was measured using the MTT assay.50 Caspase3 activity was measured using the CaspACE assay (Promega). DNA fragmentation was investigated using TUNEL assay (in situ cell death detection kit; Roche). Cells were imaged by phase contrast and fluorescence microscopy, and the ratio of stained cells to the total number of cells was calculated.
Yeast 2-hybrid analysis
The yeast host strain, PJ69-2a, was transformed with bait and prey, and reporter activities were assayed as described previously.51
Immunofluorescence microscopy
Cells were stained using the primary antibodies described and Alexa Fluor secondary antibodies (Invitrogen). Cell images were captured using a fluorescent microscope (Zeiss AxioPhot; Carl Zeiss MicroImaging Inc.) with a Plan-Neofluar 40x, 1.30 NA objective and a Plan-Apochromat 63x 1.40 NA objective, a MicroMax digital camera (Princeton Instruments), and Openlab imaging software (Improvision). To evaluate zyxin distribution in response to UV-C exposure, wild-type fibroblasts were scored for nuclear zyxin signal from at least 10 fields/treatment in 3 independent experiments. Nuclear intensity of zyxin was determined by using the corresponding DAPI signal to select nuclei for quantitation of fluorescent intensity of anti-zyxin.
Supplementary Material
Acknowledgments
The authors are grateful to Erin Jeffery, Tara Muratore-Schroeder, Tom Parsons, and Don Hunt of the Cell Migration Consortium for support of the phosphorylation mapping, to Krishna Parsawar for protein identification by mass spectrometry, and to Diana Lim for figure preparation.
Footnotes
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
This work was supported by NIH GM50877 and the Huntsman Cancer Foundation. Huntsman Cancer Institute sshared resources were supported by P30 CA42014.
Supplementary material for this article is available on the Genes & Cancer website at http://ganc.sagepub.com/supplemental.
References
- 1. Horwitz SB. Mechanism of action of taxol. Trends Pharmacol Sci. 1992;13:134-6 [DOI] [PubMed] [Google Scholar]
- 2. Gerl R, Vaux DL. Apoptosis in the development and treatment of cancer. Carcinogenesis. 2005;26:263-70 [DOI] [PubMed] [Google Scholar]
- 3. Bray K, Chen HY, Karp CM, et al. Bcl-2 modulation to activate apoptosis in prostate cancer. Mol Cancer Res. 2009;7:1487-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15-6 [DOI] [PubMed] [Google Scholar]
- 5. Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449-56 [DOI] [PubMed] [Google Scholar]
- 6. Lee JW, Juliano RL. alpha5beta1 integrin protects intestinal epithelial cells from apoptosis through a phosphatidylinositol 3-kinase and protein kinase B-dependent pathway. Mol Biol Cell. 2000;11:1973-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurenova E, Xu LH, Yang X, Baldwin AS, Jr, Craven RJ, Hanks SK, et al. Focal adhesion kinase suppresses apoptosis by binding to the death domain of receptor-interacting protein. Mol Cell Biol. 2004;24:4361-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedrich EB, Liu E, Sinha S, et al. Integrin-linked kinase regulates endothelial cell survival and vascular development. Mol Cell Biol. 2004;24:8134-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cerisano V, Aalto Y, Perdichizzi S, et al. Molecular mechanisms of CD99-induced caspase-independent cell death and cell-cell adhesion in Ewing’s sarcoma cells: actin and zyxin as key intracellular mediators. Oncogene. 2004;23:5664-74 [DOI] [PubMed] [Google Scholar]
- 10. Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem. 2000;275:22503-11 [DOI] [PubMed] [Google Scholar]
- 11. Fradelizi J, Noireaux V, Plastino J, et al. ActA and human zyxin harbour Arp2/3-independent actin-polymerization activity. Nat Cell Biol. 2001;3:699-707 [DOI] [PubMed] [Google Scholar]
- 12. Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol. 2005;171:209-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J Cell Biol. 1997;138:1139-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nix DA, Fradelizi J, Bockholt S, et al. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J Biol Chem. 2001;276:34759-67 [DOI] [PubMed] [Google Scholar]
- 15. Cattaruzza M, Lattrich C, Hecker M. Focal adhesion protein zyxin is a mechanosensitive modulator of gene expression in vascular smooth muscle cells. Hypertension. 2004;43:726-30 [DOI] [PubMed] [Google Scholar]
- 16. Kato T, Muraski J, Chen Y, et al. Atrial natriuretic peptide promotes cardiomyocyte survival by cGMP-dependent nuclear accumulation of zyxin and Akt. J Clin Invest. 2005;115:2716-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524-32 [DOI] [PubMed] [Google Scholar]
- 18. Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211-9 [DOI] [PubMed] [Google Scholar]
- 19. Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920-31 [DOI] [PubMed] [Google Scholar]
- 20. Rishi AK, Zhang L, Boyanapalli M, et al. Identification and characterization of a cell cycle and apoptosis regulatory protein-1 as a novel mediator of apoptosis signaling by retinoid CD437. J Biol Chem. 2003;278:33422-35 [DOI] [PubMed] [Google Scholar]
- 21. Hamaguchi M, Meth JL, von Klitzing C, et al. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc Natl Acad Sci U S A. 2002;99:13647-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hiraike H, Wada-Hiraike O, Nakagawa S, et al. Identification of DBC1 as a transcriptional repressor for BRCA1. Br J Cancer. 2010;102:1061-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sundararajan R, Chen G, Mukherjee C, White E. Caspase-dependent processing activates the proapoptotic activity of deleted in breast cancer-1 during tumor necrosis factor-alpha-mediated death signaling. Oncogene. 2005;24:4908-20 [DOI] [PubMed] [Google Scholar]
- 24. Tomicic MT, Christmann M, Kaina B. Apoptosis in UV-C light irradiated p53 wild-type, apaf-1 and p53 knockout mouse embryonic fibroblasts: interplay of receptor and mitochondrial pathway. Apoptosis. 2005;10:1295-304 [DOI] [PubMed] [Google Scholar]
- 25. Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725-31 [DOI] [PubMed] [Google Scholar]
- 26. Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aravind L, Koonin EV. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112-4 [DOI] [PubMed] [Google Scholar]
- 28. Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168-73 [DOI] [PubMed] [Google Scholar]
- 29. Anantharaman V, Aravind L. Analysis of DBC1 and its homologs suggests a potential mechanism for regulation of sirtuin domain deacetylases by NAD metabolites. Cell Cycle. 2008;7:1467-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grabarek Z. Structural basis for diversity of the EF-hand calcium-binding proteins. J Mol Biol. 2006;359:509-25 [DOI] [PubMed] [Google Scholar]
- 31. Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36-42 [DOI] [PubMed] [Google Scholar]
- 32. Sanchez-Carbayo M, Socci ND, Charytonowicz E, et al. Molecular profiling of bladder cancer using cDNA microarrays: defining histogenesis and biological phenotypes. Cancer Res. 2002;62:6973-80 [PubMed] [Google Scholar]
- 33. Wang W, Huper G, Guo Y, Murphy SK, Olson JA, Jr, Marks JR. Analysis of methylation-sensitive transcriptome identifies GADD45a as a frequently methylated gene in breast cancer. Oncogene. 2005;24:2705-14 [DOI] [PubMed] [Google Scholar]
- 34. Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849-56 [DOI] [PubMed] [Google Scholar]
- 35. Rishi AK, Zhang L, Yu Y, et al. Cell cycle- and apoptosis-regulatory protein-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188-98 [DOI] [PubMed] [Google Scholar]
- 36. Chan CB, Liu X, Tang X, Fu H, Ye K. Akt phosphorylation of zyxin mediates its interaction with acinus-S and prevents acinus-triggered chromatin condensation. Cell Death Differ. 2007;14:1688-99 [DOI] [PubMed] [Google Scholar]
- 37. Beausoleil SA, Jedrychowski M, Schwartz D, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scotlandi K, Perdichizzi S, Bernard G, et al. Targeting CD99 in association with doxorubicin: an effective combined treatment for Ewing’s sarcoma. Eur J Cancer. 2006;42:91-6 [DOI] [PubMed] [Google Scholar]
- 39. Hansen MD, Beckerle MC. Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J Biol Chem. 2006;281:16178-88 [DOI] [PubMed] [Google Scholar]
- 40. Gourlay CW, Carpp LN, Timpson P, Winder SJ, Ayscough KR. A role for the actin cytoskeleton in cell death and aging in yeast. J Cell Biol. 2004;164:803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kadrmas JL, Smith MA, Clark KA, et al. The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J Cell Biol. 2004;167:1019-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoshimaru M, Ray J, Sah DW, Gage FH. Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc Natl Acad Sci U S A. 1996;93:1518-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hoffman LM, Nix DA, Benson B, et al. Targeted disruption of the murine zyxin gene. Mol Cell Biol. 2003;23:70-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pomies P, Macalma T, Beckerle MC. Purification and character ization of an alpha-actinin-binding PDZ-LIM protein that is up-regulated during muscle differentiation. J Biol Chem. 1999;274:29242-50 [DOI] [PubMed] [Google Scholar]
- 47. Coon JJ, Syka JE, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. Biotechniques. 2005;38:519, 521,, 523 [DOI] [PubMed] [Google Scholar]
- 48. Schroeder MJ, Webb DJ, Shabanowitz J, Horwitz AF, Hunt DF. Methods for the detection of paxillin post-translational modifications and interacting proteins by mass spectrometry. J Proteome Res. 2005;4:1832-41 [DOI] [PubMed] [Google Scholar]
- 49. Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ausubel FM. Current protocols in molecular biology. New York: Wiley-Interscience; 2002 [Google Scholar]
- 51. James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


