Abstract
We report the complexation of a potential anticancer agent 2-methoxyestradiol (2-ME) with generation 5 (G5) poly(amidoamine) dendrimers having different surface functional groups for therapeutic applications. The complexation experiment shows that approximately 6–8 drug molecules can be complexed with one dendrimer molecule regardless the type of the dendrimer terminal groups. The bioactivity of 2-ME complexed with dendrimers was found to be significantly dependent on the surface charge of G5 dendrimers. In vitro cell biological assays show that amine, hydroxyl, and acetamide-terminated G5 dendrimers with positive, slightly positive, and close to neutral surface charges, respectively are able to deliver 2-ME to inhibit cancer cell growth. In contrast, 2-ME complexed with carboxyl-terminated G5 dendrimers with negative surface charges does not show its inherent bioactivity. Further molecular dynamics simulation studies show that the compact structure of carboxylated G5 dendrimers complexed with 2-ME does not allow the release of the drug molecules even at a pH of 5.0, which is the typical pH value in lysosome. Our findings indicate that the surface modification of dendrimers with different charges is crucial for the development of formulations of various anticancer drugs for therapeutic applications.
Introduction
Dendrimers are a new class of highly branched, monodispersed, and synthetic macromolecules with well-defined composition and architecture.1 The tailored core, interior structure, surface groups, and generation-dependent geometric properties of den-drimers make them a quite unique material for a range of applications in catalysis,2 sensors,3–5 optics,6 electronics,7,8 environmental remediation,9–11 and drug delivery.12–14 Recent advances in dendrimer-based nanomedicine show that dendrimers have been used in two different ways for drug delivery applications: (1) dendrimers can be used as a platform to covalently conjugate drug molecules for cancer therapeutics;15–19 and (2) dendrimers or functionalized dendrimers can also be used to physically encapsulate or complex drug molecules inside their interior to improve the water solubility and bioavailability of the drugs.13,20–27 The former approach involves a covalent attachment of drugs onto dendrimer surfaces, which offers stable dendrimer-drug conjugates. However, the conjugation generally involves multi-step organic reactions and the covalent conjugation chemistry has to be optimized in order for the drug molecules to be cleaved and released at the specific biological conditions. The latter approach is relatively simple; however, the in vivo stability of the dendrimer-drug complexes could be a challenging issue. Both approaches have received much attention for the development of various drug formulations.
The drug 2-ME is present in the serum of women during the ovulatory and luteal phases of the menstrual cycle and during pregnancy. As a metabolite of 17-β estradiol, 2-ME has been demonstrated to be a potential anticancer agent.28,29 2-ME neither displays considerable estrogenic activity at clinically efficacious doses, nor seems to promote carcinogenesis. Moreover, it has been found to be active in inhibiting tumor growth in phase I/II clinical trials.30,31 In our previous studies, we have shown that 2-ME can be encapsulated into polymer multilayer capsules through a layer-by-layer (LbL) assembly approach and display similar bioactivity to the conventional formulation of 2-ME, which is a concentrated ethanol solution.32,33 Although high payload of 2-ME can be achieved using the LbL assembly approach, the size of the final formed 2-ME particles is relatively large. It is anticipated that a nanoscale, injectable formulation of 2-ME is feasible by encapsulating or complexing it within dendrimers.
In this present study, we utilized generation 5 (G5) poly(amidoamine) (PAMAM) dendrimers with amine, hydroxyl, acetamide, and carboxyl terminal groups to complex the drug 2-ME. The influence of the dendrimer surface charge on the bioactivity of 2-ME complexed with dendrimers was investigated by testing the cytotoxicity of a tumor cell line (KB cells, a human epithelial carcinoma cell line) treated with the dendrimer-drug complexes. To further understand the therapeutic efficacy of the 2-ME drug complexed with dendrimers, molecular dynamics simulation studies were performed to simulate the molecular morphology of the complexes under the specific cellular environment. As compared with other published dendrimer work relating to drug delivery applications in literature, to our knowledge this study is the first report relating to the complexation of 2-ME using dendrimers for therapeutic applications, also the first report systematically investigating the influence of dendrimer surface charge on the bioactivity of the drug molecules complexed with dendrimers. The results generated from this study provide a basis for a rational design of functional dendrimer/drug complexes for various therapeutic applications.
Experimental section
Materials
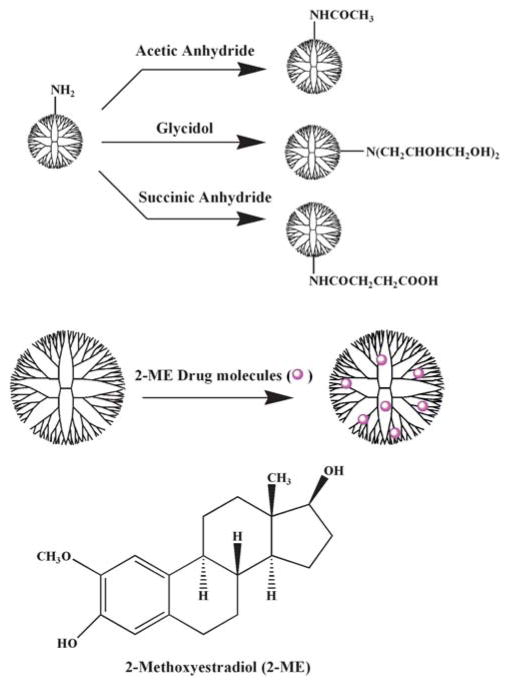
Amine-terminated PAMAM dendrimers of generation 5 (G5.NH2) with ethylenediamine core were purchased from Dendritech (Midland, MI) in methanol solution. The surfaces of G5.NH2 dendrimers were converted to acetyl, hydroxyl, and carboxyl groups by reacting with acetic anhydride, glycidol, and succinic anhydride, respectively (Scheme 1). The synthesized carboxyl, acetyl, and hydroxyl-terminated G5 PAMAM dendrimers are denoted as G5.SAH, G5.Ac, and G5.NGlyOH, respectively and were characterized using nuclear magnetic resonance, high-performance liquid chromatography (HPLC), size exclusion chromatography (SEC), and matrix-assisted laser desorption ionization-time of flight mass spectrometry. Details of the conversion procedure and characterization can be found elsewhere.34–36 The molecular weights of the G5 dendrimers are shown in Table 1. 2-ME (molecular structure is shown in Scheme 1) was purchased from Sigma. All other chemicals were obtained from Aldrich and used as received. The water used in all experiments was passed through a Millipore Milli-Q Plus 185 purification system and had a resistivity exceeding 18.2 M Ω.cm.
Scheme 1.
Schematic representation of the reactions used to synthesize G5 dendrimer derivatives and the formation of dendrimer-2-ME drug complexes.
Table 1.
Complexation capacity of 2-ME with G5 dendrimers having different terminal groups
| Dendrimers | G5.NH2 | G5.NHAc | G5.NGlyOH | G5.SAH |
|---|---|---|---|---|
| Mn | 26,010 | 30,990 | 38,382 | 40,330 |
| Polydispersity | 1.104 | 1.060 | 1.131 | 1.054 |
| Complexation capacitya | 6.2 | 8.5 | 6.7 | 7.2 |
Number of drug molecules per dendrimer.
Complexation of dendrimers with 2-ME
G5 dendrimers (10 mg) with different terminal groups (amine, acetamide, hydroxyl, and carboxyl) were dissolved in 1.5 mL water. 2-ME with 10 molar equivalents of each dendrimer derivative was dissolved in 300 μL methanol, and then mixed with the 1.5 mL dendrimer aqueous solution. The mixture solution was vigorously stirred overnight to allow the evaporation of the methanol solvent. The dendrimer-2-ME mixture solution was centrifuged (7,000 rpm for 10 min) to remove the precipitates related to non-complexed free 2-ME, which is insoluble in water. The precipitate was collected and dissolved into 1 mL methanol for HPLC analysis. The supernatant was lyophilized for 3 days to obtain the dendrimer-2-ME complexes.
HPLC analysis
The reverse phase-HPLC system used in this work consisted of a Waters Delta 600 separation module, a model 717 auto sampler equipped with a 100 μL loop, and a model 2996 PDA detector (Waters Corporation, Milford, MA). A Jupiter C5 silica-based HPLC column (250 × 4.6 mm, 300 Å) was purchased from Phenomenex (Torrance, CA). Two Phenomenex Widepore C5 safety guards (4×3 mm) were installed ahead of the Jupiter column. The mobile phase was a linear gradient beginning with 66 : 34 (v/v) water-acetonitrile (ACN) to 30 : 70 water/ACN within 20 min at a flow rate of 1 mL min−1. The injection volume was 35 μL. The detection of eluted samples was performed at 205 nm. The non-complexed 2-ME was analyzed based on a free 2-ME calibration curve.
Zeta potential analysis
Zeta-potential measurements were carried out using a Zetasizer Nano ZS system (Malvern, UK) equipped with a standard 633 nm laser. Each component of the dendrimer/drug complexes at different pH conditions (5.0, 7.0, and 10.0) was measured. The G5 dendrimers with a concentration of 1 mg mL−1 were prepared before measurement. The 2-ME drug has very limited water solubility (5.34 nM in water).37 Therefore, for 2-ME, a saturated water solution was used for the measurement.
Cell biological evaluation
KB cells (ATCC, CLL17, Rockville, MD) were continuously grown in RPMI 1640 medium supplemented with 10% heat-inactivated FBS, and 2.5 μM FA. One day before experiments, cells (1 × 104 cells per well) were plated into a 96-well plate in complete medium. The next day, 2-ME (10 (μM) in ethanol (1 μL) and 2-ME/dendrimer complexes with the same 2-ME concentration were added to cells and then incubated for 48 h at 37 °C. An MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to quantify the viability of cells. After 48 h incubation with 2-ME or 2-ME/dendrimer complexes, the metabolically active cells were then detected by adding MTT to each well. Then, the plates were read at 570 nm. Mean and standard deviation for the triplicate wells were reported. One way ANOVA statistical analysis was performed using statistical software SAS (SAS Institute Inc., Cary, North Carolina).
After treatment with 2-ME or 2-ME/dendrimer complexes, the cell morphology was also observed by phase-contrast microscopy (Leica DMIRB fluorescent inverted microscope). The magnification was set at 200 × for all samples.
Molecular dynamics (MD) simulations
G5 dendrimers with different surface functional groups were modeled and simulated at pH 5.0 simulation condition using parameters similar to those used in previous studies (pH = 7.0).38 These parameters used best mimicked explicit water environment for PAMAM dendrimers at pH 5.0, 7.0, 10.0 simulation conditions according to our previous study and the details will be followed.39 Seven 2-ME drug molecules were randomly incorporated inside the dendrimer interior after the G5 dendrimers were equilibrated. All models were built on an Onyx workstation (Silicon Graphics, Inc.; Mountain View, CA) using the Insight II software package (Accelrys, Inc.; San Diego, CA). Insight II software calculates total energy as bonded (Ubonded: empirical potential energy terms describing chemical bonds) and non-bonded (Unonbonded: Lennard-Jones and Coulomb potential energy) interactions in MD simulations. In the case of a consistent valence force field (CVFF),40 the non-bonded terms are expressed as follows:
| (1) |
where ε is the minimum energy of the Lennard-Jones potential, σ the distance yielding minimum Lennard-Jones potential, q the partial charge of the atom, D the dielectric constant (1 for vacuum), r the distance between i and j, and i,j non-bonded atom pairs. Based on previous findings, we used distance-dependent dielectric constant D = r without a long-range interaction cut-off in the simulations.39 Thus dielectric constant D was not fixed, but was changing with the distance r. After 5000 steps of steepest descent minimization, MD simulations were performed at 1000 K for 5 ps followed by 100 ps runs with 1 fsec step at 300 K, using a CVFF. The potential energies stabilized much earlier than 50 ps and the mean values were calculated in simulation from 40 ps to 100 ps. The equilibration has been confirmed by monitoring not only the potential energy but also the time evolution of the radius of gyration. The radius of gyrations, radial distribution, and surface areas of the complexes were calculated using the Decipher module in the Insight II software. We note that the force field is optimized for a poor solvent system, a vacuum. The Insight II software provides several force fields with empirical parameters for individual atoms. The combination of Lennard-Jones and Coulomb in the CVFF force field will naturally provide hydrogen bonds. Therefore, when there is a solvent such as water molecules, it calculates as if water molecules are filled in the vacuum (a poor solvent). We have found that the CVFF force field works very well (results similar to those in experiments) in our system (see below).
Results and discussion
In this study, we have demonstrated that similar to the complexation of other hydrophobic drugs using dendrimers,22–24 2-ME can also be complexed within dendrimers having different terminal groups (Scheme 1). The formation of 2-ME/dendrimer complexes is primarily based on hydrophobic interactions between dendrimers and 2-ME drug molecules. The dendrimer interior is proven to be hydrophobic,22–24 enabling effective encapsulation of hydrophobic 2-ME drug molecules.37 The payload of 2-ME within G5 dendrimers was evaluated using HPLC (Table 1). Approximately 6–9 2-ME molecules can be complexed within each G5 dendrimer molecule regardless of the type of their terminal groups. The formed 2-ME/dendrimer complexes are soluble in water and PBS buffer after lyophilization. The aqueous complex solution stored in 4 °C is stable for at least 12 months.
The surface potential changes of each component of the dendrimer/drug complexes at different pH conditions are listed in Table 2. This information is crucial for understanding the cellular interactions of the complexes. As a structually neutral molecule, the surface potential of 2-ME is close to neutral at all pH conditions. For G5.NH2, G5.NHAc, and G5.NGlyOH dendrimers, at the pH 7.0 and pH 5.0 conditions, the surface potential reasonably follows the order of G5.NH2 > G5.NGlyOH > G5.NHAc due to the surface modifications.41 The larger values at pH 5.0 for all three dendrimers compared to those at pH 7.0 should be ascribed to the pronation of a portion of the dendrimer tertiary amines.42 For G5.SAH dendrimers, the surface potential at pH 5.0 is close to zero, while at pH 7.0 and 10.0, the surface potentials are negative due to the deprotonation of the terminal carboxyl groups.
Table 2.
Surface potentials of 2-ME and G5 dendrimers with different surface functional groups in aqueous solution at different pH conditions
| Materials | Zeta potential/mV |
||
|---|---|---|---|
| pH 5.0 | pH 7.0 | pH 10.0 | |
| 2-MEa | −0.00997 ± 0.359 | −1.31 ±0.734 | −4.45 ± 1.01 |
| G5.NH2 | 39.2 ± 1.09 | 36.2 ± 1.41 | 0.358 ± 0.38 |
| G5.NHAc | 19.1 ± 1.45 | 9.18 ± 1.23 | −12.6 ± 3.08 |
| G5.NGlyOH | 29.1 ±7.12 | 22 ± 3.02 | 0.194 ±0.523 |
| G5.SAH | −0.55 ± 0.282 | −34.7 ± 1.27 | −40 ± 0.894 |
2-ME is nearly water insoluble. The saturated water solution of 2-ME was prepared for measurement.
The therapeutic efficacy of 2-ME complexed with G5 dendrimers was tested using KB cells, a human epithelial carcinoma cell line. 2-ME generally exerts its function through the induction of G2/M cycle arrest of the cells.29 The G2/M cycle starts to appear around 48 h after cell incubation. Therefore, after incubation of the 2s-ME/dendrimer complexes with cells for 48 h, an MTT assay was performed to evaluate the viability of KB cells (Fig. 1). It appeared that both free 2-ME and 2-ME/G5.NH2, 2-ME/G5.NHAc, 2-ME/G5.NGlyOH complexes caused a significant loss of cell viability in KB cells when compared with the untreated control cells (p value <0.0001 for each). In contrast, 2-ME/G5.SAH complex does not display the anticipated bioactivity. The ethanol (1 μL) used to dissolve free 2-ME drug in the culture media does not exert any favorable influence on the cell viability (p = 0.99), in agreement with our previous work.33
Fig. 1.
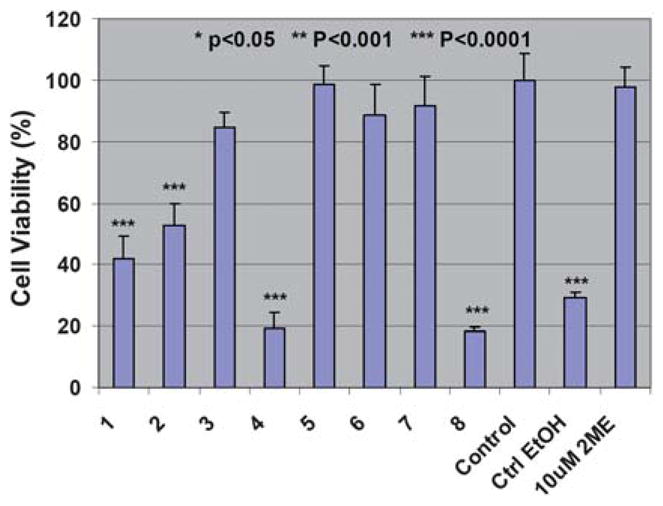
MTT assay of KB cell viability after treatment with free 2-ME (10 μM) dissolved in 1 μL ethanol, ethanol only (1 μL), 2-ME complexed with G5.NglyOH (1), G5.NHAc (2), G5.SAH (3), and G5.NH2 (4) dendrimers, and G5.NglyOH (5), G5.NHAc (6), G5.SAH (7), and G5.NH2 (8) dendrimers without the complexation of 2-ME for 48 h. The data are expressed as mean ± S. D.
To exclude the possible inherent toxicity of the dendrimers, G5 dendrimers with amine, acetyl, hydroxyl, and carboxyl terminal groups without the complexation of 2-ME were also tested. The concentrations of the G5 dendrimers tested were similar to those complexed with 2-ME used for MTT assay. It is clear that except G5.NH2 dendrimers, G5.NHAc, G5.NGlyOH, and G5.SAH are non-toxic (p < 0.0001, 0.22, 0.71, 0.99, respectively). It implies that the toxic effect of 2-ME/G5.NH2 complex may involve in both the 2-ME drug and the G5.NH2 dendrimers.41,43 However, the therapeutic activity of 2-ME/G5.NHAc and 2-ME/G5.NGlyOH complexes are solely related to the drug 2-ME.
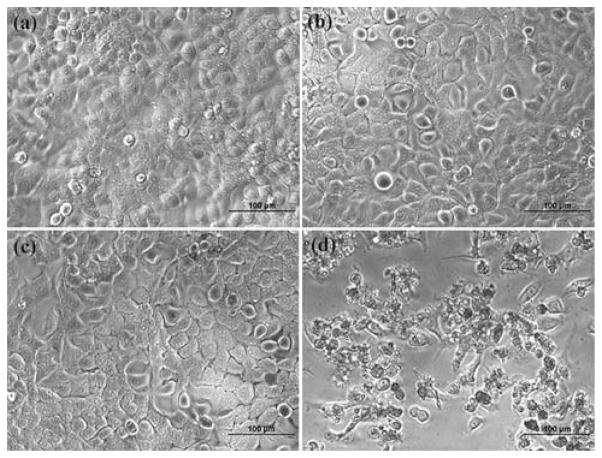
The cytotoxic effect of 2-ME drug/dendrimer complexes was further confirmed by phase contrast microscopic visualization of the cell morphology change after treatment with 2-ME with different formulations. Fig. 2 shows the morphology of the untreated KB cells, KB cells treated with 2-ME in ethanol solution, and KB cells treated 2-ME/dendrimer complexes, respectively. 2-ME in ethanol (Fig. 2e), 2-ME/G5.NGlyOH, 2-ME/G5.NHAc, and 2-ME/G5.NH2 complexes (Fig. 2a, 2b and 2 with similar 2-ME concentration (10 μM) induced similar cell morphology changes. A significant portion of the cells became rounded and non-adherent, indicative of the fact that cells have undergone apoptosis (Fig. 2). In contrast, no rounded and detached cells can be visualized in control cells without 2-ME treatment (Fig. 2f) and cells treated with 1 μL ethanol (Fig. 2g). In addition, KB cells treated with 2-ME/G5.SAH complex do not show any morphology changes when compared to the untreated control cells.
Fig. 2.
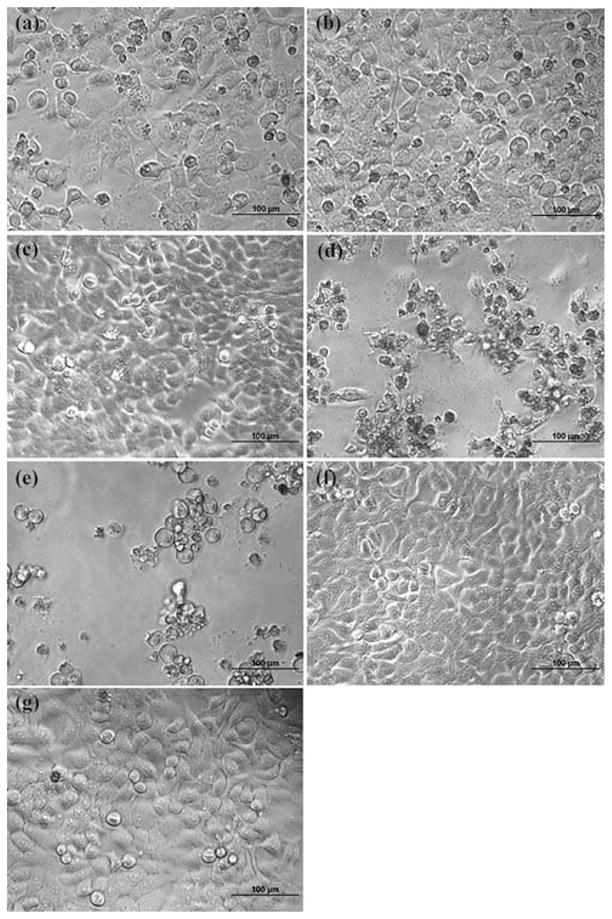
Phase-contrast photomicrographs of KB cells treated with 2-ME (10 μM) complexed with G5.NglyOH (a), G5.NHAc (b), G5.SAH (c), and G5.NH2 (d) dendrimers, respectively, KB cells treated with free 2-ME (10 μM) dissolved in 1 μL ethanol (e), the control KB cells without treatment (f), and the KB cells treated with 1 μL ethanol (g).
The KB cells were also treated with G5 dendrimers (without the complexation of 2-ME) with dendrimer concentrations similar to those in the used drug complexes. Except G5.NH2, all other dendrimers (G5.NGlyOH, G5.NHAc, and G5.SAH) do not exhibit any toxic effect (Fig. 3). This suggests that the bioactivity of 2-ME/G5.NGlyOH and 2-ME/G5.NHAc complexes are solely related to the drug 2-ME. G5.NH2 displays its inherent cytotoxicity related to the high local concentration of the surface amine groups (Fig. 3d).41 These results are consistent with the above MTT assay data.
Fig. 3.
Phase-contrast photomicrographs of KB cells treated with G5.NglyOH (a), G5.NHAc (b), G5.SAH (c), and G5.NH2 (d) dendrimers without the complexation of 2-ME.
Literature data show that the dendrimers or dendrimer-metal nanocomposites with amine, acetyl, hydroxyl, and carboxyl groups can all be internalized regardless of the dendrimer surface groups.44,45 For dendrimers not functionalized with targeting molecules, the cellular internalization process mainly involves two different mechanisms: phagocytosis and diffusion via the cell membranes. It is reasonable to understand that 2-ME/G5.NHAc and 2-ME/G5.NGlyOH complexes with close to neutral and slightly positive charge (at pH 7.0) can be internalized by cells. After the initial lysosomal uptake of the drug complexes, the 2-ME drug is then dissociated from the complexes in the lysosomes of the cells under acidic pH conditions (pH 5.0) and exerts its therapeutic effect. The non-toxic effect of 2-ME/G5.SAH complex is rather surprising (p = 0.024). We believe that this may be due to the strong interaction between the dendrimer surface carboxyl groups with its internal tertiary amines, which significantly compact the morphology of the dendritic structure, hence inhibiting the release of 2-ME drug molecules even at the acidic lysosomal environment.
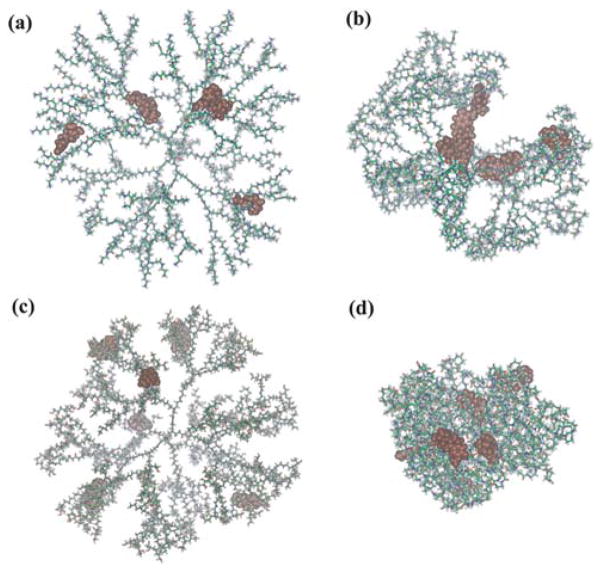
In order to delineate the molecular structures of dendrimer/drug complexes and to verify our hypothesis related to the drug release characteristics, MD simulations were performed to compare the structural difference of the dendrimer/drug complexes with different dendrimer terminal groups at the simulated lysosomal pH conditions (pH = 5.0). Table 3 lists the average characteristic values of the simulations. The radius of gyration (Rg) of G5. NH2, G5.NHAc, G5.NGlyOH, and G5.SAH dendrimers were 3.8, 3.2, 4.1, and 2.1 nm, respectively. Under acidic conditions, the protonation process leads to a relatively large geometry of G5.NH2, G5.NGlyOH, and G5.NHAc dendrimers with terminal primary (for G5.NH2 dendrimers), a portion of terminal primary or secondary amines (for G5.NGlyOH dendrimers),35 or a small amount of terminal primary amines (for G5.NHAc dendrimers)34 when comparing to G5.SAH dendrimers. The average drug positions from the center of the mass (CM) of the geometry for 2-ME/G5.NH2 and 2-ME/G5.NGlyOH complexes are farther than those of 2-ME/G5.SAH and 2-ME/G5.NHAc complexes. In addition, some drug molecules existed at a position farther than 10 nm from the CM of 2-ME/G5.NH2 and that of 2-ME/G5.NGlyOH complexes. How then can drugs within 2-ME/G5.NHAc complexes be effective when they are close to the CM? A representative configuration of the dendrimer/drug complexes with different dendrimer terminal groups is shown in Fig. 4. For the 2-ME/G5.SAH complex (Fig. 4d), it is clear that 2-ME drug molecules are completely entrapped within the compacted G5.SAH dendrimer molecule. The compact structure of G5.SAH could be related to the strong interaction between the surface carboxyl groups and the internal tertiary amine groups. In contrast, for all the other 3 types of 2-ME/dendrimer complexes with amine (Fig. 4a), acetyl (Fig. 4b), and hydroxyl (Fig. 4c) groups, the molecular structures are quite open, allowing for the release of drug molecules under acidic conditions. Therefore, in the complexes of 2-ME/G5.NH2, 2-ME/G5.NHAc, and 2-ME/G5.NGlyOH, 2-ME can be released and exert its therapeutic effect (Fig. 1 and 2). Noting that for the complex of 2-ME/G5.NH2, the dendrimer G5.NH2 shows its inherent cytotoxicity.
Table 3.
Physical characteristics of dendrimer/drug complexes with different dendrimer terminal groups after MD simulations under an acidic condition (pH 5.0)
| 2-ME/G5.NH2 | 2-ME/G5.NHAc | 2-ME/ G5.NGlyOH | 2-ME/G5.SAH | |
|---|---|---|---|---|
| Rg/nm of dendrimers | 3.8 | 3.2 | 4.1 | 2.1 |
| Number of drugs > 10 nm from CM | 2 | 0 | 1 | 0 |
| Average drug position from CM after simulations/nma | 2.8 | 1.5 | 3.9 | 1.9 |
| Average drug position from CM before simulations/nm | 2.7 | 1.6 | 2.2 | 3.1 |
Drugs released from the dendrimer are not included.
Fig. 4.
Equilibrated configurations of 2-ME/G5.NH2 (a), 2-ME/G5.NHAc (b), 2-ME/G5.NGlyOH (c), and 2-ME/G5.SAH (d) complexes after 100 ps molecular dynamics simulations. The grape-like moieties on all configurations represent 2-ME drug molecules.
The MD simulation results enhanced our understanding of the experimental results (Fig. 1 and 2). In our previous studies,39 we found that simulation results of dendrimer Rg correspond to the experimental results reasonably well. Here, we used the MD simulations to obtain the overall characteristics of each model, which are difficult to obtain in experiments such as to determine drug distribution characteristics in individual dendrimers as shown in Table 3 or configurations of dendrimer-drug complexes as shown in Fig. 4. The snapshots presented in Fig. 4 are actually representative of many ensembles for each model. Data shown in Table 3 are also average values of 300 ensembles (during the last 60 ps of simulations) and are in accordance with Fig. 4.
Using 2-ME/dendrimer complexes as a formulation could overcome the water-insolubility and improve the bioavailability of the drug. It is anticipated that, in in vivo studies, the side effect should be significantly decreased because of the protection of dendrimers. From this study, we show that 2-ME/G5.NGlyOH and 2-ME/G5.NHAc complexes with respective slightly positive and close to neutral surface charges (at pH 7.0) should be ideal delivery vehicles for enhanced antitumor therapy. 2-ME/G5.SAH and 2-ME/G5.NH2 complexes with extremely negative and positive charge at pH 7.0 are not suitable delivery vehicles, since the negative 2-ME/G5.SAH complex does not allow the delivery of 2-ME and the positive 2-ME/G5.NH2 complex exhibits the inherent toxicity of the G5.NH2 dendrimers. It indicates that the surface charge of dendrimers significantly affects the efficacy of the drug complexed with dendrimers. Further detailed studies related to the release profiles of the dendrimer/drug complexes as well as the intracellular drug retention at different time points may be necessary to delineate the relevant mechanisms.
Conclusions
In summary, we have complexed a potential antitumor agent 2-ME with dendrimers having different surface charges. The complexation approach significantly improves the water solubility of the drug. The bioactivity of the 2-ME in the drug/dendrimer complexes largely depends on the surface charge of the dendrimers. 2-ME/G5.NGlyOH and 2-ME/G5.NHAc complexes with respective slightly positive and close to neutral surface charge seem to be ideal vehicles to deliver 2-ME for antitumor therapy. The dependence of the therapeutic activity of 2-ME on the surface charge of the dendrimers used to complex the drug molecules was further delineated by MD simulations. The tunable surface chemistry of dendrimers allows for effective conjugation of cancer targeting ligands (e.g., folic acid,38,46,47 RGD peptides,48 antibodies,49 and hormones50). We envision that the 2-ME should be able to be complexed with the dendrimers modified with cancer-specific targeting ligands, thereby providing a targeted drug delivery vehicle to enhance its antitumor activity.
Acknowledgments
This project has been funded in whole or in part by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, the National Basic Research Program of China (973 Program, 2007CB936000), the National Natural Science Foundation of China (20974019 and 50925312), and Shanghai Pujiang Program (09PJ1400600). S. H. W. and J. R. B thank the support from National Institutes of Health (NIH) (under the contract # NOI-CO-97111 and NIH 1 R01 CA119409). We thank Professor Amin Cao at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences for use of his Malvern Zetasizer instrument.
Notes and references
- 1.Tomalia A, Frechet JMJ. Dendrimers and Other Dendritic Polymers. John Wiley & Sons Ltd; New York: 2001. [Google Scholar]
- 2.van Manen HJ, van Veggel FCJM, Reinhoudt DN. In: Dendrimers IV: Metal Coordination, Self-assembly, Catalysis. Vogtle F, Schalley CA, editors. Springer-Verlag; Berlin: 2001. pp. 163–199. [Google Scholar]
- 3.Kim C, Park E, Song CK, Koo BW. Synth Met. 2001;123:493–496. [Google Scholar]
- 4.Koo BW, Song CK, Kim C. Sens Actuators, B. 2001;77:432–436. [Google Scholar]
- 5.Miller LL, Kunugi Y, Canavesi A, Rigaut S, Moorefield CN, Newkome GR. Chem Mater. 1998;10:1751–1754. [Google Scholar]
- 6.Busson P, Ortegren J, Ihre H, Gedde UW, Hult A, Andersson G, Eriksson A, Lindgren M. Macromolecules. 2002;35:1663–1671. [Google Scholar]
- 7.Ma D, Lupton JM, Beavington R, Burn PL, Samuel IDW. J Phys D: Appl Phys. 2002;35:520–523. [Google Scholar]
- 8.Ma H, Chen B, Sassa T, Dalton LR, Jen AK. J Am Chem Soc. 2001;123:986–987. doi: 10.1021/ja003407c. [DOI] [PubMed] [Google Scholar]
- 9.Diallo MS, Balogh L, Shafagati A, Johnson JH, Jr, Goddard WA, III, Tomalia DA. Environ Sci Technol. 1999;33:820–824. [Google Scholar]
- 10.Diallo MS, Christie S, Swaminathan P, Balogh L, Shi X, Um W, Papelis C, Goddard WA, III, Johnson JH., Jr Langmuir. 2004;20:2640–2651. doi: 10.1021/la036108k. [DOI] [PubMed] [Google Scholar]
- 11.Diallo MS, Christie S, Swaminathan P, Johnson JH, Jr, Goddard WA., III Environ Sci Technol. 2005;39:1366–1377. doi: 10.1021/es048961r. [DOI] [PubMed] [Google Scholar]
- 12.Esfand R, Tomalia DA. Drug Discovery Today. 2001;6:427–436. doi: 10.1016/s1359-6446(01)01757-3. [DOI] [PubMed] [Google Scholar]
- 13.Patri AK, Kukowska-Latallo JF, Baker JR., Jr Adv Drug Delivery Rev. 2005;57:2203–2214. doi: 10.1016/j.addr.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 14.Gillies ER, Fréchet JMJ. Drug Discovery Today. 2005;10:35–43. doi: 10.1016/S1359-6446(04)03276-3. [DOI] [PubMed] [Google Scholar]
- 15.Kukowska-Latallo JF, Candido KA, Cao ZY, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR., Jr Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 16.Majoros IJ, Myc A, Thomas T, Mehta CB, Baker JR. Biomacromolecules. 2006;7:572–579. doi: 10.1021/bm0506142. [DOI] [PubMed] [Google Scholar]
- 17.Khandare J, Kolhe P, Pillai O, Kannan S, Lieh-Lai M, Kannan RM. Bioconjugate Chem. 2005;16:330–337. doi: 10.1021/bc0498018. [DOI] [PubMed] [Google Scholar]
- 18.Padilla De Jesus OL, Ihre HR, Gagne L, Frechet JMJ, Szoka FC., Jr Bioconjugate Chem. 2002;13:453–461. doi: 10.1021/bc010103m. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Lopina ST. J Biomater Sci, Polym Ed. 2003;14:1043–1056. doi: 10.1163/156856203769231556. [DOI] [PubMed] [Google Scholar]
- 20.Man N, Cheng Y, Xu T, Ding Y, Wang X, Li Z, Chen Z, Huang G, Shi Y, Wen L. Eur J Med Chem. 2006;41:670–674. [Google Scholar]
- 21.Morgan MT, Carnahan MA, Immoos CE, Ribeiro AA, Finkelstein S, Lee SJ, Grinstaff MW. J Am Chem Soc. 2003;125:15485–15489. doi: 10.1021/ja0347383. [DOI] [PubMed] [Google Scholar]
- 22.Morgan MT, Nakanishi Y, Kroll DJ, Griset AP, Carnahan MA, Wathier M, Oberlies NH, Manikumar G, Wani MC, Grinstaff MW. Cancer Res. 2006;66:11913–11921. doi: 10.1158/0008-5472.CAN-06-2066. [DOI] [PubMed] [Google Scholar]
- 23.Jansen JFGA, de Brabander van den Berg EMM, Meijer EW. Science. 1994;266:1226–1229. doi: 10.1126/science.266.5188.1226. [DOI] [PubMed] [Google Scholar]
- 24.Neerman MF. Anti-Cancer Drugs. 2007;18:839–842. doi: 10.1097/CAD.0b013e32809ef9d0. [DOI] [PubMed] [Google Scholar]
- 25.Dhanikula RS, Argaw A, Bouchard JF, Hildgen P. Mol Pharmaceutics. 2008;5:105–116. doi: 10.1021/mp700086j. [DOI] [PubMed] [Google Scholar]
- 26.Ooya T, Lee J, Park K. Bioconjugate Chem. 2004;15:1221–1229. doi: 10.1021/bc049814l. [DOI] [PubMed] [Google Scholar]
- 27.Kojima C, Kono K, Maruyama K, Takagishi T. Bioconjugate Chem. 2000;11:910–917. doi: 10.1021/bc0000583. [DOI] [PubMed] [Google Scholar]
- 28.Ireson CR, Chander SK, Purohit A, Perera S, Newman SP, Parish D, Leese MP, Smith AC, Potter BV, Reed MJ. Br J Cancer. 2004;90:932–937. doi: 10.1038/sj.bjc.6601591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SH, Myc A, Koenig RJ, Bretz JD, Arscott PL, Baker JR., Jr Mol Cell Endocrinol. 2000;165:163–172. doi: 10.1016/s0303-7207(00)00249-5. [DOI] [PubMed] [Google Scholar]
- 30.James J, Murry DJ, Treston AM, Storniolo AM, Sledge GW, Sidor C, Miller KD. Invest New Drugs. 2006;25:41–48. doi: 10.1007/s10637-006-9008-5. [DOI] [PubMed] [Google Scholar]
- 31.Rajkumar SV, Richardson PG, Lacy MQ, Dispenzieri A, Greipp PR, Witzig TE, Schlossman R, Sidor CF, Anderson KC, Gertz MA. Clin Cancer Res. 2007;13:6162–6167. doi: 10.1158/1078-0432.CCR-07-0807. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Wang S, Chen X, Meshinchi S, Baker JR., Jr Mol Pharmaceutics. 2006;3:144–151. doi: 10.1021/mp050078s. [DOI] [PubMed] [Google Scholar]
- 33.Wang SH, Shi X, Chen X, Baker JR., Jr Macromol Biosci. 2009;9:429–436. doi: 10.1002/mabi.200800381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majoros IJ, Keszler B, Woehler S, Bull T, Baker JR., Jr Macromolecules. 2003;36:5526–5529. [Google Scholar]
- 35.Shi X, Lesniak W, Islam MT, MuÑiz MC, Balogh L, Baker JR., Jr Colloids Surf, A. 2006;272:139–150. [Google Scholar]
- 36.Shi X, Banyai I, Rodriguez K, Islam MT, Lesniak W, Balogh P, Balogh LP, Baker JR. Electrophoresis. 2006;27:1758–1767. doi: 10.1002/elps.200500818. [DOI] [PubMed] [Google Scholar]
- 37.Guo XH, Zhang N, Cui FD, Du B, Zhang ZZ. Pharmazie. 2009;64:748–751. [PubMed] [Google Scholar]
- 38.Shi X, Wang S, Meshinchi S, Van Antwerp M, Bi X, Lee I, Baker JR., Jr Small. 2007;3:1245–1252. doi: 10.1002/smll.200700054. [DOI] [PubMed] [Google Scholar]
- 39.Lee I, Athey BD, Wetzel AW, Meixner W, Baker JR., Jr Macromolecules. 2002;35:4510–4520. [Google Scholar]
- 40.Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Proteins: Struct, Funct, Genet. 1988;4:31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- 41.Shi X, Wang S, Sun H, Baker JR., Jr Soft Matter. 2007;3:71–74. doi: 10.1039/b612972b. [DOI] [PubMed] [Google Scholar]
- 42.Cakara D, Kleimann J, Borkovec M. Macromolecules. 2003;36:4201–4207. [Google Scholar]
- 43.Hong S, Bielinska AU, Mecke A, Keszler B, Beals JL, Shi X, Balogh LP, Orr BG, Baker JR, Jr, Banaszak-Holl MM. Bioconjugate Chem. 2004;15:774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 44.Lesniak W, Bielinska AU, Sun K, Janczak KW, Shi X, Baker JR, Jr, Balogh LP. Nano Lett. 2005;5:2123–2130. doi: 10.1021/nl051077u. [DOI] [PubMed] [Google Scholar]
- 45.Quintana A, Raczka E, Piehler L, Lee I, Myc A, Majoros I, Patri AK, Thomas T, Mule J, Baker JR., Jr Pharm Res. 2002;19:1310–1316. doi: 10.1023/a:1020398624602. [DOI] [PubMed] [Google Scholar]
- 46.Majoros IJ, Thomas TP, Mehta CB, Baker JR., Jr J Med Chem. 2005;48:5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 47.Chandrasekar D, Sistla R, Ahmad FJ, Khar RK, Diwan PV. Biomaterials. 2007;28:504–512. doi: 10.1016/j.biomaterials.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 48.Hill E, Shukla R, Park SS, Baker JR., Jr Bioconjugate Chem. 2007;18:1756–1762. doi: 10.1021/bc0700234. [DOI] [PubMed] [Google Scholar]
- 49.Shukla R, Thomas TP, Peters JL, Desai AM, Kukowska-Latallo J, Patri AK, Kotlyar A, Baker JR., Jr Bioconjugate Chem. 2006;17:1109–1115. doi: 10.1021/bc050348p. [DOI] [PubMed] [Google Scholar]
- 50.Bi X, Shi X, Baker JR., Jr J Biomater Sci, Polym Ed. 2008;19:131–142. doi: 10.1163/156856208783227686. [DOI] [PubMed] [Google Scholar]