Abstract
The topics of verification and validation (V&V) have increasingly been discussed in the field of computational biomechanics, and many recent articles have applied these concepts in an attempt to build credibility for models of complex biological systems. V&V are evolving techniques that, if used improperly, can lead to false conclusions about a system under study. In basic science these erroneous conclusions may lead to failure of a subsequent hypothesis, but they can have more profound effects if the model is designed to predict patient outcomes. While several authors have reviewed V&V as they pertain to traditional solid and fluid mechanics, it is the intent of this manuscript to present them in the context of computational biomechanics. Specifically, the task of model validation will be discussed with a focus on current techniques. It is hoped that this review will encourage investigators to engage and adopt the V&V process in an effort to increase peer acceptance of computational biomechanics models.
Keywords: biomechanics, computation, validation, verification, modeling
1. Introduction
Modeling of biological systems allows simulation of the mechanical behavior of tissues to supplement experimental investigations or when experiment is not possible. It aids in defining the structure-function relationship of tissues and their constituents. Modeling plays a role in basic science as well as patient-specific applications such a diagnosis and evaluation of targeted treatments [1–5]. Regardless of the use, confidence in computational simulations is only possible if the investigator has verified the mathematical foundation of the model and validated the results against sound experimental data.
Computational biomechanics seeks to apply the principles of mechanics to living tissues. Beginning with the introduction of finite element analysis in the 1950s [6, 7], investigators used numerical algorithms to simulate structural materials in civil and aeronautical engineering applications [8, 9]. Beyond the foundation of solid mechanics, these methods were used extensively in computational fluid dynamics (CFD) and heat transfer [10–12]. As the power of the computer grew, so did the ability to tackle larger and more complex models. In the 1970s, researchers applied the principles of computational solid and fluid mechanics to biology [13–18]. Bone, ligament, cartilage, cardiac tissue and muscle exhibited complex organization and solid-fluid interactions that were not adequately described by traditional paradigms for engineering materials. Novel constitutive models were developed in an attempt to describe these materials. Again, leaps in computing power allowed the solution of more intricate problems, but adding complexity increased the potential for errors.
The issues of uncertainty in the ability of a model to describe the physics of a system did not go unnoticed. The first cohesive attempts to define methods of dealing with these problems arose in CFD [19–21]. Publications dealing with these issues in solid mechanics soon followed [22, 23]. To date no true standard has been written due to the constantly evolving state of the art, and as such these documents are thought of as guidelines.
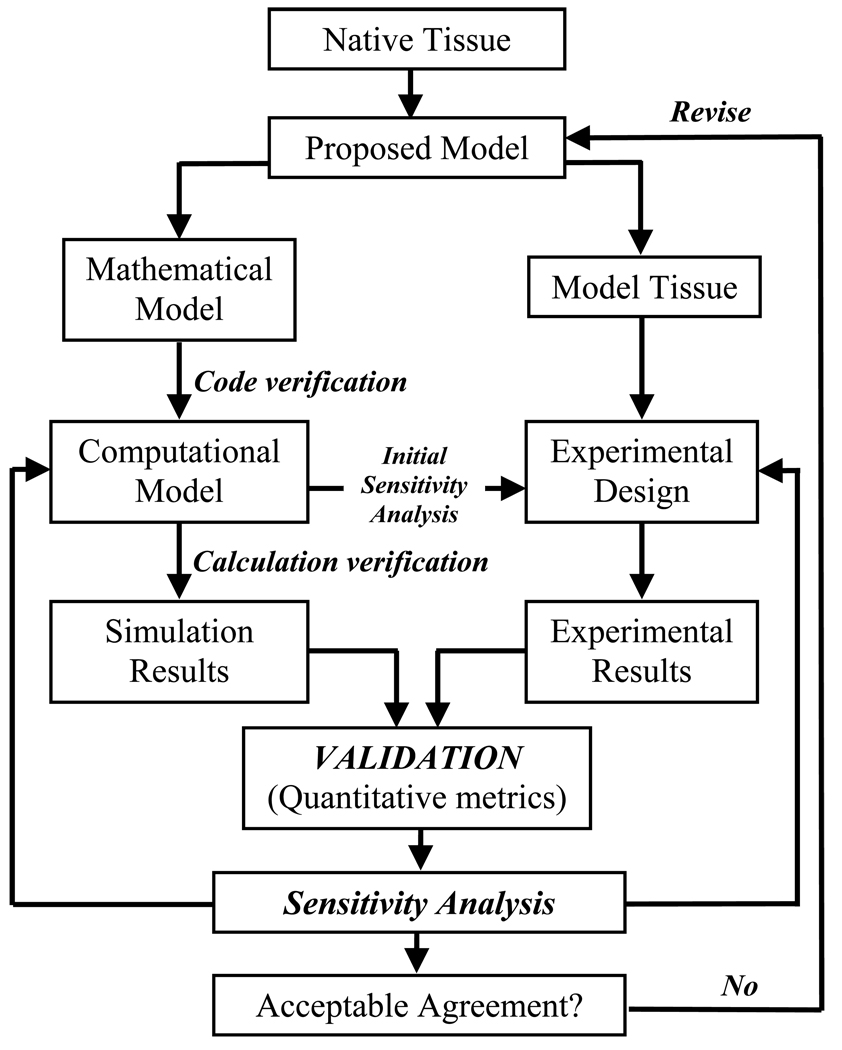
The literature refers to the areas of concern as verification and validation. Verification is defined by the ASME Committee for Verification & Validation in Computational Solid Mechanics as “the process of determining that a computational model accurately represents the underlying mathematical model and its solution” whereas validation is defined as “the process of determining the degree to which a model is an accurate representation of the real world from the perspective of the intended uses of the model” [22]. Succinctly, verification is “solving the equations right” (mathematics) and validation is “solving the right equations” (physics) [24, 25]. By definition verification must precede validation to separate errors due to model implementation from uncertainty due to model formulation [19, 20, 22, 26]. The general flow of the verification and validation process is illustrated in Figure 1.
Figure 1.
Flow of the verification and validation process in computational biomechanics. Verification solves the mathematical model and ensures it is implemented correctly (code verification) and provides acceptable solutions to benchmark problems (calculation verification). Initial computational solutions provide indicators of which parameters are critical in the model formulation (sensitivity analysis) and these can be used in the design of validation experiments. Validation is used to quantify the model’s ability to describe the experimental outcomes of the physical system given well defined boundary conditions. Sensitivity analysis is used again to determine the degree to which input parameters influence the solution output. The process is iterative until the model and validation experiments provide reasonable agreement within preset acceptance criteria. Adapted with permission from ASME Committee (PT60) on Verification and Validation in Computational Solid Mechanics (2006).
It has been argued that verification and validation are only applicable for a closed system in which all variables and their relative influence on the system are known, and natural systems never obey this simplification [27]. Oberkampf and collaborators put forth that engineering does not require “absolute truth” but instead a statistically meaningful comparison of computational and experimental results designed to assess random (statistical) and bias (systematic) errors [20]. In order to build practical confidence in any prediction the engineering approach must be used.
For the purpose of this manuscript, error is defined as the difference between a simulation or experimental value and the truth. Error can arise in a number of areas, particularly insufficiencies in the formulation of the model describing the physics of the real world or inaccurate implementation of the model into a computational code [19, 20, 22]. The intended use of the model then dictates the requirements of error analysis. Basic science may only require cursory examination of errors if the intention is to gather general information for further study. In contrast, clinical application of the predictions from a simulation necessitates extensive examination of errors, especially if patient treatment or outcome is directly impacted. The importance of recognizing and accounting for errors is critical for peer acceptance of the relevance and applicability of the model. This is apparent in the growing number of scientific journals requiring some degree of verification and validation of models presented for consideration (e.g. Annals of Biomedical Engineering [28], Clinical Biomechanics [29], etc.).
This manuscript reviews validation in computational biomechanics. Validation, as it pertains to the real world physics of biological materials, has been an elusive target due to the complexity of the tissues. Existing models provide predictions of stress or strain in a tissue, but validation may only be supported by rudimentary experiments with any level of confidence. It is the ultimate goal of validation to provide a physical foundation for comparison such that problems that are not experimentally feasible can be simulated with the belief that they provide realistic predictions. Validation is by nature a collaborative effort between experimentalists, code developers and researchers seeking to define mathematical descriptors of real-world materials. It is likely that the future will provide more firm experimental standards, commonality in reporting and continuous improvement strategies even if the goal of a “gold standard” testing regimen is never truly achievable. As this text focuses on validation, the reader is referred to a number of extensive reviews of verification and related topics in computational modeling of traditional solid, fluid and biomechanics [19–23, 26, 27, 30–36]. Verification and sensitivity studies will be covered briefly as they are inextricably tied to validation.
2. Verification and Sensitivity Studies
2.1. Verification
Verification is “the process of gathering evidence to establish that the computational implementation of the mathematical model and its associated solution are correct” [22]. A verified code yields the correct solution to benchmark problems of known solution (analytic or numeric), but does not necessarily guarantee that it will accurately represent complex biomechanical problems (the domain of validation) [19]. From this definition it is clear that verification must precede validation. The need for validation is obviated if the numerical implementation of the proposed model is not accurate in its own right.
Verification is composed of two categories, code and calculation verification. Code verification ensures the mathematical model and solution algorithms are working as intended. Typically the numerical algorithms are in the framework of finite-difference or finite-element (FE) methods, in which discretized domains are solved iteratively until convergence criteria are met. The errors that can occur include discontinuities, inadequate iterative convergence, programming errors, incomplete mesh convergence, lack of “conservation” (mass, energy, heat…), computer round-off, etc. [21, 23]. The assessment of numerical error has been studied extensively and is suggested to follow a hierarchy of test problems [19, 22]. This includes comparison to exact analytical solutions (most accurate but least likely to exist for complex problems), semi-analytic solutions with numerical integration of ordinary differential equations, and highly accurate numerical solutions to partial differential equations describing the problem domain. An example of code verification is found in Ionescu et al. [37] where a transversely isotropic hyperelastic constitutive model implementation was verified against an analytical solution for the case of equibiaxial stretch. The code was capable of predicting stresses to within 3% of an analytical solution, thus verifying the code performance. Note that this was a limited test of applicability and does not mean the model could accurately predict other responses that were not independently verified.
Calculation verification focuses on errors arising from discretization of the problem domain. Errors can arise from discretization of both the geometry and analysis time and should be verified independently. A common way to characterize discretization error in the FE method is via a mesh convergence study. A mesh is considered too coarse if subsequent refinement of the mesh results in predictions that are substantially different (i.e. solution does not asymptote). The consequence of incomplete mesh convergence is that the problem will generally be too “stiff” in comparison to an analytical solution, and increasing the number of elements will “soften” the FE solution [1, 35]. Studies of spinal segments have suggested that a change of <5% in the solution output is adequate to ensure mesh convergence is complete [31]. Mesh convergence is documented in the literature due to the prevalence in finite-element studies, and it is recommended for all discretized analyses [38–43].
Although verification is absolutely required for user-developed codes to ensure that the model is delivering the expected outputs to benchmark problems [26], use of a commercial code does not relieve the user of the need for verification. Given every conceivable problem, teams of commercial software engineers cannot ensure that all possible combinations of boundary conditions and material constraints yield accurate results [20]. Therefore the user is tasked with verification prior to use of a given model implementation.
2.2. Sensitivity studies
Many mathematical models of biological tissues are formulated based on fundamental considerations such as material symmetry, stiffness, static vs. dynamic response, etc. All material parameters, whether adopted from the literature or derived from experiments, include some degree of error [35]. This can result from the incomplete characterization of a new material in the laboratory, differences between protocols or inherent specimen-to-specimen variability. Errors are exacerbated in the case of patient-specific models where unique combinations of material properties and specimen geometry are coupled [38]. Sensitivity studies are common in computational analyses, usually focusing on the influence of experimentally derived material coefficients on the model predictions [1, 38, 39, 42, 44–49]. Multiple sources have identified the resolution of medical image data as a source of error in computational models due to deviations in the reconstruction of 3D geometry from 2D images [1, 31, 50].
Sensitivity studies are an important part of any computational study, especially those utilizing models that are previously untested [19, 20, 22, 23, 26, 30, 31, 35]. Sensitivity analysis can be performed before or after validation experiments and some argue that both are appropriate. If undertaken prior to validation, sensitivity studies can help the investigator target critical parameters [20, 30, 35]. Validation experiments can then be designed to tightly control the quantities of interest. After validation, sensitivity analyses provide assurance that the experimental results are within initial estimates and can determine if they still have a significant impact on model outputs.
Sensitivity studies provide the investigator with a measure of how error in a particular model input will impact the results of a simulation, scaling the relative importance of the inputs [23, 35]. The general procedure is to alter a single material parameter, by orders of magnitude or multiples of the standard deviation about the mean, while holding the others constant. For large scale parametric numerical analysis, Monte Carlo simulations can been used to evaluate combinations of parameters [32]. Simulations that are not altered significantly with variations in an input parameter are said to be insensitive to changes in the given input. To the contrary, a parameter that dramatically influences the output should be investigated to ensure proper characterization.
3. Validation
3.1. Validation
Validation is the process of ensuring that a computational model accurately represents the physics of the real world system [19, 22]. While some consider validation of natural systems to be impossible [27], the engineering viewpoint suggests the “truth” about the system is a statistically meaningful prediction that can be made for a specific set of boundary conditions [20, 26, 29]. This does not suggest that in vitro experimental validation (in a controlled laboratory environment) represents the in vivo case (within the living system) since the boundary conditions are likely impossible to mimic. It means that if a simplified model cannot predict the outcome of a basic experiment, it is probably not suited to simulate a more complex system.
Validation is often the most laborious and resource-dependent aspect of computational analysis, but if done properly can ensure that the model predictions are robust [20, 21, 26]. These costs may pale in comparison to the repercussions of a false prediction if the intended use of the model is critical. The level of validation required is directly tied to the intended use of the model and the supporting experiments should be tailored accordingly.
A general validation methodology is to determine the outcome variables of interest and prioritize them based on their relative importance. Oberkampf suggests using the PIRT (Phenomenon Identification and Ranking Table) [20, 51]. The PIRT guidelines scale each variable based on its impact within the system and determine if the model adequately represents the phenomena in question. It then identifies if existing experimental data are able to validate the model or if additional experiments are required. Finally, PIRT provides a framework to assess validation metrics which quantify the predictive capability of the model for the desired outcome variable.
The central question is one of time, cost and the complexity of the experiments needed to validate the simulation and the ramifications of erroneous conclusions. Is the model the most appropriate representation of the physical system? Will a simpler model be adequate, or will it be unable to capture details only available in more sophisticated formulations? Can experiments provide realistic data for the critical outcome variables or are the intended uses of the model incapable of laboratory examination?
3.2. Types of validation
The two predominant types of validation are direct and indirect [31]. Direct validation performs experiments on the quantities of interest, from basic material characterizations to hierarchal systems analysis. Though they may seem trivial, the most basic validation experiments are often the most beneficial as they provide fundamental confidence in the model’s ability to represent constituents of the system [30]. The goal is to produce an experiment that closely matches a desired simulation so each material property and boundary condition can be incorporated. Limitations include reproduction of the physical scale or an inability to generate data for the specific model output that is most desired. Typically these relate to the regeneration of the complex boundary conditions associated with in vivo systems as quantified by in vitro experiments.
Indirect validation utilizes experimental results that cannot be controlled by the user, such as from the literature or results of clinical studies. Experimental quality control, sources of error, and the degree of variability are typically not known if the data are not collected by the analyst. Indirect validation is clearly less favored than direct validation, but may be unavoidable. The required experiments may be cost prohibitive, difficult to perform, or may simply be unable to quantify the value that is sought by the model.
3.3. Experimental design considerations
The investigator should consider how the model will be tested directly. Does it include one constituent or many, what quantities are being measured, and can individual components be tested independently from one another as well as in their combined state? The governing committees both suggest building the testing protocol through three general stages: 1) finite benchmark problems of the constituents 2) subsystems 3) complete system analysis [19, 22]. Often only the first stage is possible, but the others may be used to guide further analysis even if direct experimental validation is not possible. When reporting experimental data, the results should be presented as means and standard deviations along with the number of independent measures collected [19, 20, 22, 23].
A significant consideration in computational biomechanics is how well in vitro testing can mimic the in vivo environment. Boundary conditions may be easily manipulated on the laboratory bench, but data may not represent conditions within the living system. This can negatively affect computational predictions since isolated tissues may behave differently than when part of the composite biological system. An example is found in the work of Gardiner et al. [45], where in situ ligament strains were determined on a subject-specific basis. Finite-element predictions of strain using averaged data fit the experimental data poorly compared to the subject-specific models, emphasizing the need for well-defined boundary conditions.
As computing power increases, subject-specific simulations will become more commonplace. Experimental validation needs to account for the influence of geometric and parameterized simplifications (generic population vs. subject-specific). The analysis of individual models may not provide better results given the time and effort required to formulate all different test cases. Validation also may not be possible in the clinical setting where researchers and clinicians seek to predict the outcomes of diagnosis and treatment. This is a tradeoff of model generation versus the confidence that can result from direct experimentation. Again, what is the desired outcome of validation and is it achievable based on available tools?
3.4. Validation metrics
Validation metrics quantify the differences between the experimental results and the simulation and can include all assumptions and estimates of errors [20, 22, 30]. Qualitative observations require user interpretation and provide no universal scale for comparison. Graphical comparisons such as scaled fringe plots are quasi-quantitative since they find a basis in human interpretation. Metrics must be quantitative in nature and are suggested to follow the design flow of the PIRT in order to scale and capture the most relevant parameters [20].
Metrics can be deterministic, using graphical comparisons to relate the model to experiments [20]. Deterministic metrics rely on graphs and plots, which are inherently qualitative. Statistical analysis such as regression and correlation can strengthen quantitative conclusions [20, 30] but must be thoroughly examined since variation in the data directly affects the strength of their relationships [52]. Alone these are not complete, so experimental uncertainty metrics and numerical error metrics are employed [20]. Experimental uncertainty metrics quantify the inaccuracies built into the experimental apparatus and their impact on experimental data. The tolerances of the devices should be provided, and accuracy should be determined by the user as a secondary check that the devices are within specification. Accuracy of any device should be reported as the mean ± 1.96 standard deviations as this is the statistically relevant confidence interval within which specifications should fall [20, 22]. Numerical error metrics include the influence of computational error on the model domain. Numerical error is quantified by manipulating the solution strategy or input parameters to test the impact on the solution. These may include the use of implicit versus explicit time integration or altered convergence criteria [30].
Metrics may also be non-deterministic. These are the most comprehensive since they include the deterministic quantifications as well as estimates of all input parameters as probability distributions [20]. Material properties derived from experiments include error. The probability distribution is the statistical definition of the material properties as a function of that error, factoring in the variance and number of samples tested. This allows the metric to be truly quantitative since it builds in the effects of experimental uncertainty and numerical error simultaneously. Oberkampf provides an excellent illustration of this phenomenon in section 2.4.4 of his text [20].
3.5. Examples of validation in computational biomechanics
Many studies provide examples of validation, but few encompass the fully scaled approach of non-deterministic metrics. This is likely due to the time and cost constraints that come with experimental validation, but also the relative infancy of the field of computational biomechanics. As a rapidly growing field, many studies validate models with the intention that predictions are for use in subsequent analysis and experimentation. Direct applicability on patient outcome is often an extended goal [2–5]. Given the disparity in intended use of models, the level of validation varies greatly. It is the intention to provide the reader with examples of validation in computational biomechanics, though no “gold-standard” currently exists. The following discussion samples computational biomechanics papers from various disciplines including cell, cardiovascular, soft tissue, bone, and implant mechanics.
In most cases validation was performed by comparison to commercially available software, typically finite element solvers such as ABAQUS (Dassault Systems, SIMULIA, Warwick, RI) or NIKE3D (Lawrence Livermore National Laboratory, Livermore, CA). Some conducted their work using custom computational frameworks, but no study explicitly called out code verification. The development and use of the code begets confidence as simple problems are solved successfully. Unfortunately, when code verification is not confirmed, the reader is left to assume the author has performed due diligence in ensuring proper code functionality. No matter how trivial, it is recommended that every author provide documented assurance that the code has been verified.
As for numerical verification, most studies performed mesh convergence analysis. The typical mention of convergence analysis concluded with the number of elements and nodes, as well as element types used [42, 43, 46, 53] while others added the refined mesh quality as an iterative percentage change in a solution parameter [1, 38, 39, 54]. The presentation of mesh quality as a metric provides the reader a baseline discretization error and is recommended for future studies.
Ideally, validation experiments target critical parameters to provide confidence that the inputs to and outputs from a model accurately define the physics of the system. Almost universally, the input parameters were derived in previous studies. Commonly accepted material properties were used in the case of well characterized structural materials like metals and polymers [40, 43, 54]. For biological tissues, mean values were used as inputs and sensitivity analysis tested the influence of their variance on the system. The most basic exploratory models of cell and vascular response contained one or two materials, so sensitivity analysis was simplified considerably [47, 53, 55, 56]. Complex computational studies of bone, cartilage and ligament deformation involving multiple materials, loading scenarios and geometries scaled up the degree of sensitivity analysis [1, 38, 39, 42]. Analyses of meniscus and intervertebral disc performed stochastic and parametric optimizations to determine the influential parameters as well as scale them relative to one another [46, 57]. While these reports used sensitivity analysis a posteriori, it should not be overlooked as a building block to aid in the design of validation experiments.
Validation primarily tested the composite systems, regardless of the level of complexity. The boundary conditions generally agreed well between experiment and simulation, since extension of model applicability was not of immediate concern in the basic system validation. A few models were tested with the intention of future clinical use in patient diagnostics, implantation and outcome [1, 42, 46, 47, 54, 55, 58], but require further analysis before extension into the clinical environment.
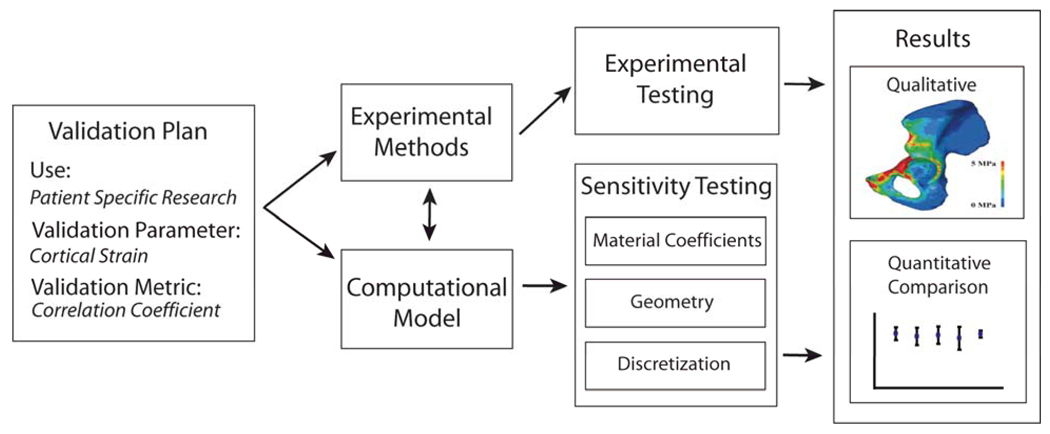
An example of the flow of validation and sensitivity analysis can be found in a study of cortical bone strains in the human pelvis (Fig. 2) [38]. This work used direct mechanical testing to simulate body weight loading through the hip joint while measuring deformation on the cortical bone with strain gauges. The target parameters and metrics were defined a priori and a dual computational and experimental approach allowed the influence of the unknown material properties to be evaluated with sensitivity analysis, identifying critical parameters for investigation in the experiments.
Figure 2.
Validation of pelvic cortical bone strains by Anderson et al [38]. A well defined validation plan where the model use, parameters and metrics were defined a priori and provided the initial problem statement. A computational model was constructed to assist the development of the experimental model. Sensitivity tests were performed on the computational models in parallel to the experimental testing. Comparisons and conclusions were made based on both qualitative and quantitative assessments of measured cortical bone strains and simulated pelvic loading.
Given the diversity and complexity of the systems, validation experiments were well suited for the intended use of the models. The investigators took precautions to apply the models to well defined experimental methods, employing techniques like atomic force microscopy [56], magnetic resonance imaging [47] and traditional mechanical testing batteries [1, 38, 40, 59, 60]. The techniques were capable of providing the defined measure that was sought as the model output, and errors associated with data collection were minimized where possible. Where errors could not be reduced, metrics were employed to capture their impact on the simulations.
The use of metrics was as varied as the models that employed them. Most studies used graphical comparison such as fringe plots and line graphs [1, 38, 42, 43, 47, 53, 55, 56, 58, 60, 61]. These assessments illustrated agreement of model predictions with experiments, using quantities such as stress or strain. As discussed earlier, they are subjective. This is generally acceptable in exploratory studies but should be supported by non-deterministic metrics if conclusions are to be drawn from the simulation.
Traditional forms of metrics were used in the majority of the studies that were sampled. These included change as a percent of nominal [43, 47], root-mean-square error [1, 40, 46], and correlation coefficients and coefficients of determination [38, 42, 58, 60–62]. Each of these may be deterministic or non-deterministic depending on the inclusion of sampling error into the calculation.
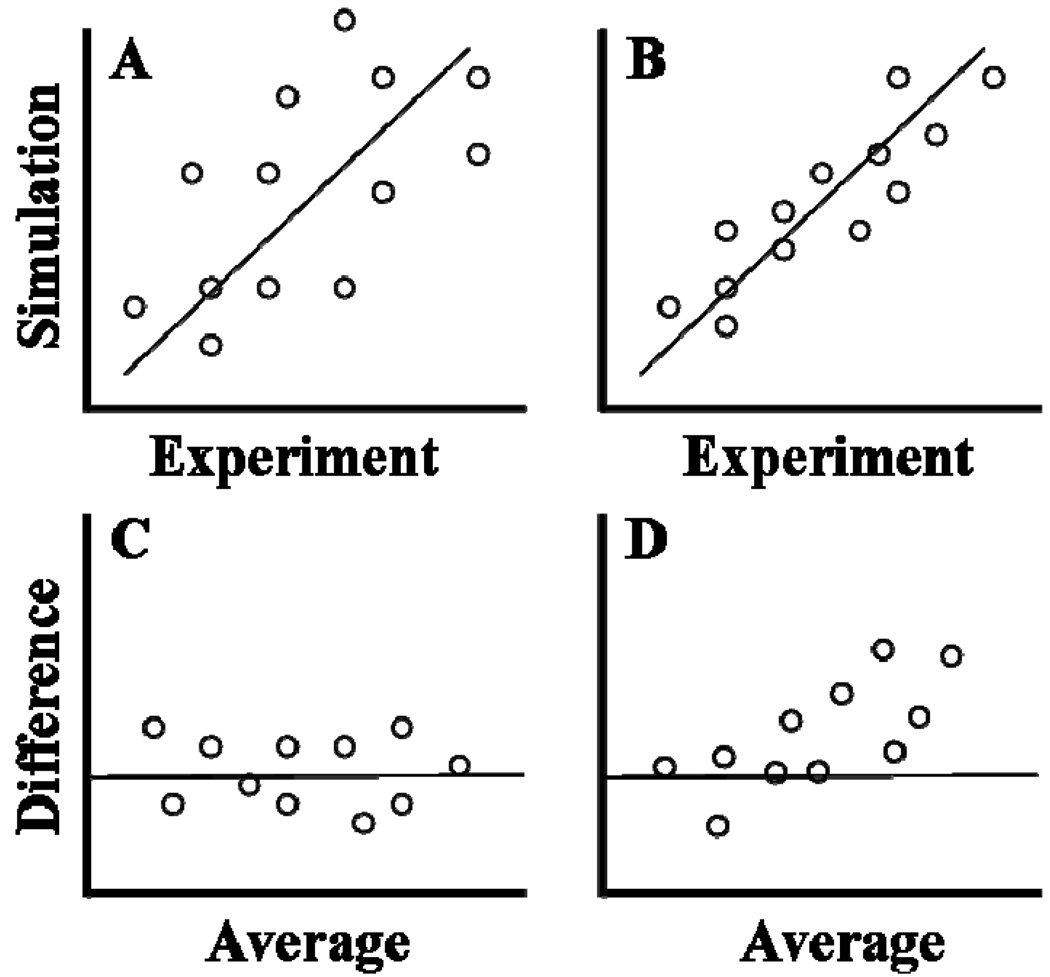
Correlation coefficients (R) and coefficients of determination (R2) are often shown on a scatter plot comparing simulation versus experimental values. While ideal comparison yields a regression line with a slope of one, it does not capture the variance of the respective data sets. This means high variation between individual data as compared to the measurement error within the experiment begets artificially high correlation. Bland and Altman [52] note that “the correlation coefficient is not a measure of agreement; it is a measure of association”. They suggest an alternate form of analysis which accounts for repeatability (the difference between two measurements on a single subject) and reproducibility (the difference between two measurements on a single subject after a change in setup). The typical experimental (E) vs. simulation (S) scatter plot is the starting point. Data are then plotted as the average of the methods ((E + S)/2) on the x-axis and the difference between the methods (E-S) on the y-axis. A new regression line is then tested against a null hypothesis that the difference (E-S) is zero, capturing variation that inflates traditional correlation. The approach is illustrated in Figure 3.
Figure 3.
Scatter plot regression vs. Bland-Altman plots. Top panes (A, B) show typical scatter plots of experimental versus simulation data. Though both show a regression line of slope = 1, they are derived from populations with higher (A) and lower (B) variance. Pane C shows a Bland-Altman plot where the difference between experiment and simulation (E-S) is plotted versus the average range (E+S/2). Deviation above or below the line (E-S = 0) indicates the relative scale of agreement between experiment and simulation. Pane D illustrates how the difference (E-S) may vary as a function of their average, indicating bias toward a given range of experimental and simulation conditions.
3.6. Examples of validation with clinical implications
A recent push has generated models with increased clinical significance. This is typically performed by use of patient-specific geometric and material data that can be extracted from imaging data (e.g. CT, MRI) to create a patient-specific biomechanical model. An example of this application was put forth by Anderson et al. in which the contact pressure in acetabular hip cartilage was simulated using finite element models and experimentally validated using a cadaveric human pelvis [1]. One stated intent was to provide a model for obtaining “clinically meaningful data in terms of improving the diagnosis and treatment of hip OA…” [1]. Likewise, Fernandez et al. generated knee models for the purpose of pathologic diagnosis and surgical planning [63]. Finite element models of the knee were generated from patient-specific data and were used to perform two different surgical simulations but no validation was reported.
As patient-specific models becomes more common, it will be a fundamental requirement that they are validated in light of their intended clinical impact. If medical professionals cannot be convinced of the predictive capabilities, the models will not be applied. Therefore, validation should be provided with future studies aimed at direct clinical application. This should include experimental validation of individual tissues and the composite system, sensitivity analyses of the intended parameters, and thorough statistical evaluation of the predictive nature of the model.
There is a growing interest in the development of surgical planning software. For instance, the software suite MedEdit uses patient-specific data in the planning of bone fixation after traumatic fracture [64]. It allows the surgeon to interactively add, edit and remove bone fixation hardware from patient-specific bone models and then perform FE simulations in hopes of finding an optimal surgical procedure. If there is intent for the software to obtain FDA approval, the need for detailed validation becomes especially important.
There is additional interest in using patient-specific models to aid in the design of medical implants. Vartziotiz et al. used FE simulations to aid the design of custom hip implants based on patient-specific geometry, while Ridzwan and Ruben used FE models coupled with topology optimization algorithms to generate an optimal hip implant shape [65–67]. Similar efforts have sought to use computational modeling in the design of knee and spinal implants [43, 68]. Ideally, each patient could be analyzed and fitted with custom hardware based on model predictions of their specific morphology and gait patterns, improving the success rate of surgical intervention.
Although the prospect of using simulations in clinical applications is exciting, caution must be exercised. Many studies that use finite element analysis have done little to establish the validity of their methods via thorough experimentation. If it can not be established that these new methods can increase the level of care clinicians are able to provide, then their usefulness will be limited.
A study on a subject-specific tibia model provides a useful example [40]. The purpose of the model was to “evaluate new and modified designs of joint prostheses and fixation devices” [40]. The models and experimental validations were performed for both a normal and knee prosthesis implanted tibia. Numerous loading conditions were applied, making the validation more comprehensive. Additionally, important model assumptions (such as selection of material properties) were clearly stated. These practices facilitate the transition from research based modeling to clinical application.
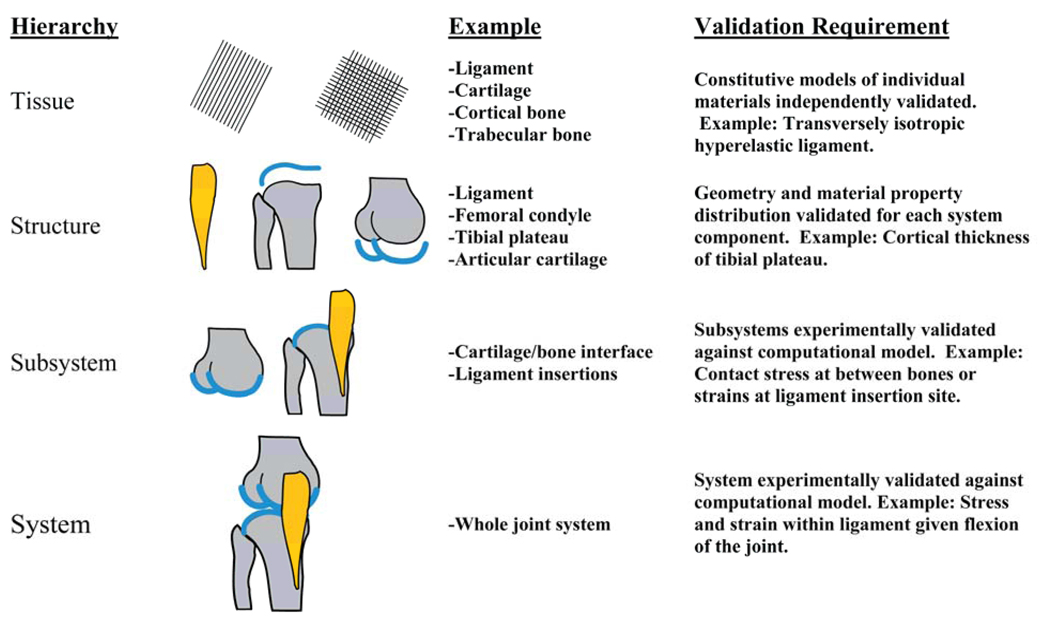
As computational models become more complex, a consistent methodology for model validation is desirable. Oberkampf suggests a validation hierarchy to describe the levels of model construction [20] (Figure 4). The first step is the need to validate a specific constitutive model for each unique tissue. Validation at this level typically involves material testing and extraction of material coefficients from experimental data. The next level involves validating the model of an entire structure (articular cartilage and bone). This includes assessing the assignment of material properties and validating the model geometry. At the subsystem level, two or more structures are combined into a single model (bone to bone contact, ligament and bone). Finally, the combination of two or more subsystems (articular cartilage-bone-ligament) results in a system level model. These are the most challenging to validate, but also the most clinically relevant.
Figure 4.
Oberkampf’s validation hierarchy [20], illustrated for use in computational biomechanics. The hierarchy starts at the tissue level and progresses to the structural, subsystem, and system levels. The validation hierarchy is illustrated here with a knee model including bone, articular cartilage, and the medial collateral ligament. Suggested validation requirements are shown for each level of model complexity.
Patient-specific models for use in diagnosis, surgical simulation and implant design are expected to provide useful tools for clinicians. However, like any tool, each model must come with a well defined set of criteria that defines the model’s scope, predictive capability, strengths, weakness and intended uses. Good validation practices, clearly defined scope and model assumptions, and predictive capability are important for acceptance of biomechanical models in the clinic.
4. Conclusions
This review discussed verification and validation used in computational biomechanics. The inclusion of V&V is required for credibility of a proposed model, and there will no doubt be continued improvement in the methodologies used. Given the diversity of applications in this small sampling of the literature, it is no wonder that a standard for V&V is an elusive goal. Provided that investigators continue to employ critical scientific reasoning and sound experimental practice to these problems, computational models will become fundamental tools to address future research questions and clinical applications.
Acknowledgements
Financial support from NIH #R01AR047369, R01EB006735, R01GM083925, R01AR053344, and R01AR053553 is gratefully acknowledged.
References
- 1.Anderson AE, Ellis BJ, Maas SA, Peters CL, Weiss JA. Validation of finite element predictions of cartilage contact pressure in the human hip joint. J Biomech Eng. 2008;130(5):051008. doi: 10.1115/1.2953472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Reilly MA, Whyne CM. Comparison of Computed Tomography Based Parametric and Patient-Specific Finite Element Models of the Healthy and Metastatic Spine Using a Mesh-Morphing Algorithm. Spine. 2008;33(17):1876–1881. doi: 10.1097/BRS.0b013e31817d9ce5. [DOI] [PubMed] [Google Scholar]
- 3.Reinbolt JA, Haftka RT, Chmielewski TL, Fregly BJ. A computational framework to predict post-treatment outcome for gait-related disorders. Medical Engineering and Physics. 2008;30(4):434–443. doi: 10.1016/j.medengphy.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Sermesant M, Peyrat JM, Chinchapatnam P, Billet F, Mansi T, Rhode K, Delingette H, Razavi R, Ayache N. Toward Patient-Specific Myocardial Models of the Heart. Heart Failure Clinics. 2008;4(3):289–301. doi: 10.1016/j.hfc.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Viceconti M, Davinelli M, Taddei F, Cappello A. Automatic generation of accurate subject-specific bone finite element models to be used in clinical studies. J Biomech. 2004;37(10):1597–1605. doi: 10.1016/j.jbiomech.2003.12.030. [DOI] [PubMed] [Google Scholar]
- 6.Courant R. Variational methods for the solution of problems of equilibrium and vibrations. Bull. Am. Math Soc. 1943;49:1–43. [Google Scholar]
- 7.Martin HC. Introduction to matrix methods of structural analysis. New York: McGraw-Hill; 1966. [Google Scholar]
- 8.Argyris JH. Energy theorems and structural analysis; a generalised discourse with applications on energy principles of structural analysis including the effects of temperature and nonlinear stress-strain relations. London: Butterworth; 1960. [Google Scholar]
- 9.Turner MJ, Clough RW, Martin HC, Topp LJ. Stiffness and deflection analysis of complex structures. J. Aero. Sci. 1956;23(9):805–823. [Google Scholar]
- 10.Taylor RL, Brown CB. Darcy flow solutions with a free surface. J. Hydr. Div. ASCE. 1967;93:25–33. [Google Scholar]
- 11.Taylor RL, Doherty WP, Wilson EL, Structural Engineering L. Three-Dimensional, Steady State Flow of Fluids in Porous Solids. Structural Engineering Laboratory, University of California. [Distributed by National Technical Information Service]; 1969. [Google Scholar]
- 12.Wilson EL, Nickell RE. Application of the finite element method to heat conduction analysis. Nuclear Engineering and Design. 1966;4:276–286. [Google Scholar]
- 13.Davids N, Mani MK. Effects of turbulence on blood flow explored by finite element analysis. Comput Biol Med. 1972;2(4):311–319. doi: 10.1016/0010-4825(72)90018-2. [DOI] [PubMed] [Google Scholar]
- 14.Belytschko T, Kulak RF, Schultz AB, Galante JO. Finite element stress analysis of an intervertebral disc. J Biomech. 1974;7(3):277–285. doi: 10.1016/0021-9290(74)90019-0. [DOI] [PubMed] [Google Scholar]
- 15.Pao YC, Ritman EL, Wood EH. Finite-element analysis of left ventricular myocardial stresses. J Biomech. 1974;7(6):469–477. doi: 10.1016/0021-9290(74)90081-5. [DOI] [PubMed] [Google Scholar]
- 16.Rybicki EF, Simonen FA. Mechanics of oblique fracture fixation using a finite-element model. J Biomech. 1977;10(2):141–148. doi: 10.1016/0021-9290(77)90077-x. [DOI] [PubMed] [Google Scholar]
- 17.Hakim NS, King AI. A computer-aided technique for the generation of a 3-D finite element model of a vertebra. Comput Biol Med. 1978;8(3):187–196. doi: 10.1016/0010-4825(78)90019-7. [DOI] [PubMed] [Google Scholar]
- 18.Vinson CA, Gibson DG, Yettram AL. Analysis of left ventricular behaviour in diastole by means of finite element method. Br Heart J. 1979;41(1):60–67. doi: 10.1136/hrt.41.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.AIAA Guide for the verification and validation of computational fluid dynamics simulations. American Institute of Aeronautics and Astronautics; 1998. [Google Scholar]
- 20.Oberkampf WL, Trucano TG, Hirsch C. Verification, Validation, and Predictive Capability in Computational Engineering and Physics. Sandia National Laboratories; 2003. pp. 3–78. [Google Scholar]
- 21.Stern F, Wilson RV, Coleman HW, Paterson EG, Iowa Inst Of Hydraulic Research Iowa C. Verification and Validation of CFD Simulations. IOWA INST OF HYDRAULIC RESEARCH IOWA CITY; 1999. [Google Scholar]
- 22.Guide for verification and validation in computational solid dynamics. American Society of Mechanical Engineers; 2006. [Google Scholar]
- 23.Roache PJ. Verification and validation in computational science and engineering. Hermosa Publishers; 1998. [Google Scholar]
- 24.Blottner FG. Accurate Navier-Stokes results for the hypersonic flow over a spherical nosetip. AIAA Journal of Spacecraft and Rockets. 1990;27:113–122. [Google Scholar]
- 25.Boehm BW. Software engineering economics. Englewood Cliffs, N.J.: Prentice-Hall; 1981. p. 15. [Google Scholar]
- 26.Babuska I, Oden JT. Verification and validation in computational engineering and science: basic concepts. Comput Mehods Appl Mech Eng. 2004;193(36–38):4057–4066. [Google Scholar]
- 27.Oreskes N, Shrader-Frechette K, Belitz K. Verification, Validation, and Confirmation of Numerical Models in the Earth Sciences. Science. 1994;263(5147):641–646. doi: 10.1126/science.263.5147.641. [DOI] [PubMed] [Google Scholar]
- 28.Author instructions for Annals of Biomedical Engineering. Annals of Biomedical Engineering; ( www.bme.gatech/.edu/abme/) [Google Scholar]
- 29.Viceconti M, Olsen S, Nolte LP, Burton K. Extracting clinically relevant data from finite element simulations. Clinical Biomechanics. 2005;20:451–454. doi: 10.1016/j.clinbiomech.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Anderson AE, Ellis BJ, Weiss JA. Verification, validation and sensitivity studies in computational biomechanics. Comput Methods Biomech Biomed Engin. 2007;10(3):171–184. doi: 10.1080/10255840601160484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones AC, Wilcox RK. Finite element analysis of the spine: Towards a framework of verification, validation and sensitivity analysis. Med Eng Phys. 2008;30(10):1287–1304. doi: 10.1016/j.medengphy.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Liu JS. Monte Carlo Strategies in Scientific Computing. Springer; 2001. [Google Scholar]
- 33.Sornette D, Davis AB, Ide K, Vixie KR, Pisarenko V, Kamm JR. Algorithm for model validation: theory and applications. Proc Natl Acad Sci U S A. 2007;104(16):6562–6567. doi: 10.1073/pnas.0611677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stern F, Wilson RV, Coleman HW, Paterson EG. Comprehensive Approach to Verification and Validation of CFD Simulations—Part 1: Methodology and Procedures. J Fluids Eng. 2001;123:793. [Google Scholar]
- 35.Weiss JA, Gardiner JC, Ellis BJ, Lujan TJ, Phatak NS. Three-dimensional finite element modeling of ligaments: technical aspects. Med Eng Phys. 2005;27(10):845–861. doi: 10.1016/j.medengphy.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Wilson RV, Stern F, Coleman HW, Paterson EG. Comprehensive Approach to Verification and Validation of CFD Simulations—Part 2: Application for Rans Simulation of a Cargo/Container Ship. J Fluids Eng. 2001;123:803. [Google Scholar]
- 37.Ionescu I, Weiss JA, Guilkey J, Cole M, Kirby RM, Berzins M. Ballistic injury simulation using the material point method. Stud Health Technol Inform. 2006;119:228–233. [PubMed] [Google Scholar]
- 38.Anderson AE, Peters CL, Tuttle BD, Weiss JA. Subject-specific finite element model of the pelvis: development, validation and sensitivity studies. J Biomech Eng. 2005;127(3):364–373. doi: 10.1115/1.1894148. [DOI] [PubMed] [Google Scholar]
- 39.Ellis BJ, Debski RE, Moore SM, McMahon PJ, Weiss JA. Methodology and sensitivity studies for finite element modeling of the inferior glenohumeral ligament complex. J Biomech. 2007;40(3):603–612. doi: 10.1016/j.jbiomech.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 40.Gray HA, Taddei F, Zavatsky AB, Cristofolini L, Gill HS. Experimental validation of a finite element model of a human cadaveric tibia. J Biomech Eng. 2008;130(3):031016. doi: 10.1115/1.2913335. [DOI] [PubMed] [Google Scholar]
- 41.Hart RT, Hennebel VV, Thongpreda N, Van Buskirk WC, Anderson RC. Modeling the biomechanics of the mandible: a three-dimensional finite element study. J Biomech. 1992;25(3):261–286. doi: 10.1016/0021-9290(92)90025-v. [DOI] [PubMed] [Google Scholar]
- 42.Phatak N, Sun Q, Kim S, Parker DL, Sanders RK, Veress AI, Ellis BJ, Weiss JA. Noninvasive measurement of ligament strain with deformable image registration. Ann Biomed Eng. 2006 doi: 10.1007/s10439-007-9287-9. [DOI] [PubMed] [Google Scholar]
- 43.Villa T, Migliavacca F, Gastaldi D, Colombo M, Pietrabissa R. Contact stresses and fatigue life in a knee prosthesis: comparison between in vitro measurements and computational simulations. J Biomech. 2004;37(1):45–53. doi: 10.1016/s0021-9290(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 44.Donahue TL, Hull ML, Rashid MM, Jacobs CR. A finite element model of the human knee joint for the study of tibio-femoral contact. J Biomech Eng. 2002;124(3):273–280. doi: 10.1115/1.1470171. [DOI] [PubMed] [Google Scholar]
- 45.Gardiner JC, Weiss JA. Subject-specific finite element analysis of the human medial collateral ligament during valgus knee loading. J Orthop Res. 2003;21(6):1098–1106. doi: 10.1016/S0736-0266(03)00113-X. [DOI] [PubMed] [Google Scholar]
- 46.Haut Donahue TL, Hull ML, Rashid MM, Jacobs CR. How the stiffness of meniscal attachments and meniscal material properties affect tibio-femoral contact pressure computed using a validated finite element model of the human knee joint. J Biomech. 2003;36(1):19–34. doi: 10.1016/s0021-9290(02)00305-6. [DOI] [PubMed] [Google Scholar]
- 47.Steele BN, Wan J, Ku JP, Hughes TJ, Taylor CA. In vivo validation of a one-dimensional finite-element method for predicting blood flow in cardiovascular bypass grafts. IEEE Trans Biomed Eng. 2003;50(6):649–656. doi: 10.1109/TBME.2003.812201. [DOI] [PubMed] [Google Scholar]
- 48.Veress AI, Gullberg GT, Weiss JA. Measurement of strain in the left ventricle during diastole with cine-MRI and deformable image registration. J Biomech Eng. 2005;127(7):1195–1207. doi: 10.1115/1.2073677. [DOI] [PubMed] [Google Scholar]
- 49.Viceconti M, Zannoni C, Testi D, Cappello A. A new method for the automatic mesh generation of bone segments from CT data. J Med Eng Technol. 1999;23(2):77–81. doi: 10.1080/030919099294339. [DOI] [PubMed] [Google Scholar]
- 50.Yeni YN, Christopherson GT, Dong XN, Kim DG, Fyhrie DP. Effect of microcomputed tomography voxel size on the finite element model accuracy for human cancellous bone. J Biomech Eng. 2005;127(1):1–8. doi: 10.1115/1.1835346. [DOI] [PubMed] [Google Scholar]
- 51.Wilson GE, Boyack BE. The role of the PIRT process in experiments, code development and code applications associated with reactor safety analysis. Nuclear Engineering and Design. 1998;186:23–37. [Google Scholar]
- 52.Altman DG, Bland JM. Measurement in medicine: the analysis of method comparison studies. The Statistician. 1983;32(3):307–317. [Google Scholar]
- 53.Karcher H, Lammerding J, Huang H, Lee RT, Kamm RD, Kaazempur-Mofrad MR. A three-dimensional viscoelastic model for cell deformation with experimental verification. Biophys J. 2003;85(5):3336–3349. doi: 10.1016/S0006-3495(03)74753-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mortier P, De Beule M, Carlier SG, Van Impe R, Verhegghe B, Verdonck P. Numerical study of the uniformity of balloon-expandable stent deployment. J Biomech Eng. 2008;130(2):021018. doi: 10.1115/1.2904467. [DOI] [PubMed] [Google Scholar]
- 55.Chen K, Fata B, Einstein DR. Characterization of the highly nonlinear and anisotropic vascular tissues from experimental inflation data: a validation study toward the use of clinical data for in-vivo modeling and analysis. Ann Biomed Eng. 2008;36(10):1668–1680. doi: 10.1007/s10439-008-9541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unnikrishnan GU, Unnikrishnan VU, Reddy JN. Constitutive material modeling of cell: a micromechanics approach. J Biomech Eng. 2007;129(3):315–323. doi: 10.1115/1.2720908. [DOI] [PubMed] [Google Scholar]
- 57.Espino DM, Meakin JR, Hukins DW, Reid JE. Stochastic finite element analysis of biological systems: comparison of a simple intervertebral disc model with experimental results. Comput Methods Biomech Biomed Engin. 2003;6(4):243–248. doi: 10.1080/10255840310001606071. [DOI] [PubMed] [Google Scholar]
- 58.Macneil JA, Boyd SK. Bone strength at the distal radius can be estimated from high-resolution peripheral quantitative computed tomography and the finite element method. Bone. 2008;42(6):1203–1213. doi: 10.1016/j.bone.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 59.Cristofolini L, Viceconti M. Mechanical validation of whole bone composite tibia models. J Biomech. 2000;33(3):279–288. doi: 10.1016/s0021-9290(99)00186-4. [DOI] [PubMed] [Google Scholar]
- 60.Ellis BJ, Lujan TJ, Dalton MS, Weiss JA. Medial collateral ligament insertion site and contact forces in the ACL-deficient knee. J Orthop Res. 2006;24(4):800–810. doi: 10.1002/jor.20102. [DOI] [PubMed] [Google Scholar]
- 61.Dalstra M, Huiskes R, van Erning L. Development and validation of a threedimensional finite element model of the pelvic bone. J Biomech Eng. 1995;117(3):272–278. doi: 10.1115/1.2794181. [DOI] [PubMed] [Google Scholar]
- 62.Gupta S, van der Helm FC, Sterk JC, van Keulen F, Kaptein BL. Development and experimental validation of a three-dimensional finite element model of the human scapula. Proc Inst Mech Eng [H] 2004;218(2):127–142. doi: 10.1243/095441104322984022. [DOI] [PubMed] [Google Scholar]
- 63.Fernandez JW, Hunter PJ. An anatomically based patient-specific finite element model of patella articulation: towards a diagnostic tool. Biomech Model Mechanobiol. 2005;4(1):20–38. doi: 10.1007/s10237-005-0072-0. [DOI] [PubMed] [Google Scholar]
- 64.Olle K, Erdohelyi B, Halmai C, Kuba A. MedEdit: A computer assisted planning and simulation system for orthopedic-trauma surgery. 8th Central European Seminar on Computer Graphics. 2004 [Google Scholar]
- 65.Ridzwan MIZ, Shuib S, Hassan AY, Shokri AA, Ibrahim MNM. Optimization in Implant Topology to Reduce Stress Shielding Problem. J App Sci. 2006;6(13):2768–2773. [Google Scholar]
- 66.Ruben RB, Folgado J, Fernandes PR. 6th World Congress on Structural and Multidisciplinary Optimization. Brazil: Rio de Janeiro; 2005. A Three-Dimensional Model for Shape Optimization of Hip Prostheses Using a Multi-Criteria Formulation. [Google Scholar]
- 67.Vartziotis D, Poulis A, Faessler V, Vartziotis C, Kolios C. Information Technology in Biomedicine. Greece: Ioannina; 2006. Integrated Digital Engineering Methodology for Virtual Orthopedics Surgery Planning. [Google Scholar]
- 68.Harrysson OL, Hosni YA, Nayfeh JF. Custom-designed orthopedic implants evaluated using finite element analysis of patient-specific computed tomography data: femoral-component case study. BMC Musculoskelet Disord. 2007;8:91. doi: 10.1186/1471-2474-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]