Abstract
The sensitive optoelectronic properties of metal nanoparticles make nanoparticle-based materials a powerful tool to study fundamental biorecognition processes. Here we present a new and versatile method for coupling underivatized carbohydrates to gold nanoparticles (Au NPs) via the photochemically induced reaction of perfluorophenylazide (PFPA). A one-pot procedure was developed where Au NPs were synthesized and functionalized with PFPA by a ligand-exchange reaction. Carbohydrates were subsequently immobilized on the NPs by a fast light activation. The coupling reaction was efficient, resulting in high coupling yield as well as high ligand surface coverage. A colorimetric system based on the carbohydrate-modified Au NPs was used for the sensitive detection of carbohydrate-protein interactions. Binding and cross-reactivity studies were carried out between carbohydrate-functionalized Au NPs and lectins. Results showed that the surface-bound carbohydrates not only retained their binding affinities towards the corresponding lectin, but also exhibited affinity ranking consistent with that of the free ligands in solution.
1 Introduction
Metal nanoparticles coupled with biological ligands have been widely used for monitoring activities of biomolecules and their interactions with ligands.1,2 A unique characteristic of metal nanoparticles as the recognition probe is their remarkable and tunable optical property, the so-called surface plasmon resonance (SPR), determined by their size and shape, the dielectric property of the media, and the distance between particles. Colorimetric bioassays have thus been achieved based on the SPR shift when molecular interactions take place at the surface of the nanoparticles, and have been employed to study fundamental biorecognition processes including cell-cell communication, enzymatic activity, protein-protein interaction, and DNA hybridization.3–7 When the ligand-receptor interaction causes additional aggregation of nanoparticles, significant red-shift of the SPR absorption occurs producing intense color changes visible to the naked eye.8–11
Naturally occurring carbohydrates, glycoproteins, and glycolipids are present at the surface of nearly every cell in living systems, and play crucial roles in biological events as recognition sites between cells and different binding partners. They, for example, mediate various phenomena including cell growth, inflammatory responses or viral infections, and changes in glycosylation are often involved in disease states, including cancer. Efficient analysis and control of such events are of high importance. Carbohydrate-based detection platforms have recently emerged as highly useful analytical and diagnostic tools, and have demonstrated tremendous potential to superior sensitivity, selectivity, and stability.12–14 A key technology requirement in receptor/ligand-based sensing and detection is the surface conjugation chemistry that can effectively couple ligands to solid substrates. Carbohydrate structures expressed on cell surfaces are vastly diverse and complex. Despite the development of new synthetic and enzymatic protocols, obtaining large quantities of glycans with the required functionality and precise glycosylation pattern is still a major challenge. Highly desirable are effective coupling chemistries that are general and versatile, can accommodate carbohydrate diversity and give stable interfaces, and yet are simple and reproducible. Ligands attached to solid surfaces through a covalent bond are more stable than those that are physisorbed by weaker forces. Of the reported covalent coupling chemistry, the most popular involves chemisorption of thiolated carbohydrates on Au nanoparticles.15–23 Other conjugation methods include coupling N-hydroxysuccinimide (NHS)-functionalized dextran to amine-functionalized Ag NPs,24 and amine-functionalized carbohydrates to aldehyde-functionalized Au NPs.23 Coupling chemistry that does not require chemical derivatization of the carbohydrates is appealing. A few examples have been reported to conjugate underivatized carbohydrates on flat substrates for microarray construction, although the protocols have not been adopted for Au NPs. One approach used hydrazide-modified gold substrates where the hydrazide reacted with the terminal aldehyde group of the carbohydrates.25,26 A similar approach employed amine-functionalized surfaces and the coupling with carbohydrates took place by reductive amination to yield an amine conjugate.27,28 In both cases, reducing carbohydrates are necessary, and for monosaccharides, the coupled products often became acyclic and lost their binding affinities.
Herein we report a new coupling chemistry for the covalent immobilization of underivatized carbohydrates on Au NPs. The method takes advantage of the photochemistry of arylazides. Upon light activation, the azide moiety becomes converted to a reactive nitrene that most significantly inserts into CH and NH bonds, creating highly robust covalent linkages. In particular, perfluorophenylazides (PFPAs), which produce markedly enhanced insertion yields, have been widely used as photoaffinity labels to probe protein binding site structures.29–32 We have successfully employed PFPAs in surface modification, targeting polymeric materials that lack reactive functional groups for surface coupling.33–36 Carbohydrates are another class of compounds that are well-suited for the PFPA photocoupling chemistry. The design of our approach is to prepare PFPA-functionalized Au NPs which can be subsequently used to covalently couple, in principle, any carbohydrate structures by way of the insertion reactions of photochemically activated nitrene species. This coupling chemistry does not require chemical derivatization of the carbohydrate, which can be complex when multiple protection and glycosylation reactions are involved. PFPA has been successfully utilized to immobilize hyaluronan on polystyrene (PS) beads.37 PS beads with surface amino groups were treated with NHS-functionalized PFPA, and hyaluronan was then attached to the bead surface by UV irradiation. In this article, we report that PFPAs can be employed to conjugate monosaccharides and oligosaccharides on Au NPs. The surface-bound carbohydrates retained their binding affinities with the corresponding lectins, and the ranking of binding affinity was consistent with that observed for free ligands in solution. A colorimetry method was developed to determine the density of the carbohydrates attached to Au NPs. Results showed that the coupling chemistry is efficient and high yielding.
2 Experimental
2.1 Synthesis
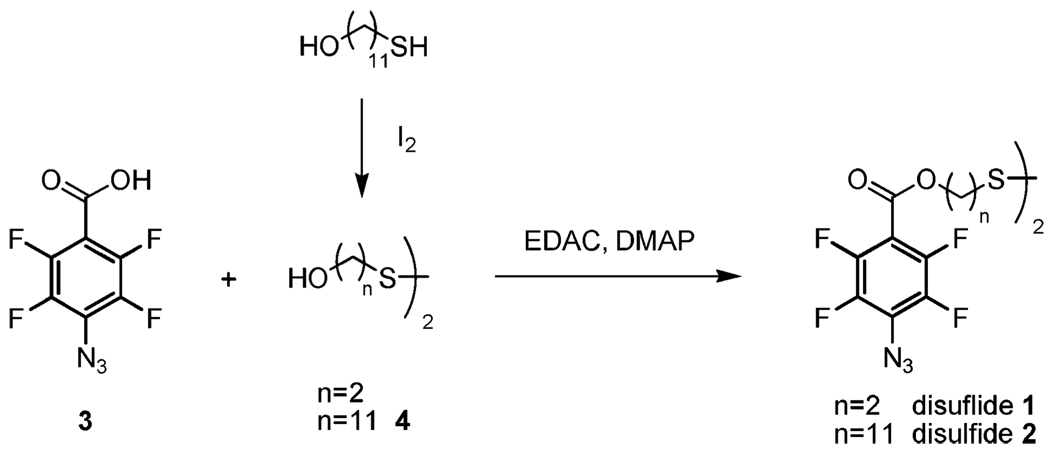
PFPA-disufide 1 was prepared following a previously reported procedure.38
11,11′-Disulfanediylbis(undecane-11,1-diyl)bis(4-azido-2,3,5,6-tetrafluorobenzoate) (PFPA-disufide 2)
11-Mercapto-1-undecanol (90 mg, 0.42 mmol) in absolute ethanol (15 mL) was titrated with a saturated solution of iodine in ethanol until the brown color of iodine persisted. The solution was concentrated to 2 mL and then water (15 mL) was added. The solution was extracted using diethyl ether (3 × 15 mL), and the combined ethereal extracts were washed with brine, dried over NaSO4, and the solvent was removed under reduced pressure to afford the disulfide 4 as a brown oil. A solution of 339 (100 mg, 0.42 mmol) in CH2Cl2 (15 mL) was cooled to 0 °C, and N,N'-dimethylaminopyridine (DMAP) (5.2 mg, 0.042 mmol) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC) (88.6 mg, 0.046 mmol) were added. The disulfide 4 was then added and the solution was stirred for 1 h, after which the solution was allowed to warm to room temperature, and stirred for 12 h. The product was recovered by extraction with CH2Cl2. The organic layer was washed with water, brine, and dried over Na2SO4. Purification of the crude product was carried out by flash column chromatography with 10/1 v/v hexane/ethyl acetate to afford PFPA-disulfide 2 as a clear oil (74.1 mg, 42%). 1H NMR (400 MHz, CDCl3): δ (ppm) 4.36 (t, J = 7.4 Hz, 2H), 2.68 (t, J = 7.2 Hz, 2H), 1.76–1.72 (m, 4H), 1.41–1.28 (m, 14H). 13C NMR (100 MHz, CDCl3): δ (ppm) 159.4, 146.0 (d, JC–F = 258 Hz), 140.4 (d, JC–F = 255 Hz), 124.7, 107.1, 63.8, 38.7, 34.4, 31.5, 29.8, 28.8, 28.1, 27.7, 25.1. Anal. Calcd for C36H44F8N6O4S2: C, 51.42; H, 5.27; N, 9.99. Found: C, 51.50; H, 5.31; N, 10.01%.
2.2 Sample preparation
The gold nanoparticles were prepared following a modified procedure of the two-phase system.40 A 0.25 mM aqueous solution (100 mL) of HAuCl4 (Aldrich) was heated to boiling and 1 wt% sodium citrate solution (1.8 mL) was added quickly under vigorous stirring. The solution was allowed to boil for an additional 5 min until the color of the solution became dark purple and finally light red. A 1.7 mM solution of PFPA-disulfide 1 or 2 in acetone (5 mL) was added slowly to the Au NP solution, and the solution was stirred for 10 hours when it turned to a burgundy color. Toluene (15 mL) was subsequently added, and the mixture was vigorously stirred for 1 hour leaving behind a light pink aqueous phase. The toluene layer was then separated, concentrated to 5 mL using a rotary evaporator at 45 °C, and then diluted with acetone (20 mL). The diluted solution was kept in a refrigerator overnight, and centrifuged at 14 000 rpm for 30 min. Precipitates were collected and re-dissolved in acetone by sonication for 1 min, before being further centrifuged. The re-dissolution and centrifugation processes were repeated 3 times to remove the excess PFPA-disulfide. The functionalized Au NPs were kept in acetone for storage. To determine the concentration of functionalized Au NPs, an aliquot of the solution was centrifuged, and the precipitate collected, dried, and weighed.
Carbohydrates were coupled to Au NPs according to the following general procedure using α-1,4-mannobiose as the example. A solution of PFPA-functionalized gold nanoparticles (1.5 mL) was mixed with 2.9 mM of α-1,4-mannobiose aqueous solution (0.1 mL, V-Labs) in a short flat beaker. The mixture was covered with a 280 nm long-path optical filter (WG-280, Schott Glass) and was irradiated with a 450 W medium pressure Hg lamp (Hanovia) for 5 min under vigorous stirring. Centrifugation of the solution at 14 000 rpm for 15 min separated the mannobiose-attached gold nanoparticles as precipitates. Excess mannobiose was removed by centrifugating and sonicating the nanoparticles in Milli-Q water for 3 times.
2.3 Carbohydrate density determination
A freshly-prepared anthrone solution in concentrated H2SO4 (0.5 wt%, 1 mL) was added into various concentrations of d-mannose in water (0.5 mL) in an ice bath under stirring. The solution was then heated to 100 °C and stirred for 10 min. After cooled to room temperature, the UV-vis spectra of the resulting solutions were recorded on a Perkin-Elmer Lambda 45 UV-vis spectrometer. The absorbance of the solution at 620 nm was measured and the data were plotted against the concentration of d-mannose. The result was used as the calibration curve for the calculation of the ligand density on Au NPs (see Figure 1S, ESI†). Ligand density experiments for Au NPs were carried out by dissolving freshly-prepared d-mannose-conjugated Au NPs (0.3–0.5 mg) in 0.5 mL Milli-Q water, and the solutions were treated with anthrone/H2SO4 following the same protocol described above. The weight of Au NPs was measured by drying the solution under reduced pressure for 3 hours and weighed. The final data were the mean value of 5 measurements with less than 5% variation. The un-functionalized Au NPs were treated in the same manner with anthrone/H2SO4 and the absorbance at 620 nm was used as the background deducted from the total signals measured from the d-mannose-conjugated Au NPs. The density of d-mannose immobilized was then determined using the calibration curve.
2.4 Lectin binding assay
The binding affinity of carbohydrates conjugated on Au NPs was evaluated using Concanavalin A (Con A) (from Canavalia ensiformis (Jack bean), Sigma) according to the following procedure. The binding studies with other lectins (from Griffonia simplicifolia (GS II), Arachis hypogaea (peanut) (PNA), Glycine max (soybean), Sigma) were carried out in the similar manner. In the experiment, the nanoparticles were incubated in a 10 mMpH 7.4 PBS buffer solution containing 0.1% Tween 20 and 3% bovine serum albumin (BSA) for 30 min, centrifuged, and incubated in a pH 7.4 PBS solution without BSA for another 20 min. The nanoparticles were subsequently treated with 10 µg/mL Con A solution in pH 7.4 PBS buffer (0.05 mL) containing 1 mM MnCl2 and CaCl2 for 1 hour while shaking. In cases where aggregation was induced after binding with Con A, the suspension was transferred to a centrifuge tube and centrifuged at 14 000 rpm for 15 min.
2.5 General instrumentation
1H NMR spectra were collected on a Bruker 400 MHz NMR spectrometer. Infrared spectra were measured on a Perkin-Elmer 2000 Fourier transform spectrometer. UV-vis spectra were recorded on a Perkin-Elmer Lambda 45 UV-vis spectrophotometer. TEM images were obtained on a JEOL 100CX transmission electron microscope operating at an accelerating bias voltage of 100 kV. The specimens were prepared by dropping nanoparticle suspension (10 µL) onto a 200 mesh copper grid (coated with carbon supporting film, Electron Microscopy Sciences).
3 Results and discussion
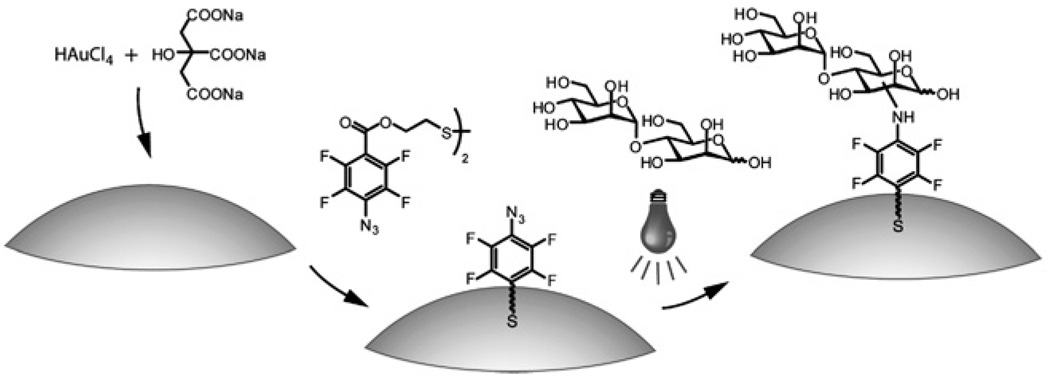
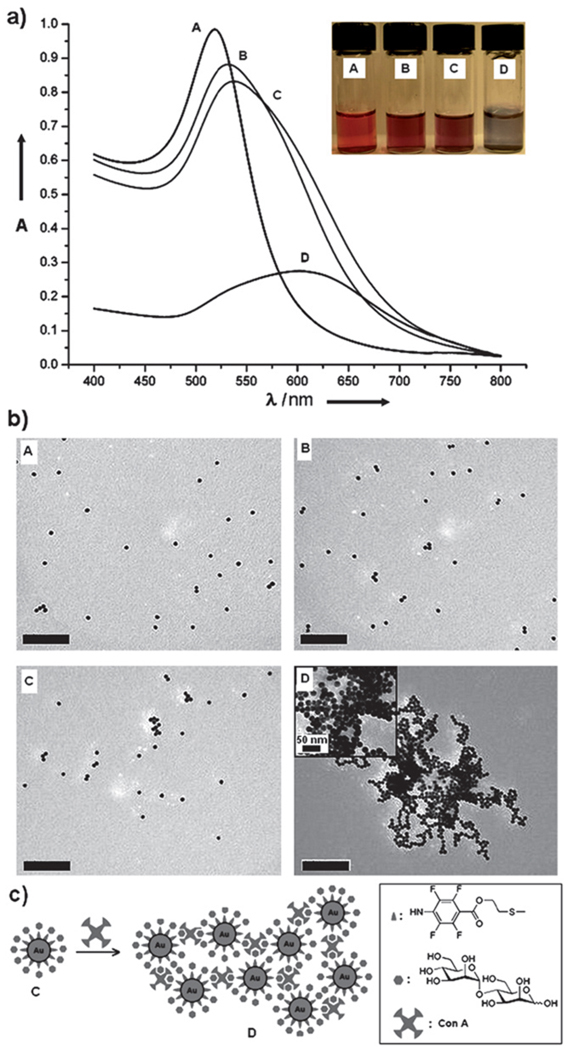
PFPA-disulfides 1 and 2 were synthesized by coupling PFPA-COOH 3 with the corresponding diol using EDAC (Scheme 1). Diol 4 was prepared by oxidizing the hydroxythiol with I2.41 The two disulfides were chosen differing in the length of the spacer linkage. A one-pot procedure was developed to simultaneously synthesize and functionalize Au NPs with PFPA. Colloidal Au NPs, ~20 nm in diameter, were prepared using the citrate reduction reaction of HAuCl4.40 The resulting citrate-stabilized Au NPs were light red in color exhibiting the surface plasmon absorption peak at ~520 nm in the UV-vis spectrum. The Au NPs were subsequently functionalized with PFPA-disulfide (Fig. 1) via a modified phase-transfer ligand-exchange reaction, after which the PFPA-functionalized Au NPs migrated to the organic phase, indicating that the hydrophilic surface of citrate-stabilized Au nanoparticles became hydrophobic. The successful functionalization of Au NPs with PFPA was confirmed by 1H NMR and FTIR (see Figures 2S and 3S in the ESI†). The subsequent coupling of carbohydrates to PFPA-functionalized nanoparticles was carried out by mixing an aqueous solution of the carbohydrate with the nanoparticles in acetone, and irradiating the mixture with >280 nm light. FTIR spectra of the resulting nanoparticles show that the characteristic –N3 absorption at 2125 cm−1 disappeared (see Figure 3S in the ESI†), indicating that the azido groups were activated. Accompanied by each step of the surface functionalization was the color change of the nanoparticle solution, shown in Fig. 2a where α-1,4-mannobiose was coupled on the Au NPs. The UV-vis spectra of PFPA-Au and mannobiose-Au both underwent red-shifts after surface functionalization (8 nm and 4 nm, respectively), likely due to the slight size growth and the change of environment around the nanoparticles.
Scheme 1.
Synthesis of PFPA disulfides 1 and 2.
Fig. 1.
Synthesis of PFPA-Au NPs and subsequent coupling of α-1,4-mannobiose.
Fig. 2.
(a) UV-vis spectra (insert: Au nanoparticle solutions) and (b) TEM micrographs (scale bar: 200 nm) of Au NPs (A), PFPA-disulfide-functionalized Au NPs (B), Au NPs with surface-coupled α-1,4-mannobiose (C), and subsequent treatment with Con A (D). (c) Schematic illustration of the interaction of mannobiose-coupled Au NPs with Con A, and the formation of Au NP aggregates.
The amount of carbohydrate ligands coupled to Au NPs was determined by a colorimetry method using anthrone/sulfuric acid.42,43 This is a well-established assay for the quantitative analysis of carbohydrates, and has been adopted in glyconanoparticle analysis.44 We investigated the ligand density on Au NPs using d-mannose. A calibration curve was obtained by treating various concentrations of d-mannose with anthrone/sulfuric acid, and the absorption at 620 nm was measured (Figure 1S, ESI†). Au NPs with d-mannose immobilized were subjected to the same assay and the absorptions at 620 nm were recorded. The amount of d-mannose attached to Au NPs was subsequently derived from the calibration curve, which averaged at 24 nmol/mg Au NPs, or 1200 molecules per Au NP (see the ESI† for detailed calculation). Assuming that d-mannose ligands are close-packed on the NP, the maximal amount of d-mannose that can occupy on each 20 nm Au NP was calculated to be 72 nmol/mg Au NPs, or 3500 molecules per Au NP (see Figure 4S, ESI† for detailed calculation). This result corresponds to a surface coverage of 34% of the photocoupled d-mannose, indicating a fairly reasonable coupling efficiency.
To further investigate the efficiency of the photocoupling reaction, various concentrations of d-mannose were used when mixing with PFPA-functionalized Au NPs during light activation. Here PFPA-disulfide 2 was used in the study. The amount of d-mannose attached to Au NPs was then determined by the anthrone/sulfuric acid assay described above, and results are summarized in Table 1. At lower ligand loading, the coupling yield was high but the surface coverage was low. As the amount of added ligand increased, the coupling efficiency decreased whereas the surface coverage increased drastically before saturating at around 80%. Note that even at low ligand loading concentration of 100 nmol/mg NPs, high surface coverage (80%) was obtained while a relatively high coupling efficiency of 57% was achieved. The result is significant that a large excess of ligand is unnecessary, which is especially beneficial to carbohydrates that are difficult or costly to obtain. Moreover, this approach provides a simple means to control the ligand density on the NPs. Nanoparticles with coupled d-mannose density varying over 3 orders of magnitude can be produced by changing the amount of the ligand initially added. Note that the amount of d-mannose coupled to Au NPs functionalized with PFPA-disulfide 2 (57.4 nmol/mg) was higher than that on NPs functionalized with 1 (31.6 nmol/mg) with the same initial ligand concentration, demonstrating that the longer spacer increased the coupling efficiency.
Table 1.
Coupling efficiency and surface coverage of d-mannose immobilized on Au NPs using PFPA-disulfide 2 as the coupling agent
| Mannose added (nmol/mg NPs) |
Mannose coupled (nmol/mg NPs) |
Coupling yield (%)a |
Surface coverage (%)b |
|---|---|---|---|
| 0.1 | 0.081 | 85 | 0.11 |
| 0.5 | 0.40 | 80 | 0.56 |
| 1 | 0.78 | 78 | 1.1 |
| 5 | 3.1 | 61 | 4.3 |
| 10 | 5.3 | 53 | 7.4 |
| 50 | 30 | 59 | 41 |
| 80 | 44 | 56 | 62 |
| 100 | 57 | 57 | 80 |
| 120 | 58 | N/A | 80 |
| 150 | 57 | N/A | 80 |
Coupling yield = mannose coupled/mannose added × 100%.
Surface coverage = mannose coupled/max. mannose computed × 100%. The max. mannose computed is 71.7 nmol/mg NPs for 20 nm Au NPs (see ESI† for detailed calculation).
The carbohydrate-functionalized NPs were subsequently subjected to binding studies with a series of lectins, i.e. carbohydrate-binding proteins, to investigate whether the coupled carbohydrates retained their binding affinities. Concanavalin A (Con A), a mannose-binding protein, was used as a model system to test the effectiveness of this coupling chemistry. At pH > 7, Con A is tetrameric, each monomer having one saccharide binding site specific for mannose, and to a lesser extent, glucose.13 Upon treating the mannobiose-Au NPs with Con A, rapid and drastic color change occurred (D, Fig. 2a insert), and the UV-vis spectrum of the resulting solution showed a large SPR red-shift of ~75 nm (D, Fig. 2a). Simultaneously observed was the cluster formation causing broadening of the SPR peak and a decrease in the absorption intensity. The aggregation of the nanoparticles is likely a result of Con A’s multiple binding sites for mannose bringing together dimannose-modified nanoparticles. Indeed, TEM micrographs showed that the nanoparticles were discrete and isolated until the addition of Con A (Fig. 2b). The tetrameric Con A acted as a cross-linker agent that agglomerated mannose strongly, forming larger sizes of nanoclusters (Fig. 2c). A control experiment was carried out where PFPA-functionalized Au NPs were treated with Con A. No SPR peak shift was observed in the UV-vis spectrum of the resulting solution (Figure 5S, ESI†). Therefore the SPR shift and agglomeration can only be attributed to the carbohydrate ligands on the Au NPs.
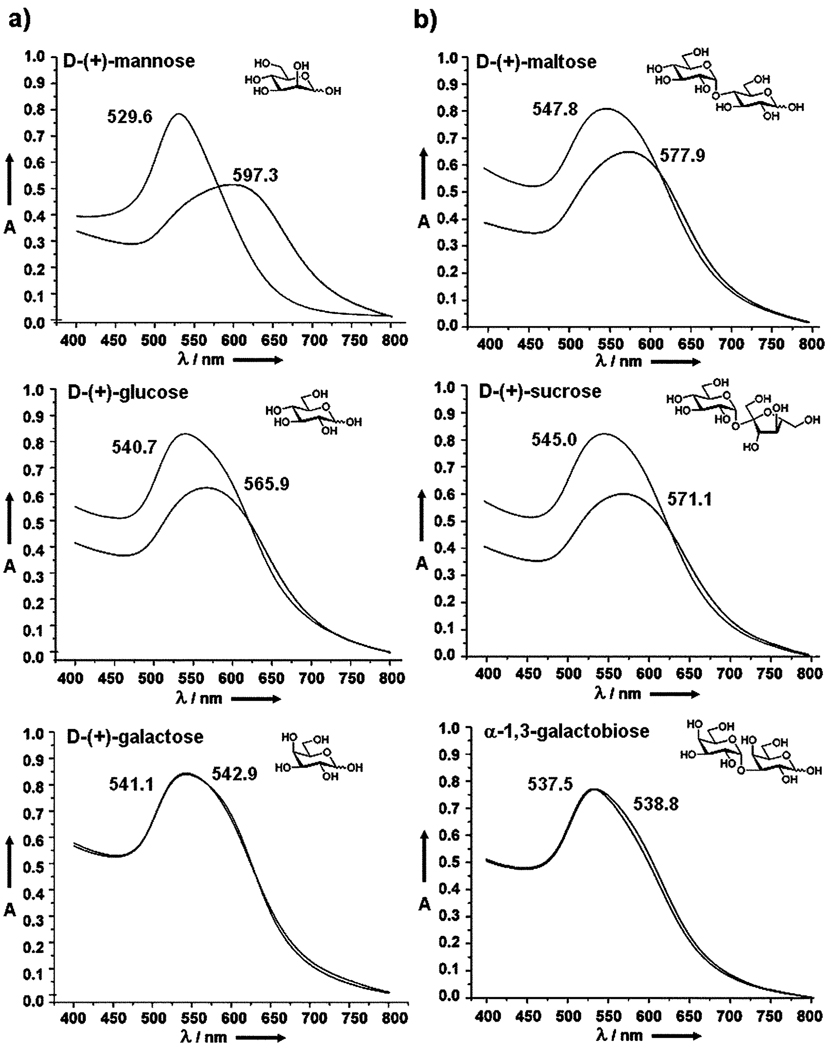
To further investigate the generality of this coupling chemistry and the specificity of surface-bound carbohydrates, monosaccharides (d-mannose, d-glucose, d-galactose) and disaccharides (maltose, sucrose, α-1,3-galactobiose) were coupled to the PFPA-functionalized Au NPs using the same experimental protocol. The resulting carbohydrate-NPs were subsequently treated with Con A and the UV-vis spectra were recorded. d-Glucose is a known ligand of Con A with a lower binding strength than that of d-mannose, and d-galactose is a non-binding ligand for Con A.45–47 The binding constants vary depending on the measurement methods. In the work of Mandal et al., the association constants (Ka) of d-glucose and d-mannose were reported to be 1.96 × 103 M−1 and 8.2 × 103 M−1, respectively, measured by isothermal microcalorimetry.48 In our studies, the UV-vis spectra of the monosaccharide-modified Au NPs showed the largest SPR red-shift for d-mannose (67.7 nm) compared to d-glucose (25.2 nm), whereas almost no change was observed for d-galactose-functionalized NPs (Fig. 3a). These results correlated well with the affinity ranking of the free monosaccharides with Con A in solution.48 Similar results were also observed for disaccharide-functionalized NPs. Maltose, having two d-glucose units, showed a red-shift of 30.1 nm whereas sucrose, containing one d-glucose unit, gave a red-shift of 26.1 nm upon binding with Con A. Almost no change in SPR absorption was observed for galactobiose which consists of two non-binding d-galactose units (Fig. 3b).
Fig. 3.
UV-vis spectra of (a) monosaccharide-, and (b) disaccharide-functionalized Au NPs before and after binding with Con A. Only one set of experimental data are shown here. The experiments were, however, repeated over 5 times and the results were consistent and reproducible.
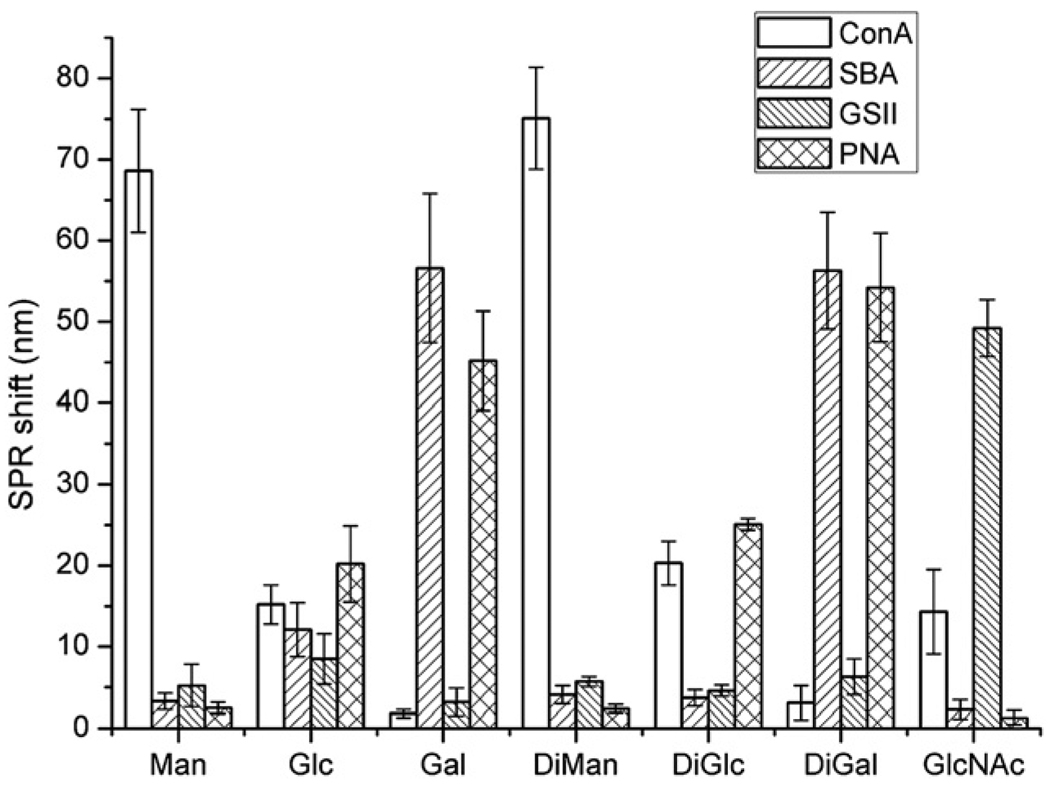
Additional cross-reactivity studies were conducted by treating Au NPs conjugated with mono- and di-saccharides (d-mannose (Man), d-glucose (Glc), d-galactose (Gal), α-1,4-mannobiose (DiMan), β-1,3-glucobioses (DiGlc), α-1,3-galactobiose (DiGal) and N-acetyl-d-glucosamine (GlcNAc)), with Con A and 3 other lectins (GSII, PNA and SBA). The SPR peak shifts were determined from the UV-vis spectra and are shown in Fig. 4. The affinity ranking derived from the SPR peak shift directly correlates with reported solution binding affinity between each carbohydrate and lectin.12,13 For example, larger shifts were observed for the carbohydrate-lectin pairs of Gal-SBA, Gal-PNA, DiGal-SBA, DiGal-PNA, and GlcNAc-GSII, which was consistent with our previous study using a carbohydrate microarray where strong interactions were also observed for these carbohydrate-lectin pairs.38
Fig. 4.
SPR peak shifts of carbohydrate-functionalized Au NPs after treating with various lectins. Each data reading was an average of 5 samples.
Conclusions
In summary, we have developed a general method for coupling underivatized carbohydrates to gold nanoparticles. The method is based on the photochemically induced CH insertion reactions of PFPAs, and it does not require chemical derivatization of the carbohydrate structures. Furthermore, the coupling reaction is fast, taking place in minutes instead of hours which is needed in most thermally-initiated conjugation reactions. The coupling efficiencies were high, and surface coverage of over 80% was obtained. The coupled carbohydrates effectively retained their recognition abilities as demonstrated by the strong interactions with their corresponding carbohydrate-binding proteins. In addition, the binding affinities of surface-bound carbohydrates with various lectins were consistent with those of the free carbohydrates with the corresponding lectins in solution. The sensitive SPR signal was conveniently used to monitor the surface chemistry occurring on the nanoparticles, especially in examining the interactions of surface-bound carbohydrates with their binding proteins where large SPR red-shifts were observed causing visible color changes of the Au NP solutions. The method developed can be readily applied to other carbohydrate structures, and we have successfully coupled oligosaccharides and polysaccharides using the same approach. This general coupling chemistry together with the convenient optical detection offers an attractive platform for label-free, rapid, and sensitive detection of carbohydrate-based molecular recognition.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of General Medical Science (NIGMS) under NIH Award Numbers 1R01GM080295 and 2R15GM066279, ONAMI ARL Center, and in part by the European Commission (MRTN-CT-19561).
Footnotes
Electronic supplementary information (ESI) available: Characterization of PFPA-functionalized Au NPs, including 1H NMR, FT-IR; blank experiments; mannose ligand density estimation and measurement. See DOI: 10.1039/b917900c
Notes and references
- 1.El-Kouedi M, Keating CD. In: Nanobiotechnology. Niemeyer CM, Mirkin CA, editors. Weinheim: Wiley-VCH; 2004. pp. 429–443. [Google Scholar]
- 2.Homola J. In: Springer Series on Chemical Sensors and Biosensors. Urban G, editor. vol. 4. Heidelberg: Springer-Verlag GmbH; 2006. pp. 232–243. [Google Scholar]
- 3.Liu R, Liew R, Zhou J, Xing B. Angew. Chem., Int. Ed. 2007;46:8799–8803. doi: 10.1002/anie.200702773. [DOI] [PubMed] [Google Scholar]
- 4.Oh E, Lee D, Kim Y-P, Cha SY, Oh D-B, Kang HA, Kim J, Kim H-S. Angew. Chem., Int. Ed. 2006;45:7959–7963. doi: 10.1002/anie.200601948. [DOI] [PubMed] [Google Scholar]
- 5.Tsai CS, Yu TB, Chen CT. Chem. Commun. 2005:4273–4275. doi: 10.1039/b507237a. [DOI] [PubMed] [Google Scholar]
- 6.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Müller UR. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, Xia Y. Analyst. 2003;128:686–691. doi: 10.1039/b212437h. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Y, Zhao H, Zhu N, Lin Y, Yu P, Mao L. Angew. Chem., Int. Ed. 2008;47:8601–8604. doi: 10.1002/anie.200804066. [DOI] [PubMed] [Google Scholar]
- 10.Kamat PV. J. Phys. Chem. B. 2002;106:7729–7744. [Google Scholar]
- 11.Okamoto T, Yamaguchi I. J. Phys. Chem. B. 2003;107:10321–10324. [Google Scholar]
- 12.Lee YC, Lee RT. Acc. Chem. Res. 1995;28:321–327. [Google Scholar]
- 13.Lis H, Sharon N. Chem. Rev. 1998;98:637–674. doi: 10.1021/cr940413g. [DOI] [PubMed] [Google Scholar]
- 14.Ngundi MM, Kulagina NV, Anderson GP, Taitt CR. Expert Rev. Proteomics. 2006;3:511–524. doi: 10.1586/14789450.3.5.511. [DOI] [PubMed] [Google Scholar]
- 15.Huang CC, Chen CT, Shiang YC, Lin ZH, Chang HT. Anal. Chem. 2009;81:875–882. doi: 10.1021/ac8010654. [DOI] [PubMed] [Google Scholar]
- 16.Schofield CL, Field RA, Russell DA. Anal. Chem. 2007;79:1356–1361. doi: 10.1021/ac061462j. [DOI] [PubMed] [Google Scholar]
- 17.de la Fuente JM, Barrientos AG, Rojas TC, Rojo J, Canada J, Fernandez A, Penades S. Angew. Chem., Int. Ed. 2001;40:2257–2261. doi: 10.1002/1521-3773(20010618)40:12<2257::AID-ANIE2257>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 18.Hernaiz MJ, de la Fuente JM, Barrientos AG, Penades S. Angew. Chem., Int. Ed. 2002;41:1554–1557. doi: 10.1002/1521-3773(20020503)41:9<1554::aid-anie1554>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y-P, Park S, Oh E, Oh Y-H, Kim H-S. Biosens. Bioelectron. 2009;24:1189–1194. doi: 10.1016/j.bios.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Ojeda R, de Paz JL, Barrientos AG, Martin-Lomas M, Penades S. Carbohydr. Res. 2007;342:448–459. doi: 10.1016/j.carres.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Lin C-C, Yeh Y-C, Yang C-Y, Chen G-F, Chen Y-C, Wu Y-C, Chen C-C. Chem. Commun. 2003:2920–2921. doi: 10.1039/b308995a. [DOI] [PubMed] [Google Scholar]
- 22.Thygesen M, Sauer J, Jensen KJ. Chem.–Eur. J. 2009;15:1649–1660. doi: 10.1002/chem.200801521. [DOI] [PubMed] [Google Scholar]
- 23.Otsuka H, Akiyama Y, Nagasaki Y, Kataoka K. J. Am. Chem. Soc. 2001;123:8226–8230. doi: 10.1021/ja010437m. [DOI] [PubMed] [Google Scholar]
- 24.Earhart C, Jana NR, Erathodiyil N, Ying JY. Langmuir. 2008;24:6215–6219. doi: 10.1021/la800066g. [DOI] [PubMed] [Google Scholar]
- 25.Zhi Z-L, Powell AK, Turnbull JE. Anal. Chem. 2006;78:4786–4793. doi: 10.1021/ac060084f. [DOI] [PubMed] [Google Scholar]
- 26.Lee M-R, Shin I. Org. Lett. 2005;7:4269–4272. doi: 10.1021/ol051753z. [DOI] [PubMed] [Google Scholar]
- 27.Seo JH, Adachi K, Lee BK, Kang DG, Kim YK, Kim KR, Lee HY, Kawai T, Cha HJ. Bioconjugate Chem. 2007;18:2197–2201. doi: 10.1021/bc700288z. [DOI] [PubMed] [Google Scholar]
- 28.Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat. Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 29.Scriven EFV, editor. Azides and Nitrenes: Reactivity and Utility. New York: Academic Press; 1984. [Google Scholar]
- 30.Platz MS. Acc. Chem. Res. 1995;28:487–492. [Google Scholar]
- 31.Brunner J. Annu. Rev. Biochem. 1993;62:483–514. doi: 10.1146/annurev.bi.62.070193.002411. [DOI] [PubMed] [Google Scholar]
- 32.Pandurangi RS, Karra SR, Kuntz RR, Volkert WA. Photochem. Photobiol. 1997;65:208–221. doi: 10.1111/j.1751-1097.1996.tb02427.x. [DOI] [PubMed] [Google Scholar]
- 33.Yan M, Ren J. Chem. Mater. 2004;16:1627–1632. [Google Scholar]
- 34.Liu L, Engelhard MH, Yan M. J. Am. Chem. Soc. 2006;128:14067–14072. doi: 10.1021/ja062802l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gann JP, Yan M. Langmuir. 2008;24:5319–5323. doi: 10.1021/la7029592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan M. Polym. News. 2002;27:6–12. [Google Scholar]
- 37.Joester D, Klein E, Geiger B, Addadi L. J. Am. Chem. Soc. 2006;128:1119–1124. doi: 10.1021/ja0537474. [DOI] [PubMed] [Google Scholar]
- 38.Pei Y, Yu H, Pei Z, Theurer M, Ammer C, Andre S, Gabius HJ, Yan M, Ramstrom O. Anal. Chem. 2007;79:6897–6902. doi: 10.1021/ac070740r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keana JFW, Cai SX. J. Org. Chem. 1990;55:3640–3647. [Google Scholar]
- 40.Turkevich J, Stevenson PC, Hollier J. Discuss. Faraday Soc. 1951;11:55–75. [Google Scholar]
- 41.Danehy JP, Doherty BT, Egan CP. J. Org. Chem. 1971;36:2525–2530. [Google Scholar]
- 42.Koehler LH. Anal. Chem. 1952;24:1576–1579. [Google Scholar]
- 43.Pons A, Roca P, Aguiló C, Garcia F, Alemany M, Palou A. J. Biochem. Biophys. Methods. 1981;4:227–231. doi: 10.1016/0165-022x(81)90060-9. [DOI] [PubMed] [Google Scholar]
- 44.Chien Y, Jan M, Adak A, Tzeng H, Lin Y, Chen Y, Wang K, Chen C, Chen C, Lin C. ChemBioChem. 2008;9:1100–1109. doi: 10.1002/cbic.200700590. [DOI] [PubMed] [Google Scholar]
- 45.Van Landschoot A, Loontiens FG, De Bruyne CK. Eur. J. Biochem. 1980;103:307–312. doi: 10.1111/j.1432-1033.1980.tb04316.x. [DOI] [PubMed] [Google Scholar]
- 46.Gupta D, Dam TK, Oscarson S, Brewer CF. J. Biol. Chem. 1997;272:6388–6392. [PubMed] [Google Scholar]
- 47.Schwarz FP, Puri KD, Bhat RG, Surolia A. J. Biol. Chem. 1993;268:7668–7677. [PubMed] [Google Scholar]
- 48.Mandal DK, Kishore N, Brewer CF. Biochemistry. 1994;33:1149–1156. doi: 10.1021/bi00171a014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.