Abstract
Misregulation of the methyl-CpG-binding protein 2 (MECP2) gene has been found to cause a myriad of neurological disorders including autism, mental retardation, seizures, learning disabilities, and Rett syndrome. We hypothesized that mutations in other members of the methyl-CpG-binding domain (MBD) family may also cause autistic features in individuals. We evaluated 226 autistic individuals for alterations in the four genes most homologous to MECP2: MBD1, MBD2, MBD3, and MBD4. A total of 46 alterations were identified in the four genes, including ten missense changes and two deletions that alter coding sequence. Several are either unique to our autistic population or cosegregate with affected individuals within a family, suggesting a possible relation of these variations to disease etiology. Variants include a R23M alteration in two affected half brothers which falls within the MBD domain of the MBD3 protein, as well as a frameshift in MBD4 that is predicted to truncate almost half of the protein. These results suggest that rare cases of autism may be influenced by mutations in members of the dynamic MBD protein family.
Keywords: Autism, Rett syndrome, MeCP2
Introduction
Autism is a neurodevelopmental disorder with a triad of typical features: (1) impairments in social relatedness, (2) problems in communication, and (3) the presence of repetitive stereotyped behaviors or restricted interests [1]. Autism has an incidence of approximately one in every 1,000 individuals in the general population and has a skewed male to female ratio of about 4:1 [2]. Autism is part of a spectrum of disorders which includes Asperger’s syndrome, pervasive developmental disorder not otherwise specified, childhood disintegrative disorder, and Rett syndrome. Combined, autism spectrum disorders occur at a rate of approximately one in 150 individuals [3]. A high concordance of autism in monozygotic twins signifies a strong genetic component and heritability of the disorder [4–6]; however, linkage and association studies have been largely unable to uncover a single causative gene that accounts for an appreciable percentage of autism cases [7]. This is thought to result from one of two scenarios: (1) distinct, rare variants in an unknown number of genes whose strong effect results in a clinical phenotype or (2) common variants that have little effect by themselves, but acting in concert with modifiers or additional loci, may converge to promote the manifestation of autism. The first hypothesis is demonstrated by distinct cases of autism that have been shown to stem from single gene mutations, such as those in NLGN3, NLGN4, and SHANK3 [8, 9]. Evidence supporting the latter idea comes from studies that have identified a common CNTNAP2 variant in families with male affected individuals and from genome-wide association studies that recognized a novel region of interest on chromosome 5 [10–13].
While there are many autism candidate genes, the autism spectrum disorder Rett syndrome results almost exclusively from alterations in a single gene, methyl-CpG-binding protein 2 (MECP2) [14]. Rett syndrome differs from classic autism in several ways, the most obvious being that a majority of patients are female. Due to MEPC2’s location on the X chromosome, many mutations are heterozygous viable in females but hemizygous lethal in males. Rett patients typically acquire the hallmark feature of repetitive hand wringing or clasping [15, 16]. Rett syndrome is also characterized by developmental regression, a feature only seen in a subset of autistic patients [1, 17]. On the other hand, Rett patients are similar to autistic individuals in that they demonstrate increased anxiety in novel situations and apathy toward people and objects around them [18].
Point mutations, deletions, and duplications in MECP2 cause more than 85% of Rett syndrome cases [14, 19]. While these mutations have been found throughout the gene in Rett patients, they tend to occur in domains of functional importance: the methyl-CpG-binding domain (MBD) or the transcriptional repression domain (TRD) [20]. Current evidence demonstrates that the MeCP2 protein modulates gene expression through chromatin remodeling, acting both as an enhancer and a repressor of gene activity [21].
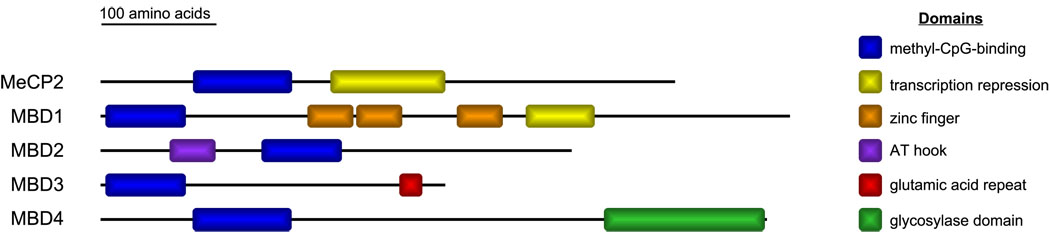
MeCP2 is a member of the MBD family of proteins. Along with MeCP2, there are four other closely related MBD proteins designated in humans: MBD1, MBD2, MBD3, and MBD4 (Fig. 1) [22]. MBD1, MBD2, MBD4, and MeCP2 each have the ability to bind DNA at methylated CpG sites while MBD3 requires additional factors in order to be recruited to DNA [23]. All of the MBD proteins have demonstrated transcriptional repression activity. The MBD4 protein is unique in that it has preferential binding for G–T mismatches and acts to repair DNA damage [24]. The MBD proteins are also capable of simultaneously binding the same promoter regions, implying either a functional interdependence or redundancy between these proteins [25, 26]. The latter can be seen where MBD2 and MBD3 act in a mutually exclusive manner to form distinct Mi-2/NuRD chromatin-remodeling complexes [27]. Therefore, it is possible that dysfunction in one of these MBD proteins could titrate away core components of the Mi-2/NuRD complex, leading to an imbalance of the protein components available to bind to the second MBD protein.
Fig. 1.
The methyl-CpG-binding domain (MBD) protein family. The five MBD proteins, each containing an MBD domain, are illustrated. Along with this domain, each protein contains at least one other functional domain that contributes to the protein's function
Significantly, mutations in the MECP2 gene have also been identified in autistic patients without classic Rett syndrome [28–30]. Specifically, our laboratory identified two autistic females carrying de novo MECP2 alterations that result in coding changes [28]. Further studies have shown that MECP2 mutations can cause a spectrum of clinical presentations including mild learning disabilities, mental retardation, schizophrenia, Angelman-like symptoms, and mental retardation presenting with infantile seizures [29–37]. These findings demonstrate that there is a broad range of phenotypes caused by misregulation of the MeCP2 protein. Moreover, a recent study demonstrated an association between markers within MECP2 and autism [38].
In addition to clinical data, there is accumulating evidence from animal models demonstrating the severe effects of MBD misregulation [39–44]. The Mecp2308/y knock-in model generated by Shahbazian and colleagues creates a truncated protein similar to that found in Rett patients, and the mice exhibit anxiety, seizures, and abnormal social and nesting behavior [40, 45, 46]. Furthermore, Mecp2308/y mice are defective in social, spatial, and contextual fear memory [47]. Another MBD model, the Mbd1−/− mouse, is also deficient in a wide range of social behaviors, showing increased anxiety, greater susceptibility to depression, and a reduced prepulse inhibition of the startle response, a feature found in autistic patients [41, 48]. Interestingly, these mice were also found to have an increase in the RNA and protein levels of serotonin receptor 2c [48]. Both of these models demonstrate that mutating or completely eliminating a single MBD protein can result in autistic features in mice.
Based on the findings of autistic patients with mutations in MeCP2 and results from animal models, we decided to investigate additional members of the MBD family to determine whether rare variations within these genes might be associated with autism. It is possible that the MBD domain may perform a similar function within each MBD protein, and when the gene is misregulated, could lead to symptoms that fall along the autism spectrum. To date, only a single study limited to a Japanese population of 65 autistic cases has evaluated the possible role of mutations in the MBD genes in autistic patients [49]. One variation of interest was identified, R269C in MBD1 [49]. In this study, we evaluated MBD1, MBD2, MBD3, and MBD4 and found multiple alterations that cause amino acid changes and potentially lead to autistic features.
Materials and methods
Patient ascertainment
We randomly selected DNA from 226 unrelated autistic individuals (195 Caucasian (CA) and 31 African-American (AA) probands) and their family members, when available, from our dataset of autism spectrum disorder families. This cohort was comprised of 180 males and 46 females.
Participants were recruited, enrolled, and sampled according to the Institutional Review Board protocols for the University of Miami and the University of South Carolina. Individuals entered the study through well-developed referral sources (e.g., clinics) as well as through family support groups, schools, and practitioners. Participants with autism were ascertained on the basis of a previously existing diagnosis of autism and met the following minimal inclusion criteria: (1) age between 3 and 21 years, (2) absence of severe sensory or significant motor impairments, and (3) absence of identified metabolic, genetic, or progressive neurological disorders. The research diagnosis for participants with autism was assigned using Diagnostic and Statistical Manual of Mental Disorders IV criteria [50], supported in most cases by the Autism Diagnostic Interview-Revised, and when available, the Autism Diagnostic Observation Schedule [51, 52]. In a minority of cases, neither measure was available, and the research diagnosis was made by our expert clinical panel. We also administered the Vineland Adaptive Behavior Scales (VABS) [53], a measure of developmental functioning to assure that the participants with autism met a basic developmental age of 18 months. This developmental threshold ensures that the diagnostic instruments are valid. Detailed family and medical histories were obtained, including information about psychiatric, developmental, and behavioral problems in family members. A complete description of the study was provided to the subjects, and written informed consent was obtained. Whole blood was collected from all participants by venipuncture.
The Caucasian control group consisted of 245 individuals that had no known indicators of behavioral, developmental, or cognitive abnormalities (100 males and 145 females). The human random control panel (Sigma-Aldrich) encompassed DNA from 96 Caucasian individuals collected in the United Kingdom (66 males and 30 females). The African-American controls were a combination of the Coriell African-American control panel (NDPT031), which was comprised of 94 individuals (25 males and 69 females) and 86 samples collected at the University of Miami (49 males and 37 females). Both of these datasets only contained individuals that were negative for evidence of neurological disease.
Statistical analysis
In order to determine our ability to detect variants within our autistic patient dataset, we used the binomial distribution to compute the probability that at least one variant allele (with frequency p) was observed in a sample of N chromosomes. We assumed sample sizes of N=390 and N=62, which represent the number of chromosomes in our affected CA and AA samples, respectively, and considered allele frequencies of 0.01 and 0.05. The probabilities were computed using the binomial cumulative density function.
Denaturing high-performance liquid chromatography
Genomic DNA was isolated from whole frozen blood according to standard protocol [54]. DNA from 226 probands was assembled into 47 distinct pools of three to five samples each. Thirty-two Caucasian control samples were assembled into five pools of three to five samples. Mutation screening was performed by denaturing high-performance liquid chromatography (DHPLC) using the WAVE™ DNA fragment analysis system (Transgenomic, Omaha). Primers were designed to amplify all exons for the MBD1, MBD2, MBD3, and MBD4 genes in each proband and control pool (Supplementary Table 1). The polymerase chain reaction (PCR) products were evaluated via agarose gel electrophoresis. For heteroduplex molecule formation, all PCR products were subjected to denaturation for 3 min followed by gradual reannealing in the thermocycler for 45 min. The temperature for successful resolution of the heteroduplex molecules was selected using Transgenomic software before proceeding to DHPLC analysis. Homozygous and heterozygous peak profiles were identified using Transgenomic software for all proband and control pools. Pools of interest for both homozygous and heterozygous profiles were broken down, and each sample within the pool was sequenced.
Sequencing
Individuals with possible variants identified by DHPLC were sequenced using Applied Biosystems Big Dye Terminator version 3.1 on the ABI 3730 sequencer, and the primers listed in Supplementary Table 1. Whenever possible, family members of the proband sample were also examined by sequencing. Additionally, a custom TaqMan allelic discrimination assay (Applied Biosystems) was designed for each novel variant. These newly identified variants were evaluated in 150 Caucasian control individuals, and when appropriate, 180 African-American individuals. Three variants were tested in the Sigma-Aldrich Caucasian human random control panel instead of the 150 CA controls. For novel variants that altered the amino acid sequence and were absent in the initial controls tested, an additional 95 Caucasian controls were sequenced across the region.
Cloning mutations
All insertion/deletion variants were verified by cloning. Exons for MBD genes were amplified by PCR; products were cloned into the Invitrogen pCR2.1-TOPO high copy plasmid vector and transformed in Top10 Escherichia coli chemically competent cells. The standard TOPO TA cloning protocol was followed with a 30-min ligation using 4µl of PCR product, 30-minute incubation on ice, and 75µl of transformation plated on Luria–Bertani medium (LB)/ampicillin/X-gal. Individual colonies were selected and grown in LB ampicillin medium, and plasmid DNA was extracted using the Qiagen Miniprep Spin kit. The plasmid DNA samples were sequenced on ABI 3100 using BigDye 3.1 Sequencing kit (Applied Biosystems) and M13 forward and reverse primers (Integrated DNA Technologies).
Prediction of splice factor binding sites
Sequences containing variations within intronic and exonic regions in this study were entered into the Human Splicing Analyzer version 2.3 to reveal potential binding sites for splicing factors (http://www.umd.be:2300/) [55].
Results
We identified 198 autistic individuals carrying genetic alterations at 46 different locations in the four MBD genes that we studied (Table 1, Fig. 2). Twenty-five of these changes represent novel variations, while twenty-one are single nucleotide polymorphisms (SNPs) that have been previously identified and incorporated into the National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=snp). The frequencies of previously recognized SNPs in both AA and CA populations can be found in Supplementary Table 2. Of the novel changes that were identified, 20 were single nucleotide alterations, and five were insertion/deletions. For these 25 new variants, we tested 150 control Caucasian individuals (300 chromosomes), and when appropriate, an additional 180 African-American control individuals (360 chromosomes) to evaluate the relative frequency of each variation in the general population. Fourteen of the novel single nucleotide changes and three of the deletions were not found in 150 Caucasian or 180 African-American controls. When a novel variant was identified that altered the amino acid sequence, it was tested in an additional 95 Caucasian controls (190 chromosomes). We did not detect the MBD1 R269C variation that was previously reported in a study of Japanese autistic patients, suggesting differences in variations due to ethnicity [49].
Table 1.
MBD variations identified in autistic probands and control individuals
| Gene | Autistic individuals |
Controls (frequency) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Position | Location | Nucleotide | Amino acid | SNP/novel | Probands | Male/female | Ethnicity | CA | AA |
| MBD1 18q21.1 | Exon 4 | 46,057,352 | G>A | – | Rs140686 | 5 | 4/1 | 1 AA, 4 CA | ||
| Exon 5 | 46,057,065 | G>A | R147K | Novel | 1 | 1/0 | 1 CA | 0/245 | ||
| Intron 10 | 46,055,301 | C>T | – | Novel | 2a | 2/0 | 2 CA | 6/149 (0.04) | ||
| Exon 12 | 46,054,177 | OG | P401A | Rs125555 | 1 | 0/1 | 1 CA | |||
| Exon 12 | 46,054,095 | G>A | R428H | Novel | 1 | 1/0 | 1 AA | 0/242 | 0/180 | |
| Exon 12 | 46,053,968 | G>A | – | Novel | 1 | 1/0 | 1 CA | 0/245 | ||
| Intron 13 | 46,053,335 | T>C | – | Novel | 1 | 0/1 | 1 AA | 0/150 | 0/179 | |
| Exon 14 | 46,053,234 | A>T | K558N | Novel | 1 | 1/0 | 1 CA | 0/239 | ||
| Intron 15 | 46,051,935 | C>T | – | Novel | 1 | 1/0 | 1 AA | 0/146 | 0/178 | |
| Exon 17 | 46,050,015 | C>T | – | rs72923678 | 4 | 4/0 | 4 CA | |||
| Exon 17 | 46,049,627 | G>T | – | rs11663629 | 39 | 29/10 | 13 AA, 26 CA | |||
| MBD2 18q21.2 | Exon 1 | 50,005,135 | C>T | – | Novel | 4 | 4/0 | 4 CA | 3/150 (0.02) | |
| Exon 1 | 50,005,114 | T>C | – | Novel | 4 | 4/0 | 4 CA | 1/142 (0.01) | ||
| Intron 2 | 49,985,346 | G>A | – | rs16957946 | 5a | 5/0 | 1 AA, 4 CA | |||
| Exon 7 | 49,935,242 | A>G | – | rs7614 | 3 | 2/1 | 3 CA | |||
| Exon 7 | 49,934,805 | G>A | – | rs62099763 | 1 | 1/0 | 1 CA | |||
| Exon 7 | 49,934,755 | G>A | – | rs1259938 | 85a | 63/22 | 4 AA, 81 CA | |||
| MBD3 19p13.3 | Exon 1 | 1,543,563 | G>T | R23M | Novel | 1 | 1/0 | 1 AA | 0/243 | 0/172 |
| Intron 1 | 1,543,502 | C>T | – | Novel | 1 | 1/0 | 1 AA | 0/234 | 3/172 (0.017) | |
| Intron 4 | 1,532,272 | G>A | – | Novel | 3 | 2/1 | 3 CA | 0/146 | ||
| Exon 5 | 1,532,144 | C>T | – | Novel | 1 | 1/0 | 1 CA | 0/150 | ||
| Exon 6 | 1,529,372-1,529,374 | delGAG | E281del | Novel | 1 | 1/0 | 1 AA | 0/187b | 3/167 (0.018) | |
| Exon 6 | 1,529,321 | C>T | – | Novel | 2 | 2/0 | 2 AA | l/190b (0.005) | 6/162 (0.037) | |
| Exon 7 | 1,529,191 | A>G | – | Novel | 1 | 1/0 | 1 CA | 0/148 | ||
| Exon 7 | 1,529,075-1,529,077 | delGCG | – | Novel | 3a | 2/1 | 1 AA, 2 CA | 1/142 (0.007) | 10/178 (0.056) | |
| Exon 7 | 1,529,140 | C>T | – | Novel | 1 | 1/0 | 1 CA | 0/144 | ||
| Exon 7 | 1,528,326 | C>T | – | Novel | 1 | 1/0 | 1 AA | 0/140 | 0/178 | |
| Exon 7 | 1,528,325 | G>A | – | Novel | 1 | 1/0 | 1 CA | 0/145 | ||
| Exon 7 | 1,528,208 | C>T | – | rs1053151 | 151a | 118/33 | 28 AA, 123 CA | |||
| Exon 7 | 1,528,117 | G>A | – | rs9585 | 154a | 120/34 | 31 AA, 123 CA | |||
| Exon 7 | 1,528,025-1,528,045 | del20insCTG | – | Novel | 1 | 0/1 | 1 AA | 0/137 | 0/176 | |
| Exon 7 | 1,527,932 | A>G | – | rs16994495 | 51a | 40/11 | 6 AA, 45 CA | |||
| Exon 7 | 1,527,928 | A>G | – | rs2238588 | 51a | 40/11 | 6 AA, 45 CA | |||
| Exon 7 | 1,527,924 | G>C | – | rs12884 | 51a | 40/11 | 6 AA, 45 CA | |||
| Exon 7 | 1,527,834 | G>A | – | Novel | 1 | 1/0 | 1 CA | 6/94b (0.064) | ||
| Exon 7 | 1,527,785-1,527,805 | Deletion | – | Novel | 1 | 1/0 | 1 AA | 0/145 | 0/163 | |
| MBD4 3q21.3 | Intron 2 | 130,639,226 | T>C | – | rs140692 | 38 | 29/9 | 11 AA, 27 CA | ||
| Intron 2 | 130,638,887 | OG | – | rs3138340 | 5 | 5/0 | 4 AA, 1 CA | |||
| Exon 3 | 130,638,360 | G>A | A273T | rs10342 | 27 | 19/8 | 2 AA, 25 CA | |||
| Exon 3 | 130,638,232-130,638,235 | delAAGA | E314fsX316 | Novel | 1 | 0/1 | 1 AA | 0/242 | 0/163 | |
| Exon 3 | 130,638,153 | T>C | S342P | rs2307289 | 9a | 9/0 | 8 AA, 1 CA | |||
| Exon 3 | 130,638,141 | G>A | E346K | rs140693 | 3 | 3/0 | 3 CA | |||
| Exon 3 | 130,638,104 | T>C | I358T | rs2307298 | 5a | 3/2 | 5 CA | |||
| Intron 3 | 130,637,926 | T>G | – | rs3138341 | 5 | 5/0 | 4 AA, 1 CA | |||
| Exon 5 | 130,635,394 | A>G | N467S | Novel | 2 | 1/1 | 2 CA | 0/244 | ||
| Exon 6 | 130,634,779 | C>T | – | rs140696 | 38 | 29/9 | 11 AA, 27 CA | |||
These variations were also identified in the DHPLC Caucasian control samples
The Caucasian Sigma control samples were used for these variations
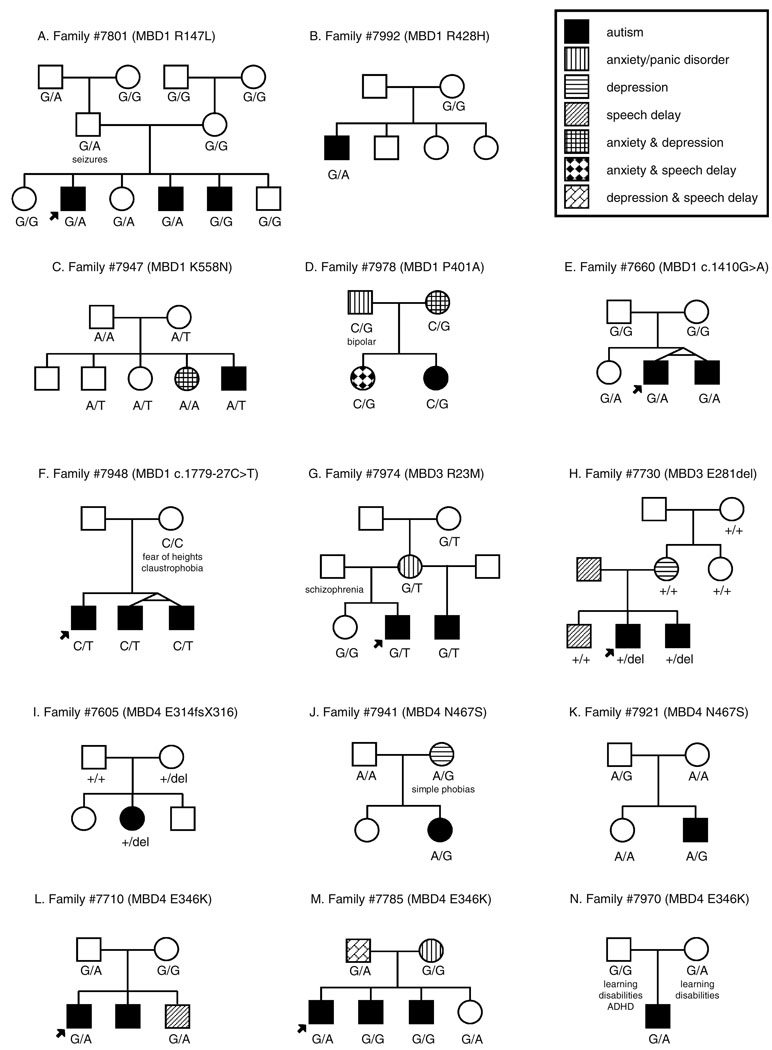
Fig. 2.
Pedigrees of autistic families identified with mutations in MBD genes. Alterations identified in MBD1 (a–f), MBD3 (g–h), and MBD4 (i–n) are shown. Each nucleotide change is marked under the individual in whom it occurs. In families with multiple affected individuals, the initially identified autistic individual (proband) is signified with an arrow
With our cohort of 226 autistic individuals, we calculated that we would have a 98.0% probability of detecting rare variants that occurred at a 1% frequency in our 390 CA chromosomes. This probability increases to 99.9% for alleles with a 5% frequency. We would also have a 95.8% likelihood of detecting rare alleles occurring at a 5% frequency in our 62 AA chromosomes.
MBD1
Eleven SNPs were detected in MBD1, four known and seven novel. Three of the novel SNPs as well as rs12555 resulted in nonsynonymous amino acid changes (R147K, R428H, K558N, and P401A, respectively). The novel alteration c.440G>A (R147K) was identified in the Caucasian family no. 7801 (Fig. 2a). This alteration was found in two of three affected brothers and an unaffected sister. Interestingly, the two affected individuals carrying the variant appeared to be more severely affected clinically than the brother without the variant; while all three brothers were found to have speech difficulties, the proband developed speech later than the brother without the variant (III:5), and the third brother (III:4) remained nonverbal at 45 months and had irregular breathing and gait (Table 2). The proband also demonstrated regression in language as well as social interest and play skills, all before 5 years of age. This alteration was inherited from the paternal grandfather; while the father was not reported to have autism, he did have a history of seizures. The variant was not identified in 245 Caucasian controls.
Table 2.
Clinical features of affected individuals identified with variations in MBD genes
| Variant | Family | Individual | Allele | Age at exama |
Age to walka |
1st wordsa |
Phrase speecha |
Verbal | VABS | Regression | Midline movements |
Seizures | Irregular breathing |
Unusual gait |
Head circumference in cm (age3) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MBD1 R147L | 7801 | Male proband | G/A | 126 | 11 | 60 | 96 | Y | - | Y | N | N | N | Y | 57.0 (114) |
| Brother (III:4) | G/A | 45 | 14 | - | - | N | 68 | N | N | N | Y | Y | 52.5 (36) | ||
| Brother (III:5) | G/G | 62 | 12 | 36 | 48 | Y | - | N | N | N | N | N | 50.5 (21) | ||
| MBD1 P401A | 7978 | Female proband | C/G | 92 | 13 | 22 | 72 | Y | 63 | Y | N | Y | N | Y | - |
| MBD1R428H | 7992 | Male proband | G/A | 40 | 13 | - | - | N | - | N | N | N | N | Y | 52.5 (46) |
| MBD1K558N | 7947 | Male proband | A/T | 140 | 15 | 36 | 40 | Y | 54 | N | N | N | N | N | 53.2 (148) |
| MBD1 c.1410G>A | 7660 | Male proband | G/A | 123 | - | 36 | 54 | Y | 35 | - | - | - | - | - | - |
| Identical twin brother | G/A | 178 | 11 | 48 | 57 | Y | 31 | N | N | Y | N | Y | - | ||
| MBD1 c.l779-27C>T | 7948 | Male proband | C/T | 192 | 18 | 36 | 60 | Y | - | N | N | N | N | Y | - |
| MBD3 R23M | 7974 | Male proband | G/T | 98 | 12 | 78 | - | N | 43 | N | Y | N | N | N | 54.0 (91) |
| Half brother | G/T | 41 | 11 | 36 | 36 | Y | 93 | N | N | N | N | N | 48.26 (18) | ||
| MBD3 E281del | 7730 | Male proband | +/del | 209 | - | - | - | - | - | - | - | - | - | - | - |
| Brother (III:3) | +/del | 176 | - | 84 | - | N | - | Y | N | N | N | N | - | ||
| MBD4 E314fsX316 | 7605 | Female proband | +/del | 142 | 13 | 48 | 60 | N | - | Y | N | N | N | Y | 53.0 (132) |
| MBD4 E346K | 7710 | Male proband | G/A | 56 | 17 | 39 | 42 | N | 72 | N | N | N | N | Y | 55.9 (140) |
| Brother (II:2) | - | 60 | - | - | - | - | - | - | - | - | - | - | 56.1 (109) | ||
| MBD4 E346K | 7785 | Male proband | G/A | 81 | 8 | 36 | 36 | Y | - | N | N | N | N | N | - |
| Brother (II:2) | G/G | 53 | 10 | 36 | - | N | - | Y | N | N | N | Y | - | ||
| Brother (II:3) | G/G | 49 | 10 | - | - | N | - | Y | Y | N | N | Y | - | ||
| MBD4 E346K | 7970 | Male proband | G/A | 32 | 13 | - | - | N | 73 | Y | N | N | N | N | 49.5 (31) |
| MBD4 N467S | 7941 | Female proband | A/G | 109 | 18 | 57 | 57 | N | 34 | Y | N | N | N | N | 56.0 (184) |
| MBD4 N467S | 7921 | Male proband | A/G | 63 | - | - | - | Y | 48 | - | - | - | - | - | - |
Endash the trait was not evaluated
Age measured in months
The second novel missense mutation, c.1283G>A (R428H), was identified in a single affected individual in the African-American family no. 7992 and was not transmitted maternally (Fig. 2b). This male proband was nonverbal at the time of examination (40 months, Table 2). The third novel nonsynonymous change occurred at c.1674A>T (K558N) and was passed maternally in family no. 7947 to a single affected son as well as an unaffected son and daughter (Fig. 2c, Table 2). Neither of these novel SNPs were identified in control individuals. The final nonsynonymous variant, rs125555, causes a c.1201C>G alteration and results in a P401A change. This variant was found in a singleton Caucasian family (no. 7978) and was carried by two daughters—one diagnosed with autism and the other with anxiety and speech delay (Fig. 2d). The autistic proband presented with regression in language and social interest prior to 5 years of age as well as seizures and an unusual gait (Table 2). The allele is also present in both the mother, who presented with anxiety and depression, and the father, who presented with anxiety/panic and bipolar disorders.
A possible case of germline mosaicism was identified in MBD1. A novel synonymous change at c.1410G>A was found within exon 12 in the Caucasian family no. 7660 in affected identical twin brothers and one unaffected sister which was not identified in either of the parents (Fig. 2e, Table 2). Previous genetic testing with this family did not demonstrate Mendelian inconsistencies, ruling out nonpaternity as the cause for unusual results and suggesting instead germline mosaicism at this locus. While this variation does not produce a change in amino acid sequence, the alteration is predicted to affect binding sites of splicing factors (Supplementary Table 2). A second novel noncoding variant of interest was identified within intron 15 in the African-American family no. 7948 (Fig. 2f). The c.1779-27C>T change (chr18:46,051,935) is concordant with disease in three affected brothers, two of whom are identical twins. While the mother does not carry the change, the father was unavailable for testing. Neither variant was present in Caucasian controls nor was the c.1779-27C>T alteration present in African-American controls.
MBD2
We identified two novel and four previously reported SNPs within MBD2. Five of the six changes occurred in untranslated regions of the gene, and the remaining alteration did not result in a change of amino acid sequence or generate a readily identified cryptic splice site.
MBD3
Variations discovered in the MBD3 gene included five known SNPs, ten novel SNPs, and four insertions/deletions. Only three variants occurred in the coding region, two of which altered the amino acid sequence of MBD3. Of particular interest is a c.68G>T (R23M) change identified in two affected half brothers in an African-American family (no. 7974, Fig. 2g). This variation falls within the methyl-CpG-binding domain and changes the charged arginine to a nonpolar methionine. The alteration was inherited from the maternal grandmother, suggesting a possible sex-specific effect. Both brothers were late to acquire speech and only one developed functional language (Table 2). The mother was diagnosed with anxiety/panic disorder. This variation was not identified in either the African-American or Caucasian controls.
MBD4
Alterations in our autistic population were discovered within the MBD4 gene at eight previously identified SNPs and two novel variants—a deletion and a single nucleotide change. Six of these variants result in a coding change. Of special interest is the deletion, c.942_945delAAGA (E314fsX316), transmitted maternally in a single female proband in the African-American family no. 7605 (Fig. 2i). The affected daughter demonstrated regression in play, social skills, and communication to the extent of becoming nonverbal (Table 2). This alteration is predicted to cause a frameshift within exon 3 at amino acid 314, encoding two aberrant amino acids and then a premature stop codon. While retaining the methyl-CpG-binding domain, this truncation is expected to remove almost half of the protein’s 580 amino acids including the DNA glycosylase domain. No Caucasian or African-American controls were identified with the alteration.
Five out of the nine single nucleotide variants result in missense changes within MBD4. The novel c.1400A>G (N467S) variant is interesting due to its location within the functional DNA glycosylase domain [24]. This alteration occurred in two Caucasian singleton families, one being inherited from mother to daughter (family no. 7941, Fig. 2j) and the other passing from father to son (family no. 7921, Fig. 2k). The novel variation changes an asparagine to a serine and was not found in 244 Caucasian control individuals. The remaining four missense changes occur at c.817G>A (A273T), c.1024T>C (S342P), c.1036G>A (E346K), and c.1073T>C (I358T). Each change alters either the polarity or charge of the original residue. The E346K alteration (rs140693) was found in three families and was absent in the DHPLC control DNA (Fig. 2l–n). While this variant is not concordant with disease in the families, it is interesting that in family no. 7710, the unaffected sister who also inherited the E346K change was found to have a delayed speech acquisition. The S342P and I358T variants were each identified in controls, suggesting that these alterations are likely not related to autism.
Discussion
While it is unlikely that every alteration identified in this study is causative of autism, several variants appear especially interesting in their relationship to this disease. Specifically, there are two alterations which may alter the amino acid sequence sufficiently to interfere with protein function. The first is the R23M alteration in MBD3. This alteration falls within the methyl-CpG-binding domain of the protein and replaces the basic arginine with a nonpolar methionine. While this amino acid is not completely conserved in the other members of the MBD family, another basic amino acid, lysine, is found in the same position for the MeCP2, MBD1 and MBD2 proteins. An alteration of the charge at this position could interfere with the function of the MBD domain. It is also interesting that the variation is inherited from the maternal grandmother through a mother presenting with an anxiety disorder to two affected half brothers. This leaves open the possibility that there is a sex-specific effect for this variation.
The second variation of interest is E314fsX316 within MBD4. This maternally inherited allele is predicted to cause a frameshift at amino acid 314, encode two aberrant amino acids, and then a stop codon, resulting in a prematurely truncated protein. The mutant protein would contain the MBD domain, but the DNA glycosylase domain would be absent. While it is likely that much of the abnormal RNA would be removed by nonsense-mediated decay [56], it is conceivable that any truncated protein generated with an intact MBD domain may be able to interfere in the normal function of all of the MBD proteins. Indeed, Bader and colleagues recently demonstrated in vitro that a MBD4 protein truncated at amino acid 313 could inhibit the glycosylase activity of the wild-type MBD4 protein as well as uracil DNA glycosylase [57]. An alternative scenario is that an irregular phenotype could simply result from haploinsufficiency of the wild-type protein. This change warrants further investigation.
There are additional examples of amino acid changes detected in the MBD proteins that could result in functional changes. There are three nonsynonymous changes, MBD1 R147K, MBD1 K558N, and MBD4 E346K, which either remove or create lysine residues. Lysine is an amino acid subject to dramatic posttranslational modifications including methylation, acetylation, sumoylation, ubiquitination, and neddylation [58]. The MBD proteins have demonstrated that they are susceptible to such modifications [59–62]. Altering this key amino acid could dramatically influence the function of a protein, affect protein regulation, or interfere with protein–protein interactions. Another example of a variation that may affect posttranslational modification is S342P in MBD4. The disruption of a serine could remove a potential site of phosphorylation or glycosylation. Likewise for the creation of a serine site, as occurs in the MBD4 N467S variation within the glycosylase domain. Other variants which alter the polarity and charge of the residues, as seen in missense mutations in MBD1 and MBD4, could interfere with a binding site on the protein or cause misfolding.
It is also possible that some of the detected MBD variations could result in RNA processing errors by destroying splice sites, creating cryptic splice sites, or even changing the ratios of alternative isoforms by eliminating or generating binding sites for splicing factors (Supplementary Table 3). One example of this is the MBD1 intronic variant c.1585-12T>C, just 12 bp upstream of exon 14. The alteration causes a change in a highly conserved nucleotide, potentially breaking a consensus site for the SRp55 splicing factor and creating new binding sites.
Surprisingly, there appears to be a higher occurrence of variations with potential effects on amino acid function within the African-American autistic patients as compared to Caucasian patients. African-Americans comprise only 31 of the 226 (13.7%) families participating in this study, yet these families represent a disproportionate number of those with nonsynonymous alterations (24%). This could be due, in part, to the higher genetic diversity found in African populations [63, 64]. Furthermore, missense changes that were most abundant varied between the ethnicities, with the MBD4 A273T (rs10342) change being the most common for Caucasian families and the MBD4 S342P (rs2307289) alteration being predominantly found in African-American families. These results are consistent with the HapMap data for these known SNPs in the AA and CA populations. In addition, four of the five deletions identified in this study occurred only in African-American families, three of which were not identified in any control individuals. The remaining MBD3 GCG deletion was identified in both AA and CA affected and controls while the MBD3 GAG deletion was found exclusively in AA affected and controls, suggesting that both deletions are most likely not associated with autism. Nonetheless, these racial differences in variant frequencies add to previous findings in which African-Americans with autism showed subtle differences in phenotype and association to SNPs in GABAergic genes [65, 66].
In the families with nonsynonymous mutations, psychiatric disorders were reported (on family history interview) in first degree relatives who carry a variation. This is consistent with previous reports of an increased prevalence of neuropsychiatric disorders including schizophrenia, depression, and anxiety in the parents of autistic children [67–70]. Inheritance of MBD variants from parents diagnosed with psychiatric disorders was found in families numbers 7978, 7974, 7941, and 7785 (Fig. 2d, g, j, m). Moreover, two families have additional offspring who carry the same variant found in their affected sibling and has psychiatric problems without a diagnosis of autism. The sister in family no. 7978 was diagnosed with anxiety and speech delay, and a brother in family no. 7710 also presented with delayed speech (Fig. 2d, l). This lends support to the idea that the variants identified here could contribute to autism and other psychological defects. Indeed, some variants were identified in both affected and unaffected individuals within a family and multiplex families were not always concordant for the variant being identified. In some instances, as outlined above, the variants appear in people negative for autism, but demonstrating clinical characteristic found in autism. Such is the case for family no. 7974 where a mother with anxiety/panic disorder passes along the MBD3 R23M variant to two autistic half brothers. The variation in phenotypic presentation may also be related to incomplete penetrance of genetic factors. This can be demonstrated by the fact that there is an increase in the concordance of autism in monozygotic twins as compared to dizygotic twins, but there is not 100% concordance in MZ twins [4–6]. Therefore, although genetics plays a strong role in autism etiology, it is not the sole component influencing presentation of disease. Lack of perfect concordance should not be used as a measure to rule out causal mutations within complex diseases. Other factors such as environmental or epigenetic events may also contribute to the intricacy of autism.
It is intriguing that we observed clinical similarities in select patients to the Rett syndrome phenotype. While, by definition, Rett syndrome and autism overlap clinically, there are features common to Rett syndrome which are less frequent in the general autistic population. For example, regression is found in most classical manifestations of Rett, but only in a minority of autistic individuals [1, 17]. Yet, we identified eight cases with a regression in language, in some cases accompanied by losses in social responsiveness or play skills. Furthermore, select individuals were reported to have irregular breathing, unusual gait, and seizures, all traits are commonly found in Rett patients. Also of note were two individuals identified with unusual midline movements, reminiscent of the classic Rett feature where girls clasp their hands together at the center of their body [15, 16]. Perhaps, these patients are presenting clinically in a manner more similar to Rett individuals due to the alterations identified in this study within the MBD genes. It is also interesting that the MBD1 R147K alteration identified in family no. 7801 was found in the two more severely affected of three autistic brothers as well as an unaffected sister. This could suggest that this alteration results in a sex-specific effect that may contribute to the autism phenotype without being the sole causative factor.
While there is mounting evidence from human and mouse studies implicating MBD genes in autism, few patients have been identified with alterations in MBD1, MBD2, MBD3, and MBD4 [28, 29, 39, 45, 48, 71]. This seeming inconsistency may be clarified by evaluating the murine phenotypes. While the Mbd1−/− mouse exhibits deficiencies in a wide range of social behaviors, it is able to live an almost normal life span [48]. In contrast, Mecp2−/− mice have a severe phenotype that is usually lethal by 10 weeks of age [39, 71]. Observing the relatively mild phenotype of the Mbd1 null mice in relation to the Mecp2 null mice suggests that there may be an ascertainment bias in identifying patients with recognized MECP2 mutations. MBD1 mutations may lead to such a mild phenotype that they rarely present as clinical abnormalities [48].
In summary, we identified genetic alterations at 46 unique loci in the MBD genes. Twenty-one were previously recognized SNPs, while 25 represent novel variations (five insertion/deletions and 20 SNPs). Although the majority of alterations were synonymous or noncoding variants, we identified ten nonsynonymous changes in MBD1, MBD3, and MBD4, the deletion of a single amino acid in MBD3, and a frameshift mutation in MBD4 that is predicted to truncate almost half of the protein. These results suggest that rare variants in these genes may contribute to autism susceptibility. Further studies utilizing larger cohorts will be required to determine the potential contribution of these genes to the autism clinical spectrum, but evidence to date indicates that the entire MBD family may play a role in autism etiology.
Supplementary Material
Acknowledgements
We are grateful to the patients with autism and their families for their participation, without whom, this work would not be possible. A subset of the participants was ascertained while several of the authors (RR, IK, MYR, ERM, MLC, MAPV, and JRG) were at Duke University. We also acknowledge fellow lab members for their critical review of this manuscript. This research was supported by grants from the National Institutes of Health (NS26630, NS36768, and MH080647), Autism Speaks, and by a gift from the Hussman Foundation. RR is supported by the Ramon y Cajal program.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10048-009-0228-7) contains supplementary material, which is available to authorized users.
Contributor Information
Holly N. Cukier, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
Raquel Rabionet, Genes and Disease Program, Centre de Regulació Genòmica and CIBER en Epidemiología y Salud Pública, Barcelona, Spain.
Ioanna Konidari, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA.
Melissa Y. Rayner-Evans, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
Mary L. Baltos, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
Harry H. Wright, University of South Carolina School of Medicine, Columbia, SC, USA
Ruth K. Abramson, University of South Carolina School of Medicine, Columbia, SC, USA
Eden R. Martin, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
Michael L. Cuccaro, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
Margaret A. Pericak-Vance, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA
John R. Gilbert, Email: jgilbert@med.miami.edu, John P. Hussman Institute for Human Genomics, University of Miami, 1501 NW 10th Avenue, Miami, FL 33136, USA.
References
- 1.Johnson CP, Myers SM. American academy of pediatrics council on children with disabilities: identification and evaluation of children with autism spectrum disorders. Pediatrics. 2007;120(5):1183–1215. doi: 10.1542/peds.2007-2361. [DOI] [PubMed] [Google Scholar]
- 2.Fombonne E. Epidemiological trends in rates of autism. Mol Psychiatry. 2002;7 Suppl 2:S4–S6. doi: 10.1038/sj.mp.4001162. [DOI] [PubMed] [Google Scholar]
- 3.Autism and Developmental Disabilities Monitoring Network Surveillance Year 2002 Principal Investigators, Centers for Disease Control and Prevention: prevalence of autism spectrum disorders—autism and developmental disabilities monitoring network, 14 sites, United States, 2002. MMWR Surveill Summ. 2007;56(1):12–28. [PubMed] [Google Scholar]
- 4.Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142:74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- 5.Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg DA, Hodge SE, Sowinski J, Nicoll D. Excess of twins among affected sibling pairs with autism: implications for the etiology of autism. Am J Hum Genet. 2001;69:1062–1067. doi: 10.1086/324191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma DQ, Salyakina D, Jaworski JM, Konidari I, Whitehead PL, Andersen AN, Hoffman JD, Slifer SH, Hedges DJ, Cukier HN, Griswold AJ, McCauley JL, Beecham GW, Wright HH, Abramson RK, Martin ER, Hussman JP, Gilbert JR, Cuccaro ML, Haines JL, Pericak-Vance MA. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73(3):263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang K, Haitao Z, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PMA, Kim CE, Chiavacci R, Lajonchere C, Munson J, Estes A, Korvatska O, Piven J, Sonnenblick LI, Alvarez Retuerto AI, Herman EI, Dong H, Hutman T, Sigman M, Ozonoff S, Klin A, Owley T, Sweeney JA, Brune CW, Cantor RM, Bernier R, Gilbert JR, Cuccaro ML, Wassink TH, McMahon WM, Coon H, Miller J, Nurnberger JI, State MW, Haines JL, Sutcliffe JS, Cook E, Minshew N, Buxbaum JD, Dawson G, Grant SFA, Geschwind DH, Pericak-Vance MA, Schellenberg GD, Hakonarson H. Common genetic variants on 5p14.1 associate with autism spectrum disorder. Nature. 2009;459(7246):528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 15.Hagberg B, Aicardi J, Dias K, Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann Neurol. 1983;14(4):471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 16.Hagberg B. Rett’s syndrome: prevalence and impact on progressive severe mental retardation in girls. Acta Paediatr Scand. 1985;74(3):405–408. doi: 10.1111/j.1651-2227.1985.tb10993.x. [DOI] [PubMed] [Google Scholar]
- 17.Stefanatos GA. Regression in autistic spectrum disorders. Neuropsychol Rev. 2008;18(4):305–319. doi: 10.1007/s11065-008-9073-y. [DOI] [PubMed] [Google Scholar]
- 18.Shahbazian MD, Zoghbi HY. Rett syndrome and MeCP2: linking epigenetics and neuronal function. Am J Hum Genet. 2002;71(6):1259–1272. doi: 10.1086/345360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moretti P, Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16(3):276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Buschdorf JP, Stratling WH. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J Mol Med. 2004;82(2):135–143. doi: 10.1007/s00109-003-0497-9. [DOI] [PubMed] [Google Scholar]
- 21.Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roloff TC, Ropers HH, Nuber UA. Comparative study of methyl-CpG-binding domain proteins. BMC Genomics. 2003;4(1):1. doi: 10.1186/1471-2164-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen HF, Bird A. MeCP2 and other methyl-CpG binding proteins. Ment Retard Dev Disabil Res Rev. 2002;8(2):87–93. doi: 10.1002/mrdd.10021. [DOI] [PubMed] [Google Scholar]
- 24.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401(6750):301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 25.Ballestar E, Ropero S, Alaminos M, Armstrong J, Setien F, Agrelo R, Fraga MF, Herranz M, Avila S, Pineda M, Monros E, Esteller M. The impact of MECP2 mutations in the expression patterns of Rett syndrome patients. Hum Genet. 2005;116(1–2):91–104. doi: 10.1007/s00439-004-1200-0. [DOI] [PubMed] [Google Scholar]
- 26.Matarazzo MR, De Bonis ML, Strazzullo M, Cerase A, Ferraro M, Vastarelli P, Ballestar E, Esteller M, Kudo S, D’Esposito M. Multiple binding of methyl-CpG and polycomb proteins in long-term gene silencing events. J Cell Physiol. 2007;210(3):711–719. doi: 10.1002/jcp.20879. [DOI] [PubMed] [Google Scholar]
- 27.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26(3):843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carney RM, Wolpert CM, Ravan SA, Shahbazian M, Ashley-Koch A, Cuccaro ML, Vance JM, Pericak-Vance MA. Identification of MeCP2 mutations in a series of females with autistic disorder. Pediatr Neurol. 2003;28:205–211. doi: 10.1016/s0887-8994(02)00624-0. [DOI] [PubMed] [Google Scholar]
- 29.Shibayama A, Cook EH, Jr, Feng J, Glanzmann C, Yan J, Craddock N, Jones IR, Goldman D, Heston LL, Sommer SS. MECP2 structural and 3′-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. Am J Med Genet B Neuropsychiatr Genet. 2004;128(1):50–53. doi: 10.1002/ajmg.b.30016. [DOI] [PubMed] [Google Scholar]
- 30.Coutinho AM, Oliveira G, Katz C, Feng J, Yan J, Yang C, Marques C, Ataide A, Miguel TS, Borges L, Almeida J, Correia C, Currais A, Bento C, Mota-Vieira L, Temudo T, Santos M, Maciel P, Sommer SS, Vicente AM. MECP2 coding sequence and 3′UTR variation in 172 unrelated autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144(4):475–483. doi: 10.1002/ajmg.b.30490. [DOI] [PubMed] [Google Scholar]
- 31.Wan M, Lee SS, Zhang X, Houwink-Manville I, Song HR, Amir RE, Budden S, Naidu S, Pereira JL, Lo IF, Zoghbi HY, Schanen NC, Francke U. Rett syndrome and beyond: recurrent spontaneous and familiar MECP2 mutations at CpG hotspots. Am J Hum Genet. 1999;65:1520–1529. doi: 10.1086/302690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson P, Black G, Ramsden S, Barrow M, Super M, Kerr B, Clayton-Smith J. Angelman syndrome phenotype associated with mutations in MECP2, a gene encoding a methyl CpG binding protein. J Med Genet. 2001;38:224–228. doi: 10.1136/jmg.38.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klauck SM, Lindsay S, Beyer KS, Splitt M, Burn J, Poustka A. A mutation hot spot for nonspecific X-linked mental retardation in the MECP2 gene causes the PPM-X syndrome. Am J Hum Genet. 2002;70(4):1034–1037. doi: 10.1086/339553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milani D, Pantaleoni C, D’Arrigo S, Selicorni A, Riva D. Another patient with MECP2 mutation without classic Rett syndrome phenotype. Pediatr Neurol. 2005;32(5):355–357. doi: 10.1016/j.pediatrneurol.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 35.Kankirawatana P, Leonard H, Ellaway C, Scurlock J, Mansour A, Makris CM, Dure LS, 4th, Friez M, Lane J, Kiraly-Borri C, Fabian V, Davis M, Jackson J, Christodoulou J, Kaufmann WE, Ravine D, Percy AK. Early progressive encephalopathy in boys and MECP2 mutations. Neurology. 2006;67(1):164–166. doi: 10.1212/01.wnl.0000223318.28938.45. [DOI] [PubMed] [Google Scholar]
- 36.Harvey CG, Menon SD, Stachowiak B, Noor A, Proctor A, Mensah AK, Mnatzakanian GN, Alfred SE, Guo R, Scherer SW, Kennedy JL, Roberts W, Srivastava AK, Minassian BA, Vincent JB. Sequence variants within exon 1 of MECP2 occur in females with mental retardation. Am J Med Genet B Neuropsychiatr Genet. 2007;144(3):355–360. doi: 10.1002/ajmg.b.30425. [DOI] [PubMed] [Google Scholar]
- 37.Lugtenberg D, Kleefstra T, Oudakker AR, Nillesen WM, Yntema HG, Tzschach A, Raynaud M, Rating D, Journel H, Chelly J, Goizet C, Lacombe D, Pedespan JM, Echenne B, Tariverdian G, O’Rourke D, King MD, Green A, van Kogelenberg M, Van Esch H, Gecz J, Hamel BC, van Bokhoven H, de Brouwer AP. Structural variation in Xq28: MECP2 duplications in 1% of patients with unexplained XLMR and in 2% of male patients with severe encephalopathy. Eur J Hum Genet. 2008 doi: 10.1038/ejhg.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loat C, Curran S, Lewis C, Abrahams B, Duvall J, Geschwind D, Bolton P, Craig I. Methyl-CpG-binding protein 2 polymorphisms and vulnerability to autism. Genes Brain Behav. 2008;7(7):754–760. doi: 10.1111/j.1601-183X.2008.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genetic. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- 40.Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35(2):243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci USA. 2003;100(11):6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins AL, Levenson JM, Vilaythong AP, Richman R, Armstrong DL, Noebels JL, David SJ, Zoghbi HY. Mild over-expression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13(21):2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- 43.Cukier HN, Perez AM, Collins AL, Zhou Z, Zoghbi HY, Botas J. Genetic modifiers of MeCP2 function in Drosophila. PLoS Genet. 2008;4(9):e1000179. doi: 10.1371/journal.pgen.1000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fyffe SL, Neul JL, Samaco RC, Chao HT, Ben-Shachar S, Moretti P, McGill BE, Goulding EH, Sullivan E, Tecott LH, Zoghbi HY. Deletion of Mecp2 in Sim1-expressing neurons reveals a critical role for MeCP2 in feeding behavior, aggression, and the response to stress. Neuron. 2008;59(6):947–958. doi: 10.1016/j.neuron.2008.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moretti P, Bouwknecht JA, Teague R, Paylor R, Zoghbi HY. Abnormalities of social interactions and home-cage behavior in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14(2):205–220. doi: 10.1093/hmg/ddi016. [DOI] [PubMed] [Google Scholar]
- 46.McGill BE, Bundle SF, Yaylaoglu MB, Carson JP, Thaller C, Zoghbi HY. Enhanced anxiety and stress-induced cortico-sterone release are associated with increased Crh expression in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2006;103(48):18267–18272. doi: 10.1073/pnas.0608702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26(1):319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allan AM, Liang X, Luo Y, Pak C, Li X, Szulwach KE, Chen D, Jin P, Zhao X. The loss of methyl-CpG binding protein 1 leads to autism-like behavioral deficits. Hum Mol Genet. 2008;17(13):2047–2057. doi: 10.1093/hmg/ddn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Yamagata T, Mori M, Yasuhara A, Momoi MY. Mutation analysis of methyl-CpG binding protein family genes in autistic patients. Brain Dev. 2005;27(5):321–325. doi: 10.1016/j.braindev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 50.American Psychiatric Association. Diagnostic and statistical manual of mental disorders (DSM-IV) Washington: American Psychiatric Press, Inc; 1994. [Google Scholar]
- 51.Lord C, Rutter M, DiLavore P, Risi S. Autism Diagnostic Observation Schedule-WPS. 1999 doi: 10.1007/BF02179373. WPS edition. [DOI] [PubMed] [Google Scholar]
- 52.Rutter M, LeCouteur A, Lord C. Autism Diagnostic Interview, Revised (ADI-R) 2003 [Google Scholar]
- 53.Sparrow SS, Balla D, Cicchetti D. Vineland adaptive behavior scales, interview edition. AGS Publishing, Circle Pines; 1984. [Google Scholar]
- 54.Vance JM. The collection of biological samples for DNA analysis. In: Haines JL, Pericak-Vance MA, editors. Approaches to gene mapping in complex human diseases. New York: Wiley-Liss; 1998. pp. 201–211. [Google Scholar]
- 55.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5(2):89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 57.Bader SA, Walker M, Harrison DJ. A human cancer-associated truncation of MBD4 causes dominant negative impairment of DNA repair in colon cancer cells. Br J Cancer. 2007;96(4):660–666. doi: 10.1038/sj.bjc.6603592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31(4):449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302(5646):885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 60.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302(5646):890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 61.Lyst MJ, Nan X, Stancheva I. Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J. 2006;25(22):5317–5328. doi: 10.1038/sj.emboj.7601404. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Miyake K, Nagai K. Phosphorylation of methyl-CpG binding protein 2 (MeCP2) regulates the intracellular localization during neuronal cell differentiation. Neurochem Int. 2007;50(1):264–270. doi: 10.1016/j.neuint.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 63.Salisbury BA, Pungliya M, Choi JY, Jiang R, Sun XJ, Stephens JC. SNP and haplotype variation in the human genome. Mutat Res. 2003;526:53–61. doi: 10.1016/s0027-5107(03)00014-9. [DOI] [PubMed] [Google Scholar]
- 64.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Ann Rev Genomics Hum Genet. 2008;9:403–433. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Collins AL, Ma D, Whitehead PL, Martin ER, Wright HH, Abramson RK, Hussman JP, Haines JL, Cuccaro ML, Gilbert JR, Pericak-Vance MA. Investigation of autism and GABA receptor subunit genes in multiple ethnic groups. Neurogenetics. 2006;7:167–174. doi: 10.1007/s10048-006-0045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cuccaro ML, Brinkley J, Abramson RK, Hall A, Wright HH, Hussman JP, Gilbert JR, Pericak-Vance MA. Autism in African American families: clinical-phenotypic findings. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(8):1022–1026. doi: 10.1002/ajmg.b.30535. [DOI] [PubMed] [Google Scholar]
- 67.Larsson HJ, Eaton WW, Madsen KM, Vestergaard M, Olesen AV, Agerbo E, Schendel D, Thorsen P, Mortensen PB. Risk factors for autism: perinatal factors, parental psychiatric history, and socioeconomic status. Am J Epidemiol. 2005;161(10):916–925. doi: 10.1093/aje/kwi123. discussion 926-8. [DOI] [PubMed] [Google Scholar]
- 68.Daniels JL, Forssen U, Hultman CM, Cnattingius S, Savitz DA, Feychting M, Sparen P. Parental psychiatric disorders associated with autism spectrum disorders in the offspring. Pediatrics. 2008;121(5):e1357–e1362. doi: 10.1542/peds.2007-2296. [DOI] [PubMed] [Google Scholar]
- 69.Mazefsky CA, Williams DL, Minshew NJ. Variability in adaptive behavior in autism: evidence for the importance of family history. J Abnorm Child Psychol. 2008;36(4):591–599. doi: 10.1007/s10802-007-9202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace AE, Anderson GM, Dubrow R. Obstetric and parental psychiatric variables as potential predictors of autism severity. J Autism Dev Disord. 2008;38(8):1542–1554. doi: 10.1007/s10803-007-0536-4. [DOI] [PubMed] [Google Scholar]
- 71.Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27(3):327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.