Abstract
Intrinsically disordered proteins (IDPs) lack well-defined structure but are widely represented in eukaryotic proteomes. Although the functions of most IDPs are not understood, some have been shown to have molecular recognition and/or regulatory roles where their disordered nature might be advantageous. Anhydrin is an uncharacterized IDP induced by dehydration in an anhydrobiotic nematode, Aphelenchus avenae. We show here that anhydrin is a moonlighting protein with two novel, independent functions relating to desiccation tolerance. First, it has a chaperone-like activity that can reduce desiccation-induced enzyme aggregation and inactivation in vitro. When expressed in a human cell line, anhydrin localizes to the nucleus and reduces the propensity of a polyalanine expansion protein associated with oculopharyngeal muscular dystrophy to form aggregates. This in vivo activity is distinguished by a loose association of anhydrin with its client protein, consistent with a role as a molecular shield. In addition, anhydrin exhibits a second function as an endonuclease whose substrates include supercoiled, linear, and chromatin linker DNA. This nuclease activity could be involved in either repair of desiccation-induced DNA damage incurred during anhydrobiosis or in apoptotic or necrotic processes, for example, but it is particularly unexpected for anhydrin because IDP functions defined to date anticorrelate with enzyme activity. Enzymes usually require precise three-dimensional positioning of residues at the active site, but our results suggest this need not be the case. Anhydrin therefore extends the range of IDP functional categories to include catalysis and highlights the potential for the discovery of new functions in disordered proteomes.
Keywords: anhydrobiosis, endonuclease, intrinsically unstructured protein, molecular shield, natively unfolded protein
Desiccation tolerance, or anhydrobiosis, is a phenomenon long recognized across all biological kingdoms, but the molecular mechanisms underlying it are still poorly understood. Recent studies have highlighted highly hydrophilic proteins as a major component of the response to extreme water stress, the best-characterized examples being the late embryogenesis abundant (LEA) proteins described in plants and some invertebrates (1). Almost all LEA proteins are also intrinsically disordered proteins (IDPs), i.e., they are entirely or largely unstructured in their native state, probably as a function of their extreme hydrophilicity (2).
Classically, protein function is dependent on a defined, if flexible, three-dimensional polypeptide structure. For example, an enzyme requires spatial complementarity between its active site and the transitional states of its substrate(s); mutations that critically alter active site structure abolish or impair catalytic activity. In apparent contradiction of this structure-function paradigm, IDPs are a common feature of eukaryotic proteomes: Disorder prediction programs suggest that more than 20% of proteins from higher eukaryotes are mostly disordered; bioinformatic analyses of other eukaryotes return similar predictions, with typically approximately 8% of the proteome being fully disordered (2–6).
Where known, IDP functions usually involve binding to a partner molecule (7–9); e.g., p27Kip1 is an inhibitor of cell cycle progression that interacts with cyclin–Cdk2 complexes; formation of the ternary complex induces folding in p27Kip1, although part of the molecule remains disordered (10, 11). The induction of folding by binding partner molecules is a common theme (8, 9, 12) and allows the structure-function paradigm to be extended to many IDPs, although a degree of disorder is often maintained in the bound state, a concept known as fuzziness (12). Nonetheless, some disordered domains exert their function in the unstructured state (7, 13) as entropic chains: e.g., the MAP2 projection domain that governs spacing of microtubules in the cytoplasm (14). IDPs with a protein stabilization function might behave similarly: LEA proteins reduce the formation of aggregates in a wide variety of other polypeptides subjected to environmental stress (desiccation, freezing, or heat) (15–17) or in aggregation-prone proteins containing expanded stretches of polyglutamine or polyalanine (15). However, one functional category not reported among fully disordered proteins is catalysis, possibly because precise three-dimensional positioning of residues at the active site has so far been seen as a prerequisite (2, 7).
Further investigations of desiccation tolerance are uncovering novel hydrophilic proteins that are not members of the LEA protein families; these are currently uncharacterized and their functions unknown. One of these is anhydrin (18), whose gene is induced in response to dehydration in the anhydrobiotic nematode, Aphelenchus avenae. We show here that anhydrin is entirely intrinsically disordered yet possesses two apparently independent functions: as a chaperone-like molecular shield, able to reduce aggregation of client proteins in vitro and in the nucleus, and as a novel endonuclease; the latter is particularly surprising because enzyme activity is previously undescribed in an IDP. These results illustrate the potential for discovery of new functions, and indeed multiple functions of individual proteins, within the unstructured proteomes of eukaryotes.
Results
Anhydrin Is an Intrinsically Disordered Protein.
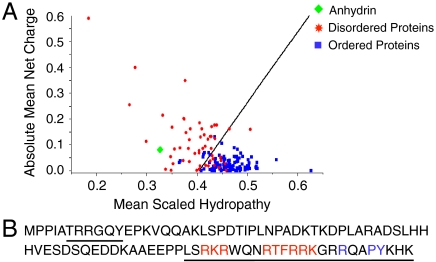
Bioinformatics suggest anhydrin is disordered: a Uversky plot of mean net charge against mean scaled hydropathy maps anhydrin with IDPs (Fig. 1A). Three independent predictors of protein disorder, VL-XT, IUPred, and FoldIndex, gave anhydrin as 96%, 100%, and 100% unfolded, respectively. Consistent with a lack of defined structure is the extreme hydrophilicity of anhydrin as indicated by a grand average hydropathy score of -1.56.
Fig. 1.
Analysis of anhydrin amino acid sequence. (A) A Uversky plot (34), predicts anhydrin to be disordered (green diamond) by comparison to a set of disordered proteins (red stars) and a set of ordered proteins (blue squares). (B) Anhydrin contains a bPY-NLS motif (blue) with adjacent basic residues (red) and predicted DNA binding regions underlined.
A recombinant form of anhydrin was produced with a molecular weight of 13321 Da by mass spectrometry, while SDS/PAGE and size exclusion chromatography gave an apparent size of 21 kDa and 26 kDa, respectively. Such anomalous mobility is often observed with IDPs: In polyacrylamide gels, mobility is retarded because of abnormally low binding of SDS to hydrophilic proteins; on size exclusion columns, IDPs are not globular and instead populate extended conformations resulting in a larger than expected Stokes radius (Fig. S1).
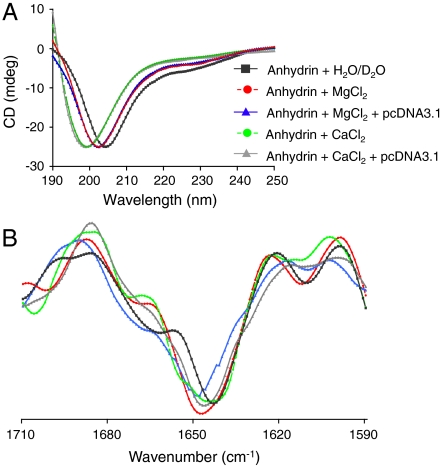
Far UV circular dichroism (CD) spectra of anhydrin in water gave a single minimum at approximately 205 nm and a negative shoulder at 220 to 230 nm confirming it as a disordered protein (Fig. 2A) (2). This was corroborated by Fourier transform infrared (FTIR) spectroscopy, which showed a broad featureless amide I band at approximately 1647 cm-1, typical of an unfolded protein (Fig. 2B and Fig. S1E). Furthermore, nuclear magnetic resonance (NMR) spectra of anhydrin showed a poor dispersion of backbone amide protons in the region 7.8–8.4 ppm, indicative of a lack of persistent secondary structure (Fig. S1F Top). When anhydrin was subjected to buffer exchange in D2O, the majority of signals arising from backbone amide protons were exchanged with deuterons from the solvent within 20 min, consistent with a lack of strong hydrogen bonding in the polypeptide backbone, and hence the absence of persistent secondary structure elements (Fig. S1F Bottom). Collectively, these data categorize anhydrin as an IDP.
Fig. 2.
Anhydrin is an intrinsically disordered protein. (A) Far UV CD spectra and (B) second-derivative FTIR spectra corresponding to data in Fig. S1E of anhydrin in water (A) or D2O (B) (black); in the presence of 10 mM MgCl2 (red); 10 mM MgCl2 and pcDNA3.1 (blue); 10 mM CaCl2 (green) or 10 mM CaCl2 and pcDNA3.1 (gray) (25∶1 anhydrin:DNA molar ratio).
Anhydrin Reduces Desiccation-Induced Enzyme Aggregation and Inactivation.
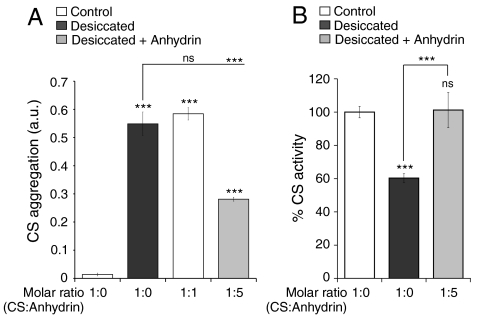
On vacuum drying, the enzyme citrate synthase (CS) aggregates markedly and suffers a concomitant decrease in activity (Fig. 3), but LEA proteins, which like anhydrin are highly hydrophilic and unstructured, are known to reduce these effects (19). We therefore hypothesized that anhydrin might possess a similar stabilizing activity in vitro. Indeed, when CS was dried in the presence of anhydrin, a significant decrease in aggregation was noted (Fig. 3A); a panel of other proteins, with the exception of the LEA protein, AavLEA1, did not reduce CS aggregation, suggesting this is not a nonspecific effect (Fig. S2). Anhydrin also relieves the detrimental effect of drying on enzyme activity (Fig. 3B).
Fig. 3.
Anhydrin reduces citrate synthase aggregation and preserves its activity. (A) Aggregation and (B) activity of 0.12 mg CS after two rounds of drying and rehydration in the absence (black bars) or presence of equimolar or five-molar excess anhydrin (gray bars); nondried controls are white bars. SD bars are shown; ns, nonsignificant and *** p < 0.001.
Anhydrin Is Located in the Nucleus and Reduces Nuclear Protein Aggregation.
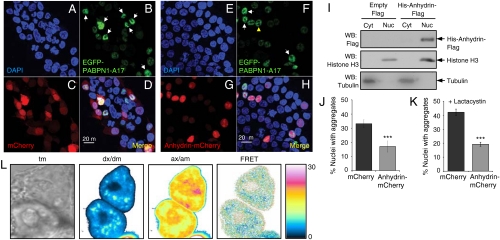
As anhydrin shows antiaggregation activity in vitro, we asked whether it has a similar function in vivo. Anhydrin’s amino acid sequence includes a bPY nuclear localization signal (bPY-NLS) (20) (Fig. 1B), and it also fulfils other criteria for nuclear import: It is basic (isoelectric point of 10.5) and is structurally disordered. Indeed, when tagged with the red fluorescent mCherry protein and expressed in the human kidney cell line, T-REx293, confocal microscopy showed anhydrin to be predominantly in the nucleus, colocalizing with DAPI staining, in contrast to cells expressing mCherry alone, which was present in both the nucleus and the cytoplasm (Fig. 4 A, C, E, and G). This result was confirmed by isolating nuclear and cytoplasmic fractions of T-REx293 cells expressing His6-anhydrin-Flag and identifying its location by Western blotting (Fig. 4I): Anhydrin was found in the nuclear fraction, along with histone H3, and was barely detectable in the cytoplasmic fraction, where tubulin localized.
Fig. 4.
Anhydrin reduces aggregation of EGFP-PABPN1-A17 in mammalian cell nuclei. (A–H) Confocal microscopy of T-REx293 cells expressing EGFP-PABPN1-A17 (B, D, F, H) with mCherry (C, D) or anhydrin-mCherry (G, H). Nuclei expressing EGFP-PABPN1-A17 are green (white arrowheads indicate aggregates and yellow arrowheads normal nuclear speckles), cells/nuclei expressing mCherry or anhydrin-mCherry are red and nuclei are stained blue with DAPI (A, D, E, H). (I) Western blot of cytoplasmic and nuclear fractions of cells transfected with either empty pCMVFLAG5a vector or His6-anhydrin-Flag construct and probed with antibodies against Flag (top), histone H3 (middle) or tubulin (bottom). (J and K) Percentage of nuclei with EGFP-PABPN1-A17 aggregates after either mCherry (black) or anhydrin-mCherry (gray) coexpression; in K, cells were treated with the proteosomal inhibitor lactacystin; results show SD and are highly significant by logistic regression analysis. ***, p < 0.001. (L) Example FRET analysis of EGFP-PABPN1-A17 (donor)/anhydrin-mCherry (acceptor) interactions in a live cell, showing DIC transmission image (tm), signal in donor channel upon excitation at donor wavelength (dx/dm), signal in acceptor channel upon excitation at acceptor wavelength (ax/am), and donor normalized and unmixed FRET transfer efficiency dFRET (labeled FRET).
Because anhydrin is a nuclear protein, we used EGFP-tagged nuclear polyadenine binding protein with 17 consecutive alanine residues (EGFP-PABPN1-A17) to test for antiaggregation activity in cells. This polyalanine expansion protein is associated with oculopharyngeal muscular dystrophy and is known to localize in the nucleus where it spontaneously aggregates (21); the EGFP-tagged aggregates are easily distinguishable from normal nuclear speckles (22) (Fig. 4B and F). Coexpression of anhydrin-mCherry with EGFP-PABPN1-A17 reduced the number of nuclei containing aggregates by approximately 50% compared to control cells coexpressing mCherry and EGFP-PABPN1-A17 (Fig. 4D, H, and J). Odds ratio analysis from multiple experiments showed that this difference was highly significant (p < 0.0001; Fig. S3A), and these results therefore support a protein antiaggregation role for anhydrin in vivo. To demonstrate that anhydrin prevents aggregate formation, rather than stimulates aggregate clearance through the proteasome, we repeated the above experiment in the presence of the proteasome inhibitor, lactacystin (21), and obtained similar results (Fig. 4K and Fig. S3B, C, and D).
Loose Interaction of Anhydrin and Client Protein in Vivo.
The ability of anhydrin to reduce aggregation of other proteins is reminiscent of that of some molecular chaperones, e.g., the small heat shock proteins, which are widely accepted to function through association with hydrophobic patches exposed in partially unfolded client proteins (23). Such interactions between molecular chaperones and their client proteins can be demonstrated by coimmunoprecipitation experiments (e.g., ref. 24). If anhydrin is a molecular chaperone, we might therefore expect it to form complexes with its substrates. Accordingly, a doubly tagged form of anhydrin carrying His6 at the N terminus and a C-terminal Flag peptide (Fig. S4A) and HA-tagged PABPN1-A17 (25) were coexpressed in T-REx293 cells. However, when His6-anhydrin-Flag was immunoprecipitated, no HA signal, which would be indicative of a tight interaction with HA-PABPN1-A17, was observed by Western blotting, although anhydrin was clearly detectable in the immunoprecipitated sample and HA-PABPN1-A17 was present in the cell lysate (Fig. S4B). The reverse experiment, where HA-PABPN1-A17 was immunoprecipitated, was also negative for interaction with His6-anhydrin-Flag (Fig. S4C). Because PABPN1-A17 self-associates (22), we validated the protocol by demonstrating coimmunoprecipitation of HA-PABPN1-A17 and EGFP-PABPN1-A17 in the same cell line (Fig. S4D).
Although lack of coimmunoprecipitation suggests any association is not strong, it is possible that anhydrin interacts more loosely with its targets. To test this, we performed quantitative Förster resonance energy transfer (FRET) experiments between EGFP-PABPN1-A17 and anhydrin-mCherry via sensitized acceptor (mCherry) fluorescence emission upon donor (EGFP) excitation. The method used (26) yields FRET efficiencies normalized by the prevailing acceptor concentration (aFRET), or the donor concentration (dFRET), the relative level of which depends on the stoichiometry of interaction (see Materials and Methods). The positive control construct (EGFP tethered to mCherry by a 7-amino acid linker) was expressed in T-REx293 cells and gave FRET levels of 15.2 ± 0.3% (here aFRET = dFRET, because the stoichiometry of interaction is 1). The negative control (coexpressed EGFP and mCherry proteins alone) did not yield significant FRET levels (0.2 ± 0.1%). For the EGFP-PABPN1-A17 and anhydrin-mCherry pair, dFRET was measured at 4.1 ± 0.9%, however, indicating an interaction between the two proteins in vivo (Fig. 4L).
Anhydrin Binds to DNA but Remains Unstructured.
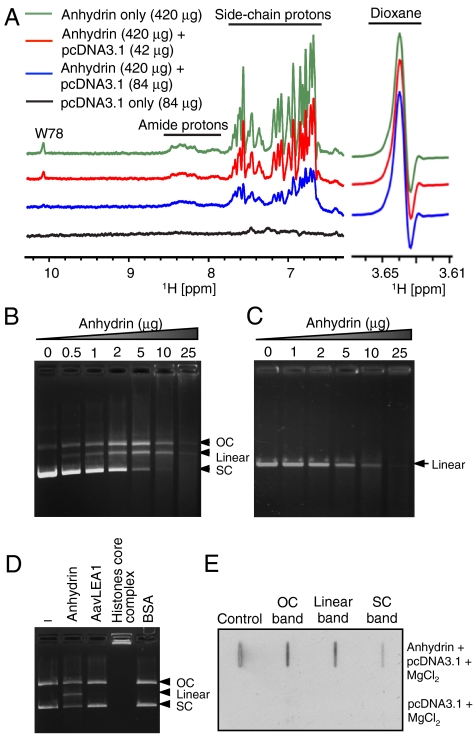
In further bioinformatic analysis, the algorithm DBS-Pred (www.netasa.org/dbs-pred) returned a 98.7% probability that anhydrin is a DNA binding protein; the consensus of four online tools predicts that both the C-terminal tail (L63-K86) and a short region near the N terminus (T6-Y11) bind DNA (Fig. 1B). To test this, we looked for interaction between anhydrin and supercoiled plasmid DNA, pcDNA3.1, using NMR (Fig. 5A). The 1D 1H-NMR spectrum of free anhydrin showed several signals arising from amide and side-chain protons such as the tryptophan (W78) ring NH. When incubated with pcDNA3.1 in the presence of 10 mM MgCl2, however, there was a progressive, DNA concentration-dependent diminution of all signals in the spectrum. This is consistent with binding of a significant proportion of anhydrin molecules to the plasmid which, because of its size, has slow diffusion properties leading to broadening of amide resonances in the complexed protein so they are no longer detected. Notably, a fraction (∼25%) of anhydrin remains unbound and in equilibrium with the NMR-invisible protein-DNA complex, for which the linewidth of the amide resonances remains sharp and hence detectable. Although NMR experiments indicate binding of anhydrin to DNA, both CD and FTIR analysis showed no increase in folding in the presence of DNA and thus anhydrin remains disordered (Fig. 2A, B and Fig. S1E).
Fig. 5.
Anhydrin binds and digests DNA. (A) The 1H-NMR spectrum of free anhydrin (420 μg, green) shows signals arising from backbone amide and side-chain protons that are diminished upon titration with plasmid DNA in a progressive manner (red 42 μg, blue 84 μg DNA added). The maintenance of height and width of the resonant peak of dioxane (right, 3.64 ppm), shown as reference, indicates instrument conditions were constant throughout and that perturbation of anhydrin NMR signals was caused by interaction with DNA. (B) pcDNA3.1 (400 ng) and (C) linear DNA (mCherry gene; 200 ng) incubated with increasing concentrations of anhydrin. (D) Activity of anhydrin compared to His6-AavLEA1, core histones and BSA. (E) Slot-blot analysis of the different forms of pcDNA3.1 digested with anhydrin (top) or untreated pcDNA3.1 (bottom) after probing with Flag antibody; “Control” is purified anhydrin. 200 ng pcDNA3.1 and 2 μg anhydrin were used above, unless otherwise stated. OC, open-circular DNA; SC, supercoiled DNA.
Anhydrin Is an Endonuclease.
To observe the effects of anhydrin on DNA, we performed agarose gel electrophoresis after incubating pcDNA3.1 with varying amounts of anhydrin (Fig. 5B). Intriguingly, we found that, with increasing concentration of anhydrin, the supercoiled (SC) form of the plasmid gradually disappeared, being replaced by open-circular (OC) plasmid and an additional band that migrated between them. At the highest protein concentration tested, these forms are themselves replaced by a smear of apparently degraded DNA fragments. The additional band ran at a position expected for linearized plasmid, and this was confirmed by electrophoresing anhydrin-treated DNA alongside pcDNA3.1 digested with restriction enzymes HindIII and BamHI, which each cut the plasmid once (Fig. S5A). Anhydrin also cleaves the plasmids pHM6 and pET28a+ (Fig. S5B), and the PCR-generated mCherry gene (Fig. 5C), showing that it recognizes a range of substrates, including linear DNA. This also explains the smear seen in Fig. 5B at higher protein concentrations. Pretreatment of anhydrin with diethylpyrocarbonate (DEPC), which modifies histidine residues, abrogated its activity (Fig. S5C). To verify that the observed endonuclease activity was not common to other hydrophilic IDPs, we tested the LEA protein, AavLEA1, and control proteins such as BSA, and found them to lack any effect on plasmid DNA. Recognized DNA binding proteins such as the core histones behave differently to anhydrin because they form DNA-protein complexes that limit the migration of plasmid in the gel (Fig. 5D). In agreement with NMR data (Fig. 5A), slot-blot analysis of OC, linear, and SC pcDNA3.1 bands after incubation with anhydrin showed that the protein is associated with all three forms of plasmid DNA (Fig. 5E).
Other experiments indicate that this novel activity is progressive, i.e., time-dependent, that anhydrin appears unchanged by its effect on DNA (Fig. S5D), and that the activity is temperature-dependent (Fig. S5E), consistent with a catalytic mechanism. The reaction buffer used contains Mg2+ ions, and incubation of SC DNA with anhydrin and Mg2+ alone was sufficient for activity; Mn2+ also supported anhydrin activity, but Ca2+ did not (Fig. S5F). Nevertheless, anhydrin still binds to DNA in the presence of Ca2+ (Fig. S5G) without gaining structure (Fig. 2A, B and Fig. S1E). Addition of ATP at concentrations in the physiological range had little effect, although higher concentrations were inhibitory for anhydrin activity (Fig. S5H), possibly due to chelation of Mg2+. To determine the specific activity of anhydrin, we defined 1U as the amount of protein required to convert > 80% of 1 μg supercoiled pcDNA3.1 to open circle or linear forms in a total reaction volume of 25 μ1 in 1 h at 37 °C, when anhydrin had a specific activity of 500U per mg protein (Fig. S5I). This is some 10-fold lower than that of T7 endonuclease I, for example (Fig. 6A), probably reflecting the lack of a highly evolved, three-dimensionally configured active site typical of conventional enzymes, but is within the range of activity levels consistent with a physiological role.
Fig. 6.
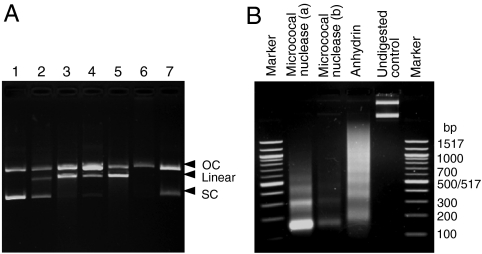
(A) Comparison with DNA-modifying enzymes: (1) 200 ng pcDNA3.1 treated with (2) anhydrin, (3) DNAse I, (4) micrococcal nuclease, (5) T7 endonuclease I, (6) topoisomerase I, and (7) DNA gyrase. (B) Anhydrin digests linker DNA. Assembled chromatin digested with micrococcal nuclease [(a) 500 ng or (b) 250 ng DNA] or anhydrin (500 ng). Marker sizes are labeled (bp). OC, open-circular DNA; SC, supercoiled DNA.
We then compared the pattern of anhydrin endonuclease activity with that of various known DNA modifying enzymes. The digestion profiles generated by limited incubation with DNase I, micrococcal nuclease, and T7 endonuclease I resembled that of anhydrin because SC plasmid DNA was converted to OC and linear forms (Fig. 6A). However, the profiles observed were not identical—for example, T7 endonuclease I is only weakly active against linear DNA (www.neb.com/nebecomm/products/faqproductM0302.asp)—indicating a different mechanism for anhydrin. Further studies will be required to define this mechanism. Finally, we investigated whether anhydrin could also digest chromatin assembled using HeLa core histones and pcDNA3.1. On incubation with anhydrin, a ladder of nucleosomes was obtained. This ladder was not as clearly defined as that obtained by micrococcal nuclease digestion, and the monomer DNA fragment size was larger—closer to 200 bp than ∼150 bp—suggesting that anhydrin cuts predominantly within linker DNA but does not efficiently attack DNA close to the nucleosome surface (Fig. 6B).
Discussion
Disordered proteins have a plasticity of function that reflects their lack of well-defined structure, and this is particularly apparent for IDPs exhibiting more than one function, i.e., those that are so-called moonlighting proteins (27, 28). For example, the tumor repressor protein, p53, can bind four different partner proteins (S100ββ, cyclin A, sirtuin, and CBP) through its unstructured C-terminal regulatory domain (29). Anhydrin, a protein associated with desiccation tolerance in the anhydrobiotic nematode, A. avenae, is also a moonlighting IDP with the striking property that it exhibits two novel functions, as either a molecular shield or an endonuclease.
Hydrophilic IDPs, implicated in anhydrobiosis in a variety of organisms, have been shown to reduce aggregation of a broad range of proteins both in vitro and in vivo (15, 17, 19). Although superficially this activity resembles that of some molecular chaperones, there are significant points of difference: Chaperones are largely well-structured and in most cases function through interaction with exposed hydrophobic regions on (partially) unfolded client proteins (23). Such interactions are sufficiently robust to recover chaperone–client complexes from cell extracts by coimmunoprecipitation (e.g., the interaction of Hsp60 with polyQ proteins) (24). However, interactions via hydrophobic interfaces are improbable for highly hydrophilic IDPs like the LEA proteins and anhydrin; rather, such activity may result from electrosteric interference where the IDP reduces the encounter frequency of aggregating protein species. We term this molecular shield activity (1, 19); a similar concept is familiar to colloid chemists in the pharmaceutical, chemical, and food industries where electrosteric stabilization of colloidal suspensions by hydrophilic, often charged, polymers is commonly used (30, 31). The interior of the cell is itself colloidal in nature (32) and conditions of reduced water activity increase protein aggregation (33); a role for hydrophilic IDPs as molecular shield proteins is therefore consistent with the need to stabilize the intracellular milieu of the anhydrobiotic cell as it dries.
Prior to the present report, the mechanism of action of hydrophilic proteins as molecular shields had not been investigated. That anhydrin does not coimmunoprecipitate with a client protein, PABPN1-A17, suggests that stable complexes do not readily form between them and differentiates anhydrin from molecular chaperones. Nevertheless, we obtained evidence from in vivo FRET experiments for a degree of association between anhydrin and PABPN1-A17 consistent with weak and/or short-lived complex formation. This indicates that anhydrin does not function exclusively as an entropic chain, by a volume exclusion effect, but rather can interact loosely with client proteins to provide an electrostatic and/or steric obstacle to close approach of other aggregating species.
The discovery that anhydrin has a catalytic function as an endonuclease is highly unexpected: The classical structure-function paradigm of proteins requires that an enzyme has an active site of defined three-dimensional conformation; consequently, catalytic activity is anticorrelated with disorder (2, 7, 13, 34), and there are no previously documented examples of fully disordered proteins acting as enzymes. Anhydrin is demonstrably disordered in solution, as indicated by bioinformatics and several experimental techniques, yet is able to convert supercoiled plasmid DNA to linear form, probably via a nicked, open circle, and to digest linear DNA to small fragments; it also cleaves linker DNA in chromatin. Anhydrin must bind DNA to effect its function, and NMR and slot-blot experiments confirm this, but CD and FTIR spectroscopy show that the protein does not thereby gain recognized secondary structure. This does not exclude the adoption of a specific spatial orientation with respect to its substrate but means that the polypeptide chain must remain extended in contact with DNA; a few other cases of IDPs remaining entirely disordered in the bound state are known (35, 36). A nuclease function for anhydrin would be of value in anhydrobiosis where desiccation is expected to cause DNA breakage, tangling, and stalled replication forks, resulting in structures that require resolution, repair, or removal during the rehydration phase. An alternative role might be in apoptosis or necrosis, where DNA breakdown is required. Of broader significance, the identification of two novel functions in a single IDP suggests that the disordered proteome is likely to be a rich source of unforeseen activities, highlighting new functions of both biological and potentially pharmaceutical significance.
Materials and Methods
Circular Dichroism and Fourier Transform Infrared Spectroscopy.
CD and FTIR were performed largely as described (16).
Nuclear Magnetic Resonance.
Lyophilized anhydrin was dissolved in water or D2O to a concentration of 50 μM; 10 mM dioxane was added as internal control. NMR experiments were recorded at 25 °C on a Bruker Avance 500 MHz spectrometer equipped with a TCI cryoprobe and employed a standard pulse sequence in Bruker’s library (p3919gp) with a watergate scheme of pulses for water suppression. Two thousand forty eight complex points were acquired with spectral width 15 ppm, and 256 scans accumulated per experiment. Spectra were processed with Bruker’s software Topsin 2.1. Where indicated, pcDNA3.1 was added to samples from a concentrated solution, and experiments were recorded in the presence of MgCl2.
Citrate Synthase Aggregation and Activity Assay.
CS assays were performed as described (19); enzyme activity was expressed as a percentage of the nondried control rate. Assays were performed in triplicate, and the standard deviation is shown; statistical relevance was determined by one-way Anova and Tukey post test using InStat3 (GraphPad Software).
Analysis of Nuclear Aggregate Formation.
Approximately 200 cells/nuclei per sample coexpressing EGFP-PABPN1-A17 (0.2 μg construct DNA) with anhydrin-mCherry or mCherry (0.6 μg) were assessed for aggregates as percentage or odds ratio, described previously (15) and in Fig. S3. Experiments were done in triplicate at least twice. Images were acquired with a Zeiss LSM510 META confocal microscope (63x, 1.4NA PlanApochromat objective) with Zeiss LSM510 v3.2 software.
FRET Measurements.
FRET between EGFP-PABPN1-A17 and anhydrin-mCherry was quantified via sensitized acceptor (mCherry) fluorescence upon donor (EGFP) excitation as described (26).
DNA Binding/Nuclease Assay.
Anhydrin (2 μg) was incubated with 200 ng of pcDNA3.1 in DNA binding buffer (20 mM HEPES pH7.4, 100 mM KCl, 10 mM MgCl2, 1 mM DTT, 4% glycerol) for 15 min at 25 °C, unless otherwise stated, and DNA was analyzed on 0.8% agarose gels.
Further detailed information is provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Alessandro Esposito for exploratory collaboration, Ben Luisi for use of the Mono S column, David Rubinsztein and Janet Davies for plasmids, Sovan Sarkar and Ron Laskey for helpful comments. S.C. is the Broodbank Trust Research Fellow and a Research Fellow of Hughes Hall, Cambridge; F.M. is a postdoctoral fellow of the Research Foundation Flanders; C.W.B. was a Marie Curie fellow (EU FP7) and gratefully acknowledges Chris Dobson for access to equipment. Funded by grants to A.T. from Leverhulme Trust (F/09 717/B), the Isaac Newton Trust, and the European Research Council (Advanced Investigator Grant 233232), and to G.S.K. and C.F.K. from the Wellcome Trust and Medical Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006276107/-/DCSupplemental.
References
- 1.Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 2.Tompa P. Structure and Function of Intrinsically Disordered Proteins. Boca Raton, FL: Taylor and Francis; 2009. [Google Scholar]
- 3.Oldfield CJ, et al. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 4.Pentony MM, Jones DT. Modularity of intrinsic disorder in the human proteome. Proteins. 2010;78:212–221. doi: 10.1002/prot.22504. [DOI] [PubMed] [Google Scholar]
- 5.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 7.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 8.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struc Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struc Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27(Kip1) cyclin-dependent-kinase inhibitor bound to the cyclin A Cdk2 complex. Nature. 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 11.Lacy ER, et al. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 12.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 14.Mukhopadhyay R, Kumar S, Hoh JH. Molecular mechanisms for organizing the neuronal cytoskeleton. Bioessays. 2004;26:1017–1025. doi: 10.1002/bies.20088. [DOI] [PubMed] [Google Scholar]
- 15.Chakrabortee S, et al. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci USA. 2007;104:18073–18078. doi: 10.1073/pnas.0706964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goyal K, et al. Transition from natively unfolded to folded state induced by desiccation in an anhydrobiotic nematode protein. J Biol Chem. 2003;278:12977–12984. doi: 10.1074/jbc.M212007200. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs D, Kalmar E, Torok Z, Tompa P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008;147:381–390. doi: 10.1104/pp.108.118208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Browne JA, et al. Dehydration-specific induction of hydrophilic protein genes in the anhydrobiotic nematode Aphelenchus avenae. Eukaryot Cell. 2004;3:966–975. doi: 10.1128/EC.3.4.966-975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388:151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee BJ, et al. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies JE, Sarkar S, Rubinsztein DC. Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum Mol Genet. 2006;15:23–31. doi: 10.1093/hmg/ddi422. [DOI] [PubMed] [Google Scholar]
- 22.Davies JE, Berger Z, Rubinsztein DC. Oculopharyngeal muscular dystrophy: Potential therapies for an aggregate-associated disorder. Int J Biochem Cell Biol. 2006;38:1457–1462. doi: 10.1016/j.biocel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Saibil HR. Chaperone machines in action. Curr Opin Struc Biol. 2008;18:35–42. doi: 10.1016/j.sbi.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Tam S, et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol. 2009;16:1279–1285. doi: 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies JE, Sarkar S, Rubinsztein DC. Wild-type PABPN1 is anti-apoptotic and reduces toxicity of the oculopharyngeal muscular dystrophy mutation. Hum Mol Genet. 2008;17:1097–1108. doi: 10.1093/hmg/ddm382. [DOI] [PubMed] [Google Scholar]
- 26.Elder AD, et al. A quantitative protocol for dynamic measurements of protein interactions by Forster resonance energy transfer-sensitized fluorescence emission. J R Soc Interface. 2009;6:S59–S81. [Google Scholar]
- 27.Jeffery CJ. Moonlighting proteins—An update. Mol Biosyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 28.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Oldfield CJ, et al. Flexible nets: Disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bright JN, Woolf TB, Hoh JH. Predicting properties of intrinsically unstructured proteins. Prog Biophys Mol Biol. 2001;76:131–173. doi: 10.1016/s0079-6107(01)00012-8. [DOI] [PubMed] [Google Scholar]
- 31.Napper DH. Polymeric stabilization of colloidal dispersions. London: Academic; 1983. [Google Scholar]
- 32.Zhou EH, et al. Universal behavior of the osmotically compressed cell and its analogy to the colloidal glass transition. Proc Natl Acad Sci USA. 2009;106:10632–10637. doi: 10.1073/pnas.0901462106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choe KP, Strange K. Genome-wide RNAi screen and in vivo protein aggregation reporters identify degradation of damaged proteins as an essential hypertonic stress response. Am J Physiol-Cell Ph. 2008;295:C1488–1498. doi: 10.1152/ajpcell.00450.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uversky V, Gillespie J, Fink A. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Cho HS, et al. Yeast heat shock transcription factor N-terminal activation domains are unstructured as probed by heteronuclear NMR spectroscopy. Protein Sci. 1996;5:262–269. doi: 10.1002/pro.5560050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.