Abstract
The growth of an organism and its size determination require the tight regulation of cell proliferation and cell growth. However, the mechanisms and regulatory networks that control and integrate these processes remain poorly understood. Here, we address the biological role of Arabidopsis translationally controlled tumor protein (AtTCTP) and test its shared functions in animals and plants. The data support a role of plant AtTCTP as a positive regulator of mitotic growth by specifically controlling the duration of the cell cycle. We show that, in contrast to animal TCTP, plant AtTCTP is not implicated in regulating postmitotic growth. Consistent with this finding, plant AtTCTP can fully rescue cell proliferation defects in Drosophila loss of function for dTCTP. Furthermore, Drosophila dTCTP is able to fully rescue cell proliferation defects in Arabidopsis tctp knockouts. Our data provide evidence that TCTP function in regulating cell division is part of a conserved growth regulatory pathway shared between plants and animals. The study also suggests that, although the cell division machinery is shared in all multicellular organisms to control growth, cell expansion can be uncoupled from cell division in plants but not in animals.
Keywords: Arabidopsis, Drosophila, organ development, cell division
In both the animal and plant kingdoms, body size is a fundamental but poorly understood attribute of biological systems. It affects important fitness variables such as mate selection, predation, and tolerance to various biotic and abiotic stresses. Attaining the correct body size is one of the most rigorous demands of multicellularity because it requires the precise coordination of multiple developmental processes such as cell proliferation, cell growth, and programmed cell death to allow the ultimate differentiation into functional organs and tissues. Molecular and genetic studies in animals and plants are beginning to elucidate the basic growth machinery and its regulation that control and integrate these processes to generate the enormous variety in organ sizes and shapes in nature (1–3).
The translationally controlled tumor protein (TCTP) is ubiquitously found in all eukaryotes. Its expression is associated with many tumors (4, 5). Animal TCTP is involved in several cellular processes, such as cell proliferation, cell growth, malignant transformation, protection against various cellular stresses, and apoptosis (4, 6). In mice, knockout of TCTP leads to embryonic lethality associated with lack of proliferation and excessive cell death (7). In Drosophila, disruption of dTCTP expression in an organ-specific manner leads to size reduction of the targeted organ due to a reduction in cell numbers and defects in cell growth (8). In plants, TCTP mRNA expression correlates with mitosis in roots and is induced gradually in periods of darkness (9). Plant TCTP protein was also proposed to have a role in long-distance movement of phloem proteins and in pollen tube growth (10, 11). These published data suggest that TCTP not only regulates organismal growth but also asserts plant-specific functions. However, the molecular and biochemical mechanisms of TCTP function are not yet well characterized.
Plants, unlike animals, have the remarkable ability to continue organogenesis throughout their entire life cycle. We took advantage of this asset to generate a full knockout Arabidopsis thaliana for TCTP and used it to demonstrate a role of TCTP in regulating mitotic growth by controlling cell cycle progression. We further show that this particular function is conserved between plants and animals.
Results and Discussion
AtTCTP Is an Essential Gene in Arabidopsis.
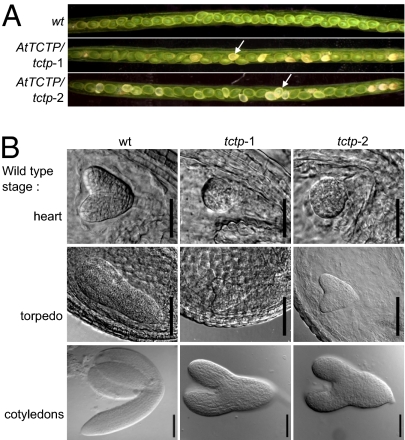
In Arabidopsis, the gene At3g16640 (AtTCTP) encodes a protein that is homologous to TCTP (11, 12). We obtained two independent T-DNA insertion lines (tctp-1 and tctp-2) and studied their phenotypes. Whereas Arabidopsis plants homozygous for these T-DNA insertions were embryonic lethal, plants heterozygous for the T-DNA insertions (AtTCTP/tctp plants) were viable, fertile, and morphologically identical to WT plants. Analyses of developing siliques from AtTCTP/tctp-1 and AtTCTP/tctp-2 heterozygous plants revealed that one fourth of seeds were white in color and were arrested in their development (Fig. 1A). This ratio of seed viability corresponded to a 3:1 Mendelian segregation, suggesting that the tctp−/− mutation is a recessive lethal trait. Importantly, all white seeds contained endosperm and developing embryos, demonstrating that fertilization did take place between tctp haploid mutant gametes. Embryo development in white seeds was compared with that of embryos from seeds showing WT phenotype in the same silique and to that of embryos from WT plants of the same age (Fig. 1B). Homozygous tctp embryos went through early developmental stages, but their development was significantly delayed and never reached the cotyledon stage. In the same silique, all tctp embryos exhibited similar development delay compared with the WT. For example, when WT embryos were at heart stage, tctp-1 and tctp-2 embryos were at globular stage. These data suggest that embryonic lethality occurs in homozygous tctp-1 and tctp-2 mutants because the delayed development leads to abortion at the time when siliques dehisce.
Fig. 1.
Loss of function of AtTCTP results in delayed embryo development. (A) Siliques produced by WT or by heterozygous AtTCTP/tctp-1 and AtTCTP/tctp-2 plants. Arrows indicate tctp homozygous seeds. These seeds were white in color and segregated at a 3:1 ratio [χ2(3:1) = 1.03, P > 0.3 for tctp-1 and χ2(3:1) = 0.90, P > 0.3 for tctp-2]. (B) Embryos from white homozygous tctp-1 and tctp-2 seeds show delayed development compared with embryos of the same age from WT seeds. Developmental stages of WT embryo are denoted as “heart”, “torpedo,” and “cotyledons”. [Scale bars, 50 μm (heart stage) and 100 μm (torpedo and cotyledons stages).]
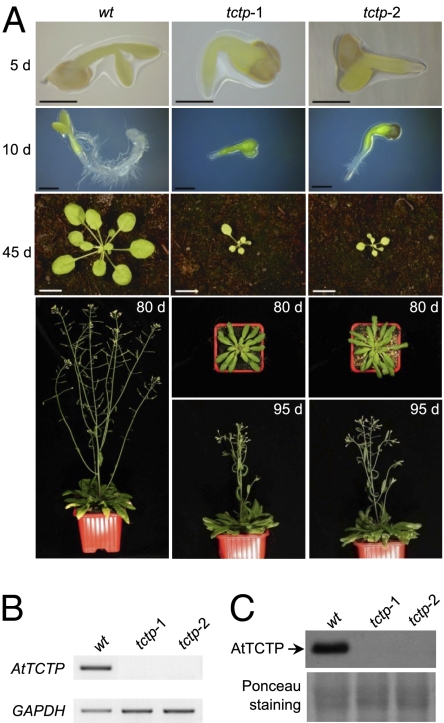
To strengthen the conclusion that delayed development was the cause of abortion, we provided tctp embryos with nutrient supplements in vitro. Under such conditions, tctp-1 and tctp-2 embryos (white seeds produced by heterozygous plants) were able to complete their development on culture medium and tctp knockout plants could be obtained (Fig. 2A). Both tctp-1 and tctp-2 plants were null alleles of AtTCTP because no trace of AtTCTP mRNA or protein could be detected (Fig. 2 B and C). The nutrient-rescued homozygous tctp plants were sterile and released no pollen from the anthers of mature flowers (Fig. S1A). Application of WT pollen to the stigmas of homozygous tctp plants led to the formation of immature siliques that contained few developing seeds (Fig. S1 B–E). These seeds contained tctp/TCTP heterozygous embryos that exhibited delayed development similar to that observed for tctp embryos, when compared with the WT, indicating that heterozygous tctp/TCTP embryos were unable to develop in a homozygous tctp background. However, because heterozygous tctp/TCTP embryos could develop in TCTP/tctp background siliques, it is likely that homozygous tctp mutant plants are unable to adequately supply embryos with the necessary nutrients during their development. This observation and the results of our nutrient-supplement rescue experiment demonstrate that because tctp embryos do not terminate their growth at the silique dehiscence stage, they collapsed, leading to the observed lethality.
Fig. 2.
Embryos homozygous for tctp-1 or tctp-2 mutations can be rescued by supplying nutrients in vitro. (A) Homozygous tctp-1 and tctp-2 embryos rescued by nutrient supplements in vitro develop into adult plants. WT plant development is used as a control. Days after germination are indicated as 5 d, 10 d, 45 d, 80 d, and 95 d. [Scale bars, 500 μm (5 and 10 d) and 1 cm (45 d).] (B) Semiquantitative RT-PCR analysis showing no expression of AtTCTP in leaves of both tctp-1 and tctp-2 plants. GAPDH was used as control. (C) Western blot analysis of AtTCTP accumulation in leaves of WT, tctp-1, and tctp-2 plants using anti-AtTCTP antibody. No AtTCTP protein accumulation is observed in tctp-1 and tctp-2 plants. Red Ponceau staining of total proteins is shown as control.
Expression of AtTCTP using 35S::AtTCTP, 35S::YFP-AtTCTP constructs or the full genomic construct (AtTCTPg-GFP) was able to complement loss of function of AtTCTP (Fig. S2), demonstrating that the embryonic lethality in tctp-1 and tctp-2 mutants is indeed associated with AtTCTP loss of function and that YFP-AtTCTP and AtTCTP-GFP fusion proteins are functional in plants.
Previous work reported that loss of function of AtTCTP (line tctp-1) was associated with impaired pollen tube growth that results in a lack of ovule fertilization, thus preventing the formation of homozygous mutant embryos (11). In contrast, we consistently observed fertilization in the two independent tctp knockout lines, tctp-1 and tctp-2, even when both tctp-1 and tctp-2 mutants were grown under different growth conditions (SI Materials and Methods). Perhaps under certain growth conditions fertilization can be blocked, as observed in Berkowitz et al. (11). However, our data support that the observed lethality in tctp knockout plants is a result of retarded development of the embryos that eventually collapse at the silique dehiscence stage. In animal studies using mouse and Drosophila, impaired TCTP function has been reported to lead to embryo lethality (7, 8), but the lack of a full knockout adult organism hampered the identification of the role of animal TCTP in developing organs. Our study generated full TCTP knockout adult organism and introduced A. thaliana as a model to study functions of TCTP in developing organs (13).
tctp Plants Display Growth Defects.
The tctp-1 and tctp-2 plants that were rescued by nutrient supplements showed a drastic reduction in size (Fig. 2A). These rescued mutant plants were severely delayed during development, flowering transition was 15 d later than in WT plants and no seed could be obtained. Similarly, plants that expressed an RNAi directed against AtTCTP (RNAi-AtTCTP) also showed retarded growth, size reduction (Fig. 3A), and 4–5 d delayed bolting when compared with the WT, a similar phenotype to that reported by Berkowitz et al. (11). RNAi-AtTCTP plants exhibited a reduction of rosette leaf size, ranging from 21% at 30 d postgermination (dpg) to 18% at 48 dpg (Fig. S3A), and a drastically reduced inflorescence stem size (Fig. 3A). However, in contrast to tctp-1 and tctp-2 knockouts, RNAi-AtTCTP lines were not embryonic lethal and could develop into adult plants. To explore the cause of such differences, we conducted RT-PCR experiments, which showed a significant reduction, but not full obliteration, of AtTCTP expression in RNAi-AtTCTP lines (Fig. S3A), suggesting that sufficient AtTCTP was produced, allowing embryos to develop. Altogether, these results demonstrate a role of AtTCTP as a ubiquitous regulator of plant growth, supporting the notion that AtTCTP may serve as a general regulator required for the development of the entire plant.
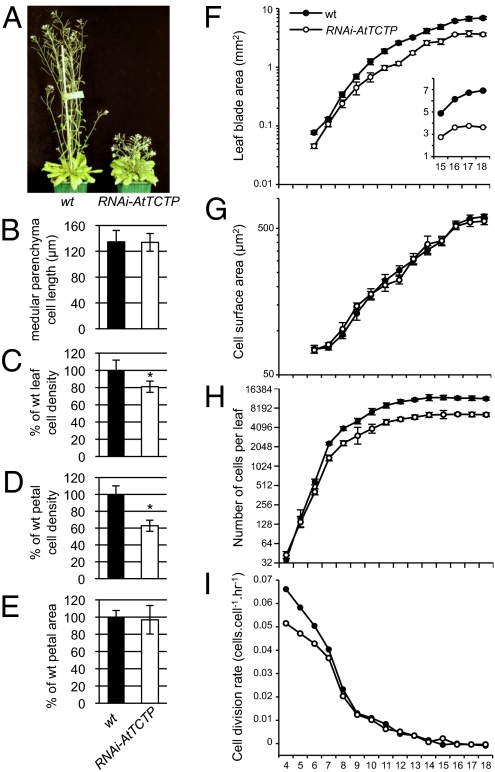
Fig. 3.
AtTCTP loss of function affects cell division. (A) RNAi-AtTCTP plants exhibit severe dwarf phenotype compared with WT plants. (B) Inflorescence stem cell length measurements show no significant difference in medullar parenchyma cell length between WT and RNAi-AtTCTP (n = 120). (C and D) A significant decrease of leaf epidermal cell density (C) and conical petal cell density (D) is observed in RNAi-AtTCTP plants compared with the WT. Asterisks indicate significant differences with PStudent < 0.001. (E) Surface area measurements of WT and RNAi-AtTCTP petals show no significant change of petal size (n = 30). (F–I) Kinematic analysis of leaf growth performed on the first leaf pair of RNAi-AtTCTP and WT. Average of leaf blade area (F), cell area (G), cell numbers (H), and cell division rate (I) are presented. A linear scaled graph of leaf blade area is inserted in F. Note that leaves are twofold smaller in the RNAi-AtTCTP line compared with the WT.
AtTCTP Is Involved in the Control of Cell Proliferation.
To explore the cause of the growth defects in the tctp mutants, we analyzed cell division and cell expansion profiles in RNAi-AtTCTP lines. The length of the medullar parenchyma cells in RNAi-AtTCTP inflorescence stems was similar to that of the WT (Fig. 3B; Fig. S4A), suggesting that the observed stem size reduction defect in RNAi-AtTCTP plants likely correlates with a reduction of cell numbers rather than cell size. Consistent with this observation, the cell density in the leaves and petals of RNAi-AtTCTP plants was decreased about 20% and 37%, respectively, compared with that of the WT (Fig. 3 C and D). However, the size of RNAi-AtTCTP petals was similar to that of the WT because of an increase in cell size in RNAi-AtTCTP petals (Fig. 3E and Fig. S4 C and D). Similarly, RNAi-AtTCTP leaves also show a reduction in cell numbers and a partial increase in final cell size (Fig. 3D and Fig. S4B). These observations suggest the activation of a compensation mechanism that enhances cell expansion in the RNAi-AtTCTP leaves and petals, where cell proliferation is impaired. For the same reason, petals of tctp-1 and tctp-2 homozygous plants rescued by nutrient supplements also showed lower cell density and larger cells (Fig. S4E). In contrast, no compensation mechanism occurred in the inflorescence stems of RNAi-AtTCTP plants (Fig. 3B and Fig. S4A). Compensation mechanisms are commonly activated to maintain normal plant organ sizes when aberrant or deficient cell divisions occur during development (14, 15). The result is marked by an increase in cell volume when cell numbers decrease. However, examples in the literature have shown that a decrease in cell numbers does not always correlate with increased cell expansion (15). Specifically, compensation is usually seen in lateral organs with a determinate fate (e.g., leaves and petals) and not in organs with an indeterminate fate, such as roots (16). This probably explains the observed lack of compensation in RNAi-AtTCTP inflorescence stems, Arabidopsis organs with indeterminate growth. Therefore, we postulate that the cell size increase in RNAi-AtTCTP leaves and petals must be indirectly related to TCTP loss of function. Analysis of the DNA content of leaf cell nuclei by flow cytometry showed no significant modifications in RNAi-AtTCTP compared with that of the WT (Fig. S5), demonstrating that the cell size increase observed in leaves of the RNAi-AtTCTP line was not correlated with endoreduplication, a common landmark of cell expansion (17).
To test our hypothesis that AtTCTP controls cell division but not cell expansion, we conducted overexpression studies with lines 35S::AtTCTP and 35S::YFP-AtTCTP. The final adult plants overexpressing these constructs have a normal development, which is in agreement with previous work (11). However, 35S::AtTCTP plantlets exhibited accelerated growth compared with the WT (Fig. S3A). For example, the growth in 35S::AtTCTP plants was about 2 d ahead at 48 d postgermination. Petal surface area and cell density measurements showed no significant difference between 35S::AtTCTP and WT (Fig. S3B), suggesting that accelerated cell proliferation is likely responsible for the observed accelerated growth in 35S::AtTCTP plants, hence consistent with our proposed role of AtTCTP in regulating mitotic growth but not postmitotic growth.
To further strengthen the conclusion that cell division is impaired in AtTCTP loss-of-function and gain-of-function plants, we analyzed hypocotyl growth. In etiolated plants, elongation of the hypocotyl occurs solely via cell expansion and does not involve cell division (18). Measurements of hypocotyl length in RNAi-AtTCTP and 35S::AtTCTP seedlings grown in the dark showed no significant difference with that of WT etiolated hypocotyls (Fig. S6), suggesting that cell elongation is likely not affected, thus further strengthening the role of AtTCTP as a positive regulator of mitotic growth but not cell expansion. Consistent with AtTCTP’s role in regulating cell proliferation, a higher accumulation of Arabidopsis AtTCTP protein was reported to be associated with young proliferating tissues such as the meristematic and division zones of the root or the embryo (11). In similar manner, we also observed that AtTCTPg-GFP plants, harboring the genomic sequence of AtTCTP containing the 5′ and 3′ UTRs, showed a strong GFP fluorescence in tissues undergoing active cell division, although low GFP fluorescence was observed in fully developed organs such as leaves or stems (Fig. S7 A and B). Interestingly, in 35S::YFP-AtTCTP plants where the 5′ and 3′ UTRs are absent, we observed a fluorescence signal in all tissues (Fig. S7 C and D), suggesting that AtTCTP is translationally controlled and that the 5′ and 3′ UTRs are required to restrict the accumulation of TCTP protein to highly dividing tissues. Consistent with these data, AtTCTP protein accumulation did not always match its mRNA expression pattern (Fig. S8). Sequence analysis identified a conserved 5′-terminal oligopyrimidine tract (5′ TOP) motif in the 5′ UTR and two AUUUA motifs in the 3′ UTR of plant AtTCTP (Fig. S7 E and F). In animals, 5′ TOP- and CG-rich regions in the 5′ UTR or AUUUA motifs in the 3′ UTR have been reported as important for the control of TCTP translation (4).
AtTCTP Regulates the Duration of Cell Cycle.
To explore further the defects in cell proliferation due to the loss of AtTCTP, we used kinematic analysis of leaf growth using plantlets grown in vitro (19). A reduction in leaf area was already observed 6 d after sowing in the RNAi-AtTCTP mutant compared with WT plants. At day 10 after sowing, the RNAi-AtTCTP mutant exhibited an approximately twofold reduction in leaf area, and the size remained constant thereafter (Fig. 3F). The reduction was due to a decrease in cell numbers, which started at day 5 and continued until day 8 when it reached a maximum twofold reduction (Fig. 3 G and H). No reduction in cell size was observed (Fig. 3G). Therefore, the difference in leaf area between RNAi-AtTCTP and WT was directly proportional to the decrease in cell numbers.
From day 4 to day 9, the cell division rate was lower in RNAi-AtTCTP leaves, suggesting slower mitotic growth at early development stages compared with the WT (Fig. 3I). Average cell cycle duration in the whole leaf was estimated as the inverse of the relative cell division rate. Between day 4 and day 7 after sowing, when growth is mediated through cell division (19), the average cell cycle duration was about 4 h longer in the RNAi-AtTCTP leaf primordia compared with the WT. For example, at day 5, cell cycle durations were 17 h in WT and 21 h in RNAi-AtTCTP plants. It should be noted that the duration of leaf expansion and that of proliferative growth were similar between the WT and the RNAi-AtTCTP plants (Fig. 3 F and H). Therefore, the observed longer cell cycle in RNAi-AtTCTP plants is likely related to the observed cell number reduction and subsequent organ size reduction associated to the loss of function for AtTCTP. Furthermore, in contrast to RNAi-AtTCTP plants grown in soil, the reduction of cell numbers in the leaves of RNAi-AtTCTP grown in vitro was not compensated by an increase in the final cell size. It appears that in vitro culturing perturbed the compensation process observed in the RNAi-AtTCTP leaf. A comparable scenario has been observed for plants mutant for the SWP gene where compensation mechanisms were observed in swp plants grown in soil but not in swp plants grown in vitro (20). Hence, the in vitro data corroborate that the observed increase in cell expansion in leaves and petals of RNAi-AtTCTP and tctp knockout lines is likely independent of TCTP function.
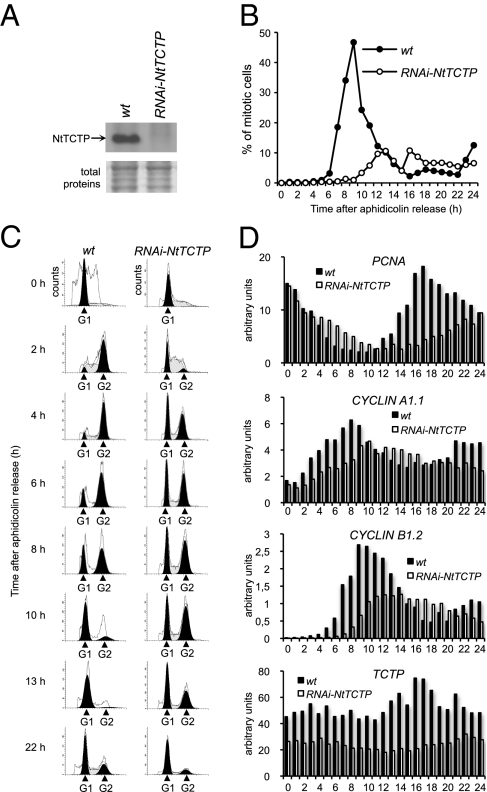
To investigate the role of plant TCTP in cell cycle progression, we generated tobacco BY-2 cells expressing an RNAi targeted to Nicotiana tabaccum NtTCTP (line RNAi-NtTCTP). The real time quantitative PCR (qRT-PCR) and Western blot analyses showed very low levels of NtTCTP mRNA and protein accumulations in the RNAi-NtTCTP cells (Figs. 4 A and D). We synchronized BY-2 cells with aphidicholin and then determined their mitotic index (the percentage of mitotic cells) (Fig. 4B). After aphidicholin release (AAR), WT BY-2 cells exhibited a mitotic peak of about 47% at 9 h AAR, whereas RNAi-NtTCTP BY-2 cells exhibited a delayed mitosis 13 h AAR and only 10% of the mutant cells were in mitosis. This mitotic index state was maintained over time in RNAi-NtTCTP cells. DNA content measurements (Fig. 4C) showed a normal progression of cell cycle over time in WT BY-2 with a rapid reduction of G1 cells 2 h AAR and a concomitant increase of G2 cells. Mitosis occurred 8 h, 10 h and 13 h AAR with a rapid reduction of G2 cells and an increase in G1 cell numbers. As expected, RNAi-NtTCTP BY-2 cells were severely delayed in cell cycle progression. The number of RNAi-NtTCTP cells at G1 remained high 2 h AAR and continued as such 8 h, 10 h, 13 h and 22 h AAR. Cells entering G2 phase were also severely delayed in RNAi-NtTCTP BY-2 with a higher number of cells at 6 h, compared with 2 h in WT BY-2. Therefore, the RNAi-NtTCTP cells exhibit a prolonged G1 phase, which results in a delay of at least 4 h for cells to reach G2/M phase.
Fig. 4.
Plant TCTP controls cell cycle duration (A) Western blot analysis of NtTCTP protein accumulation in WT and RNAi-NtTCTP BY-2 cells using anti-AtTCTP antibody. Coomassie staining of total proteins is shown as control (Bottom). (B) Mitotic index of synchronized WT and RNAi-NtTCTP BY-2 cells after aphidicholin release (AAR). Note the delay and the decrease of the mitotic peak in RNAi-NtTCTP cells compared with WT. (C) Flow cytometric analysis of DNA content in synchronized WT and mutant BY-2 cells. Note the constant G1 population after AAR in RNAi-NtTCTP cells. (D) Quantitative RT-PCR analysis of cell cycle marker genes (PCNA, cyclin A1.1, and cyclin B1.2) and NtTCTP in synchronized WT and mutant BY-2 cells.
Analysis of cell cycle marker genes revealed that in the WT BY-2 cells, proliferating cell nuclear antigen (PCNA) (21) expression decreased rapidly during S/G2 transition (Fig. 4D), whereas in RNAi-NtTCTP cells PCNA expression decreased much more slowly compared with WT BY-2. These data are in agreement with the observed delay of cells leaving G1 phase in RNAi-NtTCTP cells. In these cells, the G2/M marker cyclin A1.1 gene (21) exhibited lower levels of mRNA accumulation and delayed expression that reached a maximum between 10 and 14 h AAR. In WT BY-2, cyclin A1.1 expression was highly induced until 8 h AAR followed by a rapid decrease in its expression. Similarly, the onset of the expression of the mitosis-specific marker cyclin B1.2 (21) was delayed about 4 h in RNAi-NtTCTP BY-2. We also observed an approximately twofold reduction in cyclin B1.2 level, and its expression was more spread out over time when compared with WT BY-2 (Fig. 4D). In WT BY-2, the expression of cyclin B1.2 was at maximum 9 h AAR, which corresponds to the peak of mitotic index (Fig. 4B). Therefore, the slowing of cell cycle progression in RNAi-NtTCTP cells was associated with delayed expression of A- and B-type cyclins. The aberrant prolonging of cell cycle duration in RNAi-NtTCTP cells and in the Arabidopsis RNAi-AtTCTP line (kinematics data) strongly indicates the role of plant TCTPs as mitotic growth regulators.
Plant AtTCTP and Animal dTCTP Have Conserved Function in Controlling Mitotic Growth.
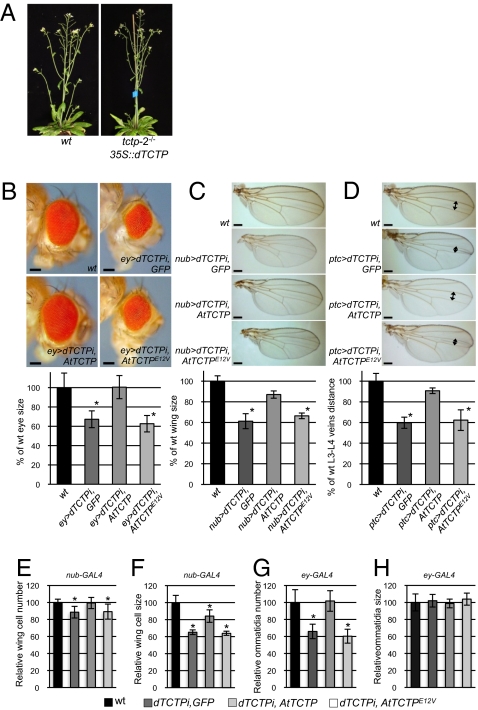
To explore whether TCTP function is conserved among plants and animals, we performed interspecies complementation experiments between Arabidopsis and Drosophila. Drosophila dTCTP was expressed under the control of the 35S constitutive promoter in tctp-2 plants. dTCTP rescued the embryonic lethality in tctp-2 and the cell proliferation defects associated with loss of function of plant AtTCTP (Fig. 5A), demonstrating that animal dTCTP performs similar function in plants and thus is involved in regulating mitotic growth.
Fig. 5.
Drosophila dTCTP complements AtTCTP loss of function in Arabidopsis and vice versa. (A) The Drosophila dTCTP (35S::dTCTP) is able to complement embryonic lethality in tctp-2 loss of function mutant. (B–D) Arabidopsis AtTCTP is able to complement loss of function of dTCTP in Drosophila. Comparison of eye surface (B), wing surface (C), and L3-L4 veins distance (D) in WT flies and in lines expressing an RNAi directed against dTCTP (dTCTPi) under the control of eyless-GAL4 (B) (ey>), nubbin-GAL4 (C) (nub>), or patch-GAL4 (D) (ptc>) drivers, along with GFP, AtTCTP, or AtTCTPE12V. Asterisks indicate significant differences with pStudent < 0.001; n = 30 for each condition. [Scale bars, 100 μm (B) and 200 μm (C and D).] (E and F) AtTCTP complements cell number defects fully (E) and cell size defects partially (F) in nub > dTCTPi fly wings. (G and H) The eye size reduction in ey > dTCTPi flies is associated with a reduction in ommatidia number (G) but not size (H). The defects in ommatidia number are fully complemented by expressing plant AtTCTP (G). Mutated AtTCTPE12V is unable to complement cell number reduction in wing and ommatidia number reduction in eye of dTCTPi flies. Asterisks indicate significant differences with pStudent < 0.001; n = 30 for each condition.
Next, we examined whether the plant AtTCTP was able to complement dTCTP loss of function in Drosophila. We used flies in which the expression of dTCTP was silenced via the expression of an anti-dTCTP RNAi under the control of organ-specific promoters in the eye (line ey > dTCTPi), in the wing (line nub > dTCTPi), or between the L3 and L4 wing veins (line ptc > dTCTPi). A GFP reporter confirmed the organ specificity of these promoters (Fig. S9). Interference with dTCTP led to a significant size reduction of the targeted organ (8) (Fig. 5 B and C). In dTCTPi flies, wing organ size reduction is thought to be the result of the combined decrease in cell proliferation and cell expansion (8). AtTCTP expression driven by nub-Gal4 (nub > AtTCTP) or by ptc-Gal4 (ptc > AtTCTP) rescued the wing size reduction phenotype in nub > dTCTPi flies and the L3 and L4 vein distance reduction phenotype in ptc > dTCTPi flies (Fig. 5 C and D). However, the rescued fly wings were smaller than the WT wings, suggesting partial rescue of the cell proliferation and/or cell expansion. Comparison of cell numbers in the wings of nub > dTCTPi flies, of nub > dTCTPi flies expressing nub > AtTCTP, and of WT flies revealed that the partial restoration of wing size via expression of AtTCTP correlates with a full restoration of cell numbers (Fig. 5E). However, a partial restoration of cell size was also observed (Fig. 5F). These data suggest that plant AtTCTP is able to fully complement the cell division but not all cell expansion, defects associated with loss of function in dTCTP wings.
Silencing dTCTP in the eye using the eyless promoter (ey > dTCTPi) resulted in eye size reduction (8) (Fig. 5B). In ey > dTCTPi flies, the eye size reduction was associated with reduced ommatidia number but not ommatidia size (Fig. 5 G and H), suggesting that cell size in the eye was not affected in ey > dTCTPi flies and that eye size reduction was only associated with a decrease in cell numbers. This is in agreement with the fact that eyless promoter drives expression in the eye imaginal disk anterior to the furrow at the time when eye cell progenitors are actively dividing (22). Expression of AtTCTP in the eye (ey > AtTCTP), where dTCTP was silenced (ey > dTCTPi), completely rescued the eye size reduction phenotype (Fig. 5B). Furthermore, expression of AtTCTP (ey > AtTCTP) fully rescued the ommatidia number defects in ey > dTCTPi Drosophila (Fig. 5 G and H), supporting the model that AtTCTP regulates cell division.
These interspecies complementation experiments provide evidence that TCTP functions as a growth regulator by controlling cell division and that this function is conserved across the animal and plant kingdoms. In AtTCTP, 38% of its amino acids are identical to its Drosophila counterpart (Fig. S10). Many of the essential amino acids and domains known to be required for animal TCTP functions are conserved in AtTCTP. The rat TCTP C-terminal domain, shown to homodimerize in the yeast two-hybrid system (23), is conserved in AtTCTP (Leu122 to Cys168). Bimolecular fluorescence complementation (BiFC) experiments (Fig. S11A) demonstrated that AtTCTP or dTCTP were able to homodimerize in vivo. Furthermore, AtTCTP was able to interact with dTCTP, suggesting that despite the overall relatively divergent protein sequences, the homodimerization domains of AtTCTP and dTCTP are structurally and functionally conserved. In Drosophila, a substitution of Glu12 to Val renders dTCTP nonfunctional (8). We found that AtTCTP harboring such a mutation (AtTCTPE12V) was unable to complement eye and wing size reduction phenotypes associated with dTCTP loss of function in flies (Fig. 5 B–H), demonstrating that the Glu12 is necessary for the correct function of TCTP in both plants and animals. In Drosophila, the dTCTP was reported to positively control the target of rapamycin (TOR) activity through interaction and activation of the Rab GTPase dRheb (8). TOR kinase is a part of a signaling complex that controls cell proliferation and growth in plants and animals (24, 25).
In Arabidopsis, at least 25 Rab GTPases share about 30–35% of their amino acids with dRheb. We found that AtTCTP is able to bind to four of these Rab GTPases (AtRABA4a, AtRABA4b, AtRABF1, and AtRABF2b) in vivo in BiFC experiments (Fig. S11B). In these experiments, AtTCTP also interacted with Drosophila dRheb, and is thus in agreement with interspecies complementation experiments demonstrating that plant and animal TCTPs act in the same regulatory pathway. Similarly, dTCTP was able to interact with dRheb and with plant AtRABA4a, AtRABA4b, AtRABF1, and AtRABF2b (Fig. S11B). GST-pull-down experiments confirmed these interactions in vitro (Fig. S11 C and D). These data suggest that, as in animals, plant TCTP may also act in the TOR pathway via Rab GTPases. Previously, structure-based modeling and analyses of genes in which expression is controlled by TOR also suggested that plant TCTP acts as a regulator of TOR (11).
In conclusion, the characterization of a viable TCTP knockout and the interspecies functional complementation studies demonstrate the function of plant AtTCTP as a cell division regulator. Although plants and animals diverged about 1.6 billion years ago, TCTPs have conserved their function in controlling mitotic organ growth. Nevertheless, the interspecies complementation experiments suggest that animal TCTP has diverged from that of plant TCTP to control cell growth. Determining and comparing the 3D structure of AtTCTP to its yeast and human counterparts (26, 27) and identifying the partners of TCTP will likely address the molecular mechanisms of the functional conservation and the differences between plant and animal TCTPs.
Materials and Methods
Constructions and Plant Lines.
A. thaliana (Col-0 accession) T-DNA-insertion knockout lines tctp-1 (SAIL_28_C03) and tctp-2 (GABI_901E08) were obtained from the Nottingham Arabidopsis Stock Centre.
Anti-AtTCTP RNAi expressing plants, 35S::AtTCTP, 35S::dTCTP and 35S::YFP-AtTCTP overexpression lines, and the AtTCTPg-GFP line (AtTCTP genomic sequence GFP-tagged) were obtained as described in SI Materials and Methods.
Embryo Rescue.
Embryos homozygous for the tctp mutation (white seeds) were dissected from heterozygous tctp-1 and tctp-2 siliques at stage 17B (28) and then rescued by nutrient supplements in vitro as described previously (29). Developed plantlets were then transferred to soil. During all steps, embryos from WT siliques were used as control.
Cell Numbers, Size Measurements, and Kinematic Analysis of Leaf Growth.
Leaves were cleared in a solution containing 80 g chloral hydrate/30 mL water and then mounted in clearing solution containing 20% glycerol. Petal surface area measurements were performed using digital images from cleared petals as described previously (12). Cell density was determined as the number of cells per surface unit from digital images (Nikon Optiphot 2 microscope with Nomarski optics) of cleared leaves and petals using ImageJ software (National Institutes of Health).
Kinematic analysis of leaf growth was performed on the two first-initiated leaves as previously described (19).
Transgene Constructions and Drosophila Lines.
The constructs pUAST::AtTCTP and pUAST::AtTCTPE12V that harbor WT or mutated AtTCTP cDNA, respectively (SI Materials and Methods), under the control of Upstream Activation Sequence (UAS) were used to generate transgenic Drosophila via embryo injections (BestGene Inc.). Drosophila lines UAS::AtTCTP, UAS::AtTCTPE12V and UAS::GFP (used as control) were crossed with UAS-dTCTP RNAi line (8). The progenies were crossed with different lines expressing a GAL4-driver: eyless-GAL4 (ey-GAL4), patch-GAL4 (ptc-GAL4), and nubbin-GAL4 (nub-GAL4) (Bloomington Drosophila Stock Center).
Drosophila eye and wing organ size was measured from digital images using ImageJ software. Wing cell numbers and cell size were calculated by counting of the number of hairs per surface area between the L3 and L4 veins (30). Ommatidia number and size in the eyes of dTCTPi flies expressing AtTCTP was assessed by measuring ommatidia density (number per surface area) and comparing it with that in WT and dTCTPi eyes.
Detailed procedures are provided as SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank A. Chauvet and S. Chamot for technical assistance, D. Ma and J. Szecsi for critical reading of the manuscript, Kwang-Wook Choi (Baylor College of Medicine, Houston, TX) and the Bloomington Drosophila stock center for fly stocks, the Salk Institute for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and the Nottingham Arabidopsis Stock Centre for providing the lines. This work was supported by Centre National de la Recherche Scientifique (Projets Exploratoires/Premier Soutien program), by Institut National de la Recherche Agronomique (Biologie Végétale department), and by ANR-Blanc (ANR-09-BLAN-0006).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007926107/-/DCSupplemental.
References
- 1.Busov VB, Brunner AM, Strauss SH. Genes for control of plant stature and form. New Phytol. 2008;177:589–607. doi: 10.1111/j.1469-8137.2007.02324.x. [DOI] [PubMed] [Google Scholar]
- 2.Krizek BA. Making bigger plants: Key regulators of final organ size. Curr Opin Plant Biol. 2009;12:17–22. doi: 10.1016/j.pbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 4.Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–385. doi: 10.1016/s1357-2725(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 5.Hinojosa-Moya J, et al. Phylogenetic and structural analysis of translationally controlled tumor proteins. J Mol Evol. 2008;66:472–483. doi: 10.1007/s00239-008-9099-z. [DOI] [PubMed] [Google Scholar]
- 6.Susini L, et al. TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ. 2008;15:1211–1220. doi: 10.1038/cdd.2008.18. [DOI] [PubMed] [Google Scholar]
- 7.Chen SH, et al. A knockout mouse approach reveals that TCTP functions as an essential factor for cell proliferation and survival in a tissue- or cell type-specific manner. Mol Biol Cell. 2007;18:2525–2532. doi: 10.1091/mbc.E07-02-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature. 2007;445:785–788. doi: 10.1038/nature05528. [DOI] [PubMed] [Google Scholar]
- 9.Sage-Ono K, Ono M, Harada H, Kamada H. Dark-induced accumulation of mRNA for a homolog of translationally controlled tumor protein (TCTP) in Pharbitis. Plant Cell Physiol. 1998;39:357–360. doi: 10.1093/oxfordjournals.pcp.a029377. [DOI] [PubMed] [Google Scholar]
- 10.Aoki K, et al. Destination-selective long-distance movement of phloem proteins. Plant Cell. 2005;17:1801–1814. doi: 10.1105/tpc.105.031419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berkowitz O, Jost R, Pollmann S, Masle J. Characterization of TCTP, the translationally controlled tumor protein, from Arabidopsis thaliana. Plant Cell. 2008;20:3430–3447. doi: 10.1105/tpc.108.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szécsi J, et al. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006;25:3912–3920. doi: 10.1038/sj.emboj.7601270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones AM, et al. The impact of Arabidopsis on human health: Diversifying our portfolio. Cell. 2008;133:939–943. doi: 10.1016/j.cell.2008.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Truernit E, Haseloff J. Arabidopsis thaliana outer ovule integument morphogenesis: Ectopic expression of KNAT1 reveals a compensation mechanism. BMC Plant Biol. 2008;8:35. doi: 10.1186/1471-2229-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukaya H. Controlling size in multicellular organs: Focus on the leaf. PLoS Biol. 2008;6:e174. doi: 10.1371/journal.pbio.0060174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferjani A, Horiguchi G, Yano S, Tsukaya H. Analysis of leaf development in fugu mutants of Arabidopsis reveals three compensation modes that modulate cell expansion in determinate organs. Plant Physiol. 2007;144:988–999. doi: 10.1104/pp.107.099325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melaragno JE, Mehrotra B, Coleman AW. Relationship between endopolyploidy and cell size in epidermal tissue of arabidopsis. Plant Cell. 1993;5:1661–1668. doi: 10.1105/tpc.5.11.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Von Arnim A, Deng XW. Light control of seedling development. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:215–243. doi: 10.1146/annurev.arplant.47.1.215. [DOI] [PubMed] [Google Scholar]
- 19.De Veylder L, et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Autran D, et al. Cell numbers and leaf development in Arabidopsis: A functional analysis of the STRUWWELPETER gene. EMBO J. 2002;21:6036–6049. doi: 10.1093/emboj/cdf614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Combettes B, et al. Study of phase-specific gene expression in synchronized tobacco cells. Methods Cell Sci. 1999;21:109–121. doi: 10.1023/a:1009880705257. [DOI] [PubMed] [Google Scholar]
- 22.Mollereau B, Domingos PM. Photoreceptor differentiation in Drosophila: From immature neurons to functional photoreceptors. Dev Dyn. 2005;232:585–592. doi: 10.1002/dvdy.20271. [DOI] [PubMed] [Google Scholar]
- 23.Yoon T, et al. Identification of the self-interaction of rat TCTP/IgE-dependent histamine-releasing factor using yeast two-hybrid system. Arch Biochem Biophys. 2000;384:379–382. doi: 10.1006/abbi.2000.2108. [DOI] [PubMed] [Google Scholar]
- 24.Deprost D, et al. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Liu D, Yao H, Wang J. Solution structure and mapping of a very weak calcium-binding site of human translationally controlled tumor protein by NMR. Arch Biochem Biophys. 2007;467:48–57. doi: 10.1016/j.abb.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Thaw P, et al. Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat Struct Biol. 2001;8:701–704. doi: 10.1038/90415. [DOI] [PubMed] [Google Scholar]
- 28.Roeder AH, Yanofsky MF. The Arabidopsis Book. Rockville, MD: The American Society of Plant Biologists; 2009. Fruit development in Arabidopsis; pp. 1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sauer M, Friml J. In vitro culture of Arabidopsis embryos. Methods Mol Biol. 2008;427:71–76. doi: 10.1007/978-1-59745-273-1_5. [DOI] [PubMed] [Google Scholar]
- 30.Adler PN, Liu J, Charlton J. Cell size and the morphogenesis of wing hairs in Drosophila. Genesis. 2000;28:82–91. doi: 10.1002/1526-968x(200010)28:2<82::aid-gene60>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.