Abstract
Tat activating regulatory DNA-binding protein (Tardbp or TDP-43), a highly conserved metazoan DNA/RNA binding protein thought to be involved in RNA transcription and splicing, has been linked to the pathophysiology of amyotrophic lateral sclerosis and frontotemporal lobar degeneration and is essential for early embryonic development. However, neither the physiological role of TDP-43 in the adult nor its downstream targets are well defined. To address these questions, we developed conditional Tardbp-KO mice and embryonic stem (ES) cell models. Here, we show that postnatal deletion of Tardbp in mice caused dramatic loss of body fat followed by rapid death. Moreover, conditional Tardbp-KO ES cells failed to proliferate. Importantly, high-throughput DNA sequencing analysis on the transcriptome of ES cells lacking Tardbp revealed a set of downstream targets of TDP-43. We show that Tbc1d1, a gene known to mediate leanness and linked to obesity, is down-regulated in the absence of TDP-43. Collectively, our results establish that TDP-43 is critical for fat metabolism and ES cell survival.
Keywords: conditional knockout mice, amyotrophic lateral sclerosis, frontotemporal dementia, energy metabolism, RNA-sequencing
Identified initially as a cellular protein that regulates human immunodeficiency viral gene transcription (1), Tat activating regulatory DNA-binding protein (Tardbp or TDP-43) is a highly conserved DNA/RNA–binding protein thought to regulate alternative splicing of cystic fibrosis transmembrane conductance regulator (CFTR) and survival of motor neuron (SMN) through binding to heterogeneous nuclear ribonucleoprotein or (UG)n repeats of these target transcripts (2–4). Biophysical studies indicate that TDP-43 forms a dimer harboring two RNA-binding domains that preferentially bind to TG/UG repeats (5). Whereas TDP-43 is normally localized to the nucleus, it is redistributed as insoluble aggregates in neuronal nuclei, perikarya and neurites in amyotrophic lateral sclerosis (ALS) (6, 7) and frontotemporal lobar degeneration (FTLD) (8). The identification of missense mutations in TARDBP in familial and sporadic ALS (9, 10) supports the idea that this nuclear protein plays a critical role in the pathogenesis of these neurodegenerative disorders. Interestingly, most of these mutations identified to date are localized to the C-terminal domain of TDP-43, a heterogeneous nuclear ribonucleoprotein–interacting region that may be critical for the normal function of the protein. Although mutant TDP-43 mice showed evidence of neurodegeneration, no TDP-43–positive cytoplasmic aggregates were observed in neurons of these mutant mice, suggesting that altered RNA metabolism rather than TDP-43 aggregates underlies the pathogenesis of ALS or FTLD (11).
To begin clarifying the molecular basis of mutant TDP-43–linked disease, it will be crucial to understand the physiological and cellular functions of TDP-43. Moreover, because increased expression of TDP-43 is also toxic to motor neurons (12, 13), it will be also important to identify a set of downstream targets of TDP-43 to facilitate our understanding of pathways that may be impacted by TDP-43. Interestingly, a recent RNAi knockdown study revealed histone deacetylase 6 (HDAC6) as a target of TDP-43 in cultured cells (14).
As TDP-43 is essential for early embryogenesis (15–17), we elected to develop a conditional Tardbp-KO mouse model to determine the physiological role of TDP-43 in the adult animal. We now show that TDP-43 is critical for fat storage in the adipocytes. Moreover, we developed a cellular Tardbp-KO model designed for the identification of a set of downstream targets of TDP-43. By using digital high-throughput RNA-sequencing (RNA-seq) analysis of our conditional Tardbp-KO embryonic stem (ES) cells, we report here the identification and verification of a set of downstream genes regulated by TDP-43, including those that impact on the survival of ES cells. Among this set of down-regulated targets is a gene expressed in skeletal muscle important for regulating leanness (18) and linked to human obesity (19) termed Tbc1d1, which is decreased in skeletal muscle of our conditional Tardbp-KO mouse model. Our findings suggest that TDP-43–dependent reduction of Tbc1d1 in skeletal muscle may be responsible for the increased fat metabolism and leanness observed in the conditional Tardbp-KO mice.
Results and Discussion
Tardbp Targeting and Validation.
To bypass embryonic lethality of the standard Tardbp-null mice, we generated a conditional Tardbp-KO mouse line by engineering a targeting vector in which the third exon of Tardbp was flanked by loxp together with a neomycin resistance gene inserted in the second intron (Fig. 1A); the disrupted Tardbp is predicted to encode a nonfunctional truncated TDP-43 variant as a result of the absence of the critical RNA-binding domain encoded by exon 3 (5) and the highly conserved C-terminal domain. We confirmed the successful targeting of Tardbp by DNA blot analysis (Fig. 1B). The neomycin cassette was subsequently deleted through a crossbreeding strategy with hACTB-flp transgenic mice (20) (floxed Tardbp mice; Fig. 1A). The floxed Tardbp mice were crossbred with a CAG-Cre transgenic mouse line that express the Cre recombinase ubiquitously (21) to generate the heterozygous Tardbp-KO (Tardbp+/−) mice (Fig. 1A). The Tardbp+/− mice were fertile and expressed similar level of TDP-43 in a variety of tissues compared with those of Tardbp+/+ mice (Fig. S1), suggesting that the level of TDP-43 is tightly controlled and compensated in the Tardbp+/− mice. However, no live-born Tardbp−/− mice were identified from ten intercrosses of Tardbp+/− mice (number of pups obtained: control/Tardbp+/−/Tardbp−/−, 23/37/0), confirming that Tardbp is essential for embryogenesis (15–17).
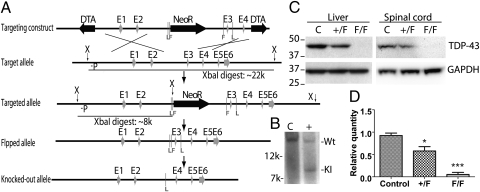
Fig. 1.
Strategy and validation for the conditional deletion of Tardbp. (A) Targeting at the Tardbp locus and removal of neomycin resistance cassette. Exon 3 is floxed and can be removed upon cre recombinase induction. (P, probe for DNA blotting; E, respective exons; F, Frt sites; L, loxP sites; DTA, diphtheria toxin selection cassette; NeoR, neomycin selection cassette.) (B) DNA blot analysis of the control and targeted ES clones; an Xba I digest produced fragments of, respectively, approximately 22 kbp and approximately 8 kbp from the WT and targeted allele when labeled with the probe P in A. (C) Tissue lysates of livers and spinal cords harvested from control, Rosa26-ErCre;Tardbp+/F, and Rosa26-ErCre;TardbpF/F mice treated with tamoxifen for 8 d were subjected to protein blot analyses using antisera against TDP-43 and GAPDH. (D) Quantification of TDP-43 level (mean ± SEM) in the liver blots shown in C (n = 3 independent animals in each group). *P < 0.05, **P < 0.01, ***P < 0.001.
To examine the physiological role of TDP-43 in postnatal mice, floxed Tardbp mice were bred with Rosa26-ErCre mice to generate inducible Tardbp-KO (ErCre;TardbpF/F) mice in which the Rosa26 enhancer/promoter will direct the expression of ErCre recombinase ubiquitously when it gains access into cell nuclei in the presence of the inducer, tamoxifen. Before tamoxifen induction, the ErCre;TardbpF/F mice were indistinguishable from ErCre;Tardbp+/F or control littermates. We first confirmed that a high percentage of recombination occurred in tamoxifen-treated ErCre;TardbpF/F mice as determined by the dramatic decrease in levels of TDP-43 protein in tissues from the mice lacking Tardbp (Fig. 1C). In contrast to the Tardbp+/− mice, the conditional Tardbp+/F mice showed a modest reduction in the levels of TDP-43 (Fig. 1 C and D). In addition, we failed to detect any N-terminal fragment of TDP-43 that could be generated as a result of the design of our targeting strategy in the brain, spinal cord, or liver of the Tardbp+/− mice (Fig. S1C).
Marked Fat Loss and High Fatty Acid Consumption in Conditional Tardbp-KO Mice.
In contrast to control, Rosa26-ErCre;TardbpF/F mice unexpectedly die by day 9 after switching to a tamoxifen-containing diet (Fig. S2). Because initial necropsy analysis of conditional Tardbp-KO mice indicated a loss of body fat, metabolic analyses (22) of these mice were performed. Upon deletion of Tardbp by diet containing tamoxifen citrate (400 mg/kg diet), body weights of all mice decreased during the first 3 d (Fig. 2A) as a consequence of reduced food intake (Fig. 2B). Whereas control mice regained some of their weights during the next 4 d, correlating with increase in food intake, the ErCre;TardbpF/F or ErCre;Tardbp+/F mice did not regain their body weights despite the increase in food consumption over this same period (cumulative weight loss by d 7, control, −3.36 ± 0.55 g, ErCre;Tardbp+/F, −4.59 ± 0.55 g, and ErCre;TardbpF/F, −5.21 ± 0.47 g, ANOVA linear post-test slope, −0.920; P < 0.05; n = 5 for each group; Fig. 2 A and B). Despite significant differences in cumulative weight loss between the control and the Tardbp-KO groups, energy intakes during tamoxifen-dependent deletion of Tardbp were similar among groups (cumulative food intake on day 7, control, 10.8 ± 1.40 kcal vs. ErCre;TardbpF/F, 9.41 ± 0.58 kcal; P > 0.05; n = 5), suggesting that decreased calorie intake was not the major cause of differences in weight loss. We next used indirect calorimetry to examine in vivo whether altered metabolism contributed to the relatively greater weight loss in the conditional Tardbp-KO mice. Both ErCre;Tardbp+/F and ErCre;TardbpF/F mice showed respiratory exchange ratios (RER; defined as VCO2 divided by VO2), indicative of pure fat oxidation (day 7, ErCre;Tardbp+/F, 0.73 ± 0.04; ErCre;TardbpF/F, 0.71 ± 0.02), versus the RER of control mice, indicating the high level of carbohydrate oxidation expected based on composition of the specialized tamoxifen-containing diet (day 7, control, 1.03 ± 0.06). The significant decrease in RER in the conditional Tardbp-KO mice by day 6 could not be explained by reduced food intake. Although the initial 3 d on tamoxifen diet did decrease energy intake in all groups of mice, energy intake was subsequently normalized. Interestingly, whereas RER was similar (approximately 0.7) on day 1 and day 7, energy intake on day 7 was much higher than on day 1 (Fig. 2 B and C, compare day 1 and day 7). Thus, our results indicate that increased fat oxidation rather than reduced energy intake is responsible for the markedly greater weight loss in the conditional Tardbp-KO mice. Consistent with the calorimetry data, quantitative NMR analysis of the carcasses showed significant decreases in whole body fat mass, but not lean mass, in the ErCre;TardbpF/+ and ErCre;TardbpF/F mice in a dosage-dependent manner (slope, −1.716; P < 0.001 by linear trend post-test; n = 4 per group; Fig. 2D). The reduction in levels of TDP-43 in ErCre;Tardbp+/F mice (Fig. 1D) exhibiting altered metabolism suggests that during the acute deletion of one Tardbp allele, the level of TDP-43 has not yet fully compensated whereas levels of TDP-43 in postnatal Tardbp+/− mice have already reached WT level (Fig. S1).
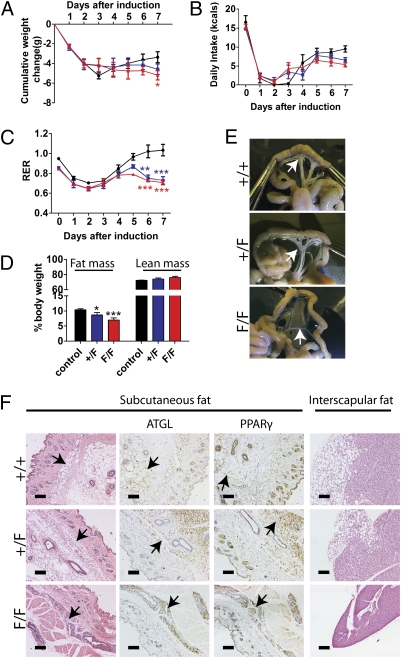
Fig. 2.
Phenotype of Tardbp conditional KO mice. (A) Cumulative body weight changes (in g, mean ± SEM) were compared in tamoxifen-treated control, Rosa26-ErCre;Tardbp+/F, and Rosa26-ErCre;TardbpF/F mice (n = 5 per group). D0, pretreatment. (B) Daily energy intakes (mean ± SEM) while on tamoxifen diet, n = 5. (C) RER (mean ± SEM) was monitored continuously from D0 (pretreatment) to day 7 (1 d before quantitative NMR measurement); n = 5. (D) Quantitative NMR measurement (mean ± SEM) of body fat in the three mouse groups as in A. Note the selective loss of fat mass in the KO mice, in contrast to lean mass. Black, blue, and red lines indicate control, Rosa26-ErCre;Tardbp+/F, and Rosa26-ErCre;TardbpF/F mice, respectively (n = 4 per group, including the two dead mice in the F/F group). (E) Mesenteric fatty tissues (arrows) in CAG-ErCre;Tardbp+/+, CAG-ErCre;Tardbp+/F, and CAG-ErCre;TardbpF/F mice. (F) H&E staining shows loss of fat content in the white (left column) and brown fat (right column). Immunohistochemical staining using the markers ATGL (second column) and PPAR-γ (third column) to visualize adipocytes found in the subcutaneous white fat. Arrows indicate adipocytes. *P < 0.05, **P < 0.01, ***P < 0.001. (Scale bar: 100 μm.)
As the Rosa26-ErCre;TDPF/F mice usually die by day 9 after switching to a tamoxifen-containing diet, a weaker driver line of CAG-ErCre mice (23) was used to confirm the observed lean phenotype and to extend the survival time of tamoxifen treated ErCre;TardbpF/F mice. Most of the CAG-ErCre;TardbpF/F or CAG-ErCre;Tardbp+/F mice survived at least 18 d after switching to the tamoxifen diet. Moreover, the reduction in levels of TDP-43 in these conditional Tardbp-KO mice correlated with the decrease in body weights (relative body weights on day 18 after treatment with tamoxifen, slope, −0.1687; P < 0.001 by linear trend post-test; Fig. S3A). Importantly, gross examination of mesenteric fat confirmed this dramatic fat loss in CAG-ErCre;TardbpF/F mice (Fig. 2E). Indeed, histological analysis of fatty tissues revealed the absence or reduction, respectively, of fatty vacuoles in adipocytes within subcutanous tissue (Fig. 2F, first column) and interscapular brown fat (Fig. 2F, fourth column) in the CAG-ErCre;TardbpF/F or CAG-ErCre;Tardbp+/F mice. Moreover, the positive immunoreactivities of two independent adipocyte markers, adipose triglyceride lipase (ATGL) and peroxisome proliferator-activated receptor–γ (PPAR-γ), showed presence of adipocytes in both white (Fig. 2F, second and third columns) and brown adipose tissues (Fig. S3B), indicating that the loss of fat content is not a result of the absence of adipocytes in these CAG-ErCre;TardbpF/F mice, but rather caused by a lack of stored fat within the adipocytes. Taken together, our results indicate that the decreased level of TDP-43 is responsible for accelerated fat loss in adipocytes of conditional Tardbp-KO mice through increased fat oxidation.
TDP-43 Is Essential for ES Cell Survival.
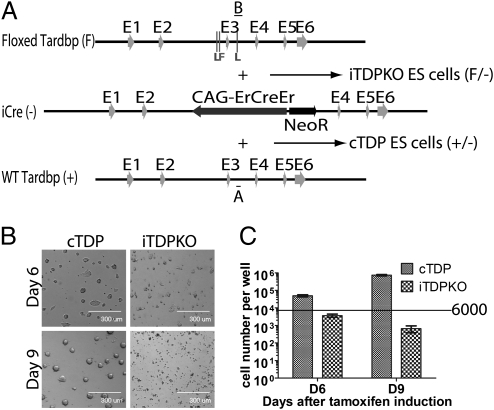
To clarify the mechanism whereby loss of TDP-43 leads to the reduction of fat content, it will be important to identify relevant downstream targets of TDP-43. Although CFTR, SMN, NF-L, and HDAC6 transcripts have been found to be bound and regulated by TDP-43, misregulation of any of these putative targets would not be predicted to provide a straightforward explanation of the lean phenotype observed in our conditional Tardbp-null mice. We elected to search for additional targets through characterization of the Tardbp-dependent transcriptome. To minimize variations in deletion efficiency among different cell types within any one tissue in conditional Tardbp mice and to enhance the deletion efficiency by creating KO cells harboring only one floxed allele, we engineered a tamoxifen inducible Tardbp-KO ES cell line (termed iTDPKO) by replacing the WT Tardbp allele of Tardbp+/F ES cells with a CAG-ErCreEr cassette (Fig. 3A). To appropriately control for the influence of tamoxifen in this iTDPKO cell model, we generated ES cells (termed cTDP) by targeting the floxed Tardbp allele of Tardbp+/F ES cells with the CAG-ErCreEr cassette (Fig. 3A) such that any effect resulting from the treatment with tamoxifen to delete Tardbp in iTDPKO cells will be nullified. Protein blot analysis revealed that TDP-43 was nearly abolished in iTDPKO cells 3 d after exposure to the inducer, 4-hydroxytamoxifen (4-HT; Fig. 4A, fourth panel). Although both iTDPKO and cTDP ES cells could grow as colonies and amplify after targeting (Fig. S4), the 4-HT–treated iTDPKO ES cells exhibited reduction in colony size coupled with increased apoptosis (Fig. 3B). Moreover, the number of surviving iTDPKO ES cells was also significantly reduced by day 6 after tamoxifen induction (4.93 ± 0.06 × 104 vs. 0.37 ± 0.08 × 104; P = 0.0191; n = 3; Fig. 3C); this difference was even greater on the ninth day after 4-HT treatment (74.33 ± 0.69 × 104 vs. 0.07 ± 0.03.5 × 104; P = 0.0086; n = 3; Fig. 3 B and C). Such dramatic reduction in number of iTDPKO cells following treatment with 4-HT can be observed in three independent iTDPKO clones. Our data suggest that TDP-43 is essential for ES cell survival and proliferation and offers an explanation for our failure to acquire Tardbp−/− pups from intercrosses of Tardbp+/− mice.
Fig. 3.
TDP-43 is required for proliferation and survival of ES cells. (A) Targeting strategies for generation of inducible Tardbp-null (iTDPKO) ES cells. iTDPKOES cells contain one floxed Tardbp allele (floxed TDP) and one disrupted Tardbp allele (iCre), whereas cTDP ES cells contain one WT allele and one disrupted Tardbp allele (iCre). PCR analysis identified an 159-bp (denoted by A) or a 305-bp (denoted by B) fragment corresponding to the WT or floxed allele, respectively. (B and C) 4-HT–treated iTDPKOES cells fail to proliferate and undergo apoptosis. Photomicrographs (B) and survival (C) (three independent pairs, mean ± SEM) of the ES cells after 6 and 9 d of treatment with 4-HT. (Scale bar: 300 μm.)
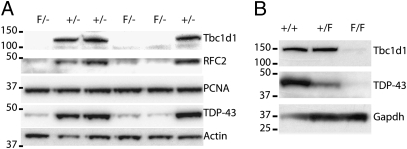
Fig. 4.
Validation of targets identified through deep sequencing analysis. (A) Protein blot analysis of Tbc1d1 and rfc2 from extracts of iTDPKO ES cells treated with 4-HT. Note the dramatic reduction in levels of TDP-43 whereas PCNA is similar in iTDPKO compared with cTDP ES cells. Antisera against actin used as a loading control. (B) Protein blot analysis of Tbc1d1 (Upper) and Tardbp (Middle) in the skeletal muscles of control, CAG-ErCre;Tardbp+/F, and CAG-ErCre;TardbpF/F mice. Note the depletion of TDP-43 and Tbc1d1 in muscle of mice lacking Tardbp. (*P < 0.05, **P < 0.01, ***P < 0.001.)
High-Throughput DNA Sequencing Revealed Downstream Targets of Tardbp.
To identify the downstream targets of TDP-43, the iTDPKO and cTDP ES cells were induced by 100 ng/mL of 4-HT for 3 d and total mRNA were isolated for RNA-seq analysis by Illumina genome analyzer. The raw reads were mapped onto the mm9 mouse genome using the public-domain Efficient Large-Scale Alignment of Nucleotide Databases, and these data were analyzed by the Partek software to identify a set of differentially expressed genes (Table S1). The dramatic reduction in level of Tardbp mRNA (Table S1) validated the deletion of Tardbp in the ES cells and the methodology of RNA-seq analysis. Significantly, protein blot analysis revealed that levels of Rfc2 and Tbc1d1, two of the top hits (Table S1), were nearly abolished in iTDPKO cells (Fig. 4A, upper two panels). Interestingly, among the top 30 hits (i.e., genes with the lowest P values and more than threefold change; Table S1), most (26 of 30) of the genes are down-regulated, suggesting that TDP-43 plays an important role in elevating RNA transcription or maintaining RNA stability. As Rfc2 is a constitutively expressed protein essential for both DNA repair and replication (24), the dramatic reduction of this protein in the highly replicating ES cells would provide an explanation for the observed lethality occurring in ES cells lacking Tardbp (Fig. 3C).
Marked Reduction of Tbc1d1 in Skeletal Muscle of Conditional Tardbp-KO Mice.
Tbc1d1, a critical protein associated with human obesity (19, 25), was drastically reduced in the Tardbp deleted iTDPKO cells (−7.35 fold, P = 7.02E−228). As it has been reported that a nonfunctional Tbc1d1mutant in the skeletal muscle is responsible for the lean phenotype in mice and thatTbc1d1is essential for Glut4 translocation to the plasma membrane of skeletal muscle cells for glucose uptake (18), a decrease inTbc1d1 in skeletal muscle might offer an explanation for the lean phenotype shown in our conditional Tardp-KO mouse model (Fig. 2). To test this notion, we assessed the levels of Tbc1d1 in the skeletal muscles of the control, CAG-ErCre;Tardbp+/F, and CAG-ErCre;TardbpF/F mice fed with tamoxifen. Protein blot analysis of muscle extracts showed depletion of Tbc1d1 in CAG-ErCre;TardbpF/F mice (Fig. 4B) that exhibited marked reduction of fat (Fig. 2 E and F and Fig. S3). In addition, correlating with the reduction of TDP-43 protein level (slope = −0.3714; P = 0.0015, linear trend post test; Fig. S5A), there is a trend toward reduction of Tbc1d1 in muscles of CAG-ErCre;Tardbp+/F mice, (slope, −0.4256; P = 0.0003, linear trend post-test; Fig. S5B), consistent with the mild loss of body weight in the CAG-ErCre;Tardbp+/F mice (Fig. S3A). As our RNA-seq analysis demonstrated that Tardbp deletion is responsible for the marked reduction of Tbc1d1 coupled with the previous report showing the functional requirement of Tbc1d1 in skeletal muscle for the maintenance of fat content in mice (18), our data are consistent with the view that the postnatal deletion of Tardbp led to the lean phenotype through reduction of level of Tbc1d1 protein in the muscle of conditional Tardbp-null mice.
In summary, our efforts here in using conditional Tardbp KO strategies revealed a physiological role for TDP-43 in regulating body fat and identified one specific target, Tbc1d1, a gene associated with human obesity (19, 25), regulated by TDP-43. These unexpected observations indicate that TDP-43 plays a crucial role in controlling energy metabolism. Although the loss of TDP-43 could lead to lethality in some cell types like ES cells, it is possible that the temporally and spatially controlled reduction of TDP-43 would provide a potential therapeutic avenue for obesity. Our conditional Tardbp-KO mice would offer a useful animal model to examine the effects of tissue specific reduction of TDP-43 to impact on obesity. Moreover, our RNA-seq analysis disclosed a cohort of downstream targets of TDP-43, a subset of which would be directly regulated by TDP-43. These findings open the possibility for discovering a series of direct targets of TDP-43, information that will provide insights not only into the cellular biology and physiology of TDP-43, but also to the pathophysiology of TDP-43 in the future. Interestingly, that down-regulation of level of Tbc1d1 in the iTDPKOES cells was validated in skeletal muscle of adult Tardbp-null mice suggest that such possibility could be observed for motor neurons or their surrounding glia cells. If so, it would be possible to identify transcripts/proteins critical for the understanding of the pathogenesis of ALS or FTLD through comparison of ES cell transcriptome with those derived from cells of the CNS in disease models associated with ALS or FTLD. That some individuals with ALS showed a hypermetabolic state (26) raised the possibility that hypermetabolism may contribute to the pathogenesis of ALS, and it will be of interest to test whether altered fat metabolism participates in motor neuron degeneration in ALS using our conditional Tardbp-KO mouse model. In parallel, CNS-specific deletion of Tardbp using our conditional mouse model will be invaluable for providing further insights into the physiological role of TDP-43 in the nervous system.
Materials and Methods
Gene Targeting and ES Cell Culture.
The Tardbp gene, isolated from a C57BL/6 genomic BAC clone (RP23-331p21; BACPAC), was characterized by a series of restriction enzyme and generated the targeting vector by DNA recombineering. In the Tardbp targeting vector, the long (7 kb) and short (2.5 kb) arms encompassing exons 1 to 4 of the Tardbp gene were inserted into the PGKneoF2L2DTA vector. The linearized Tardbp targeting vector was electroporated into v26.2 C57BL/6j embryonic stem cells (Open Biosystems), and targeted clones were screened by Southern blot analysis using a flanking probe generated by PCR using mouse DNA via primers 5′-atgtgttgggttacaggcgtgccta and 5′-tgacaactgtataatgggagatggcacag. Two independent targeted clones were injected into albino C57BL/6j mouse blastocysts to generate Tardbp chimeric mice.
The same Tardbp-targeted ES cells used for blastocyst injection were adapted to serum-free culture (2i) conditions (27) for subsequent targeting using a modified pCAG-ERT2CreERT (Addgene 13777) vector (28, 29), with identical homology arms to the Tardbp targeting vector, into the WT or floxed Tardbp allele to generate, respectively, the iTDPKO or cTDP inducible ES cell lines. The correct targeting was validated by PCR analysis using primer sets 5′-AACTTCAAGATCTGACACCCTCCCC and 5′- GGCCCTGGCTCATCAAGAACTG. The predicted PCR product for WT or floxed alleles is 159 (Fig. 3A) or 305 bp (Fig. 3 A and B), respectively. The absence of WT or floxed alleles would represent the identification of iTDPKO or cTDP ES cell clones, respectively. The targeted clones were further validated by protein blot analysis using an antisera specific for TDP-43. Three independent ES cell lines were chosen and established for subsequent analyses.
Mouse Breeding.
The targeted F1 pups were crossed with a CAG-Cre mouse line (21) to generate the standard Tardbp-KO mice lacking exon 3. The same targeted F1 pups were also bred with hACTB-flp mice (20) to remove the neomycin resistance gene cassette to generate the floxed Tardbp (Tardbp+/F) mice. DNA extracted from tail clips of mice were genotyped by PCR using the following sets of primers: 5′-AACTTCAAGATCTGACACCCTCCCC and 5′- GGCCCTGGCTCATCAAGAACTG. The resulting Tardbp+/F mice were then crossed with CAG-ErCre (Jax 004682) (23),or Rosa26-ErCre (Jax 004847) (30) mice to generate independent lines of tamoxifen-inducible Tardbp-KO mice. For genotyping of germline KO mice, the following set of primers was used: 5′-TCTTACAATGCCTGGCGTGGTG and 5′- CGTGGTTGCGCACCCTAACTATAA. All in vivo experiments were approved by The Johns Hopkins University Animal Care and Use Committee.
Metabolic Assessments in Vivo and Body Fat Composition Analysis.
ErCre;TardbpF/F mice, ErCre;Tardbp+/F mice, and controls at 4 to 6 wk of age (n = 5 per group) were used for simultaneous assessments of daily body weight change, energy intake (corrected for spillage), and whole-body metabolic profile by indirect calorimetry. Mice without ErCre or with only ErCre (without floxed Tardbp) were used as control. Mice previously maintained on standard rodent chow (no. 2018; Harlan-Teklad) were tested in an open-flow indirect calorimeter (Oxymax Equal Flow; Columbus Instruments). Data were collected for 2 d on baseline diet to confirm acclimation to the calorimetry chambers, and then for the subsequent 8 d on the gene induction diet (no. 2016; Harlan-Teklad), supplemented with tamoxifen (400 mg/kg) and sucrose (5% by weight; diet TD07262; 3.1 kcal/g, 19.7% kcal protein, 70.5% kcal carbohydrate, 9.8% kcal fat). This level of tamoxifen (31) in the diet is consistent with diet-based dosings described previously (31) and produced similar initial reductions in body weight, necessitating extension of the study for 8 d to acquire more stable, gene-based metabolic data. Rates of oxygen consumption (VO2, mL/kg/h) and CO2 production (VCO2) were measured for each chamber every 16 min throughout the study. RER (i.e., VCO2/VO2) was calculated by Oxymax software (v. 4.02) to estimate relative oxidation of carbohydrate (RER of 1.0) versus fat (RER approaching 0.7), not accounting for protein oxidation. Energy expenditure was calculated as VO2 × [3.815 + (1.232 × RER)] (22), and normalized for subject body mass (kcal/kg/h). Average metabolic values were calculated per subject, for each day, and averaged across subjects for statistical analysis. On day 8 of tamoxifen diet feeding, mice were euthanized and carcasses were subjected to quantitative NMR (Echo-MRI 100) at the Phenotyping Core at The Johns Hopkins University to determine lean and fat masses, which were expressed as percent body mass.
Histological, Immunohistochemical, and Protein Blot Analysis
The tissue fixation and processing, histological, immunohistochemical staining, biochemical analysis and tissue harvest were performed according to standard protocols previously established in our laboratories. Antibodies used for immunohistochemical or protein blot analysis were anti-Tbc1d1, anti-ATGL, and anti–PPAR-γ (Cell Signaling); anti-GAPDH, anti–α-tubulin, and anti-actin (Sigma-Aldrich); and anti-PCNA (Invitrogen), anti-Tardbp, and anti-RFC2 (Proteintech). Bands were quantified by Quantity One software and normalized by GAPDH or actin level before statistical analysis.
RNA-Seq.
Independent clones of iTDPKO and cTDP were treated with 100 ng/mL 4-HT for 72 h. Total RNA, harvested from approximately 5 × 106 cells using a miniRNA kit (Qiagen), was subjected to poly(A)+ RNA selection with oligo-dT magnetic beads (Invitrogen). Procedures recommended by Illumina were used for DNA library construction. Raw reads were mapped to the University of California Santa Cruz mm9 genome library by Efficient Large-Scale Alignment of Nucleotide Databases and the Partek genomic suite was used to generate the set of differentially expressed genes (Table S2).
Biological Pathway Analysis.
Analysis of the TDP43 KO versus control differentially expressed gene set (P < e−20) was performed using the online analysis toolkit WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt). WebGestalt was used to search for enriched biological pathways in the open-source Kyoto Encyclopedia of Genes and Genomes database. Using the entire gene set obtained from RNA-seq as a reference set, P values were calculated with the hypergeometric test. Pathways with at least four genes were selected. Red text indicates P values lower than 0.05.
Statistical Analysis.
One-way ANOVA with Dunnett post analysis were used to compare quantity measured among groups. All appropriate numbers indicate measurements from independent animals, and bars on top of each bar indicate SEM. For Fig. 2 A and D and Figs. S3A and S5 A and B, linear trend post-tests were used to show the dosage effect in the heterozygotes, although the reduction in heterozygotes was not significant in Figs. S5 A and B.
Supplementary Material
Acknowledgments
We thank X. Shan and V. Nehus for technical support; X. Shan, J. Pevsner, and J. Nathans for helpful discussions and critical reading of the manuscript; P. Soriano (Mount Sinai School of Medicine of New York University) for the knockout vector; the Biological Resource Branch at the National Institutes of Health for providing the recombineering set of reagents; and Tufts University Core Facility for the high-throughput sequencing service. This study was supported in part by the Johns Hopkins Neuropathology Consolidated Gift Fund, the Muscular Dystrophy Association (P.C.W.), the Robert Packard Center for ALS Research (P.C.W.), and National Institute of Neurological Disorders and Stroke Grant R01 NS41438 (to P.C.W.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE21993).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002176107/-/DCSupplemental.
References
- 1.Ou SH, Wu F, Harrich D, García-Martínez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 3.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 4.Bose JK, Wang IF, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18(R2):R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: The FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 9.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 11.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wils H, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiesel FC, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2009;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu LS, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2009;15:15. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 16.Sephton CF, et al. TDP-43 is a developmentally-regulated protein essential for early embryonic development. J Biol Chem. 2009;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kraemer BC, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chadt A, et al. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet. 2008;40:1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- 19.Meyre D, et al. R125W coding variant in TBC1D1 confers risk for familial obesity and contributes to linkage on chromosome 4p14 in the French population. Hum Mol Genet. 2008;17:1798–1802. doi: 10.1093/hmg/ddn070. [DOI] [PubMed] [Google Scholar]
- 20.Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 22.Lusk G. The Elements of the Science of Nutrition. 4th Ed. Philadelphia: Saunders; 1928. [Google Scholar]
- 23.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 24.Reynolds N, Fantes PA, MacNeill SA. A key role for replication factor C in DNA replication checkpoint function in fission yeast. Nucleic Acids Res. 1999;27:462–469. doi: 10.1093/nar/27.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stone S, et al. TBC1D1 is a candidate for a severe obesity gene and evidence for a gene/gene interaction in obesity predisposition. Hum Mol Genet. 2006;15:2709–2720. doi: 10.1093/hmg/ddl204. [DOI] [PubMed] [Google Scholar]
- 26.Dupuis L, Oudart H, René F, Gonzalez de Aguilar JL, Loeffler JP. Evidence for defective energy homeostasis in amyotrophic lateral sclerosis: Benefit of a high-energy diet in a transgenic mouse model. Proc Natl Acad Sci USA. 2004;101:11159–11164. doi: 10.1073/pnas.0402026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badea TC, Wang Y, Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci. 2003;23:2314–2322. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiermayer C, Conrad M, Schneider M, Schmidt J, Brielmeier M. Optimization of spatiotemporal gene inactivation in mouse heart by oral application of tamoxifen citrate. Genesis. 2007;45:11–16. doi: 10.1002/dvg.20244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.