Abstract
The nonclassical MHC class I-related (MHC-I) molecule HFE controls cellular iron homeostasis by a mechanism that has not been fully elucidated. We examined the regulation of HFE by K5, the E3 ubiquitin ligase encoded by Kaposi's sarcoma-associated herpesvirus (KSHV/HHV8), that is known to down-regulate classical MHC-I. K5 down-regulated HFE efficiently, using polyubiquitination of the membrane proximal lysine in the HFE cytoplasmic tail (K331), to target the molecule for degradation via ESCRT1/TSG101-dependent sorting from endosomes to multivesicular bodies (MVBs)/lysosomes. In the primary effusion lymphoma cell line BC-3, which carries latent KSHV, HFE was degraded rapidly upon virus reactivation. HFE was ubiquitinated on lysine-331 in unactivated BC-3 cells, conditions where K5 was not detectable, consistent with an endogenous E3 ubiquitin ligase controlling HFE expression. The results show regulated expression of HFE by ubiquitination, consistent with a role in cellular iron homeostasis, a molecular mechanism targeted by KSHV to achieve a positive iron balance.
Keywords: hemochromatosis, iron, immunology
Iron is an essential cofactor for host cells and pathogenic microorganisms, but causes cellular damage when in excess. Molecular mechanisms have evolved to maintain optimal iron levels in host cells while restricting the supply to pathogens, which also require iron for growth and replication (1, 2).
The major histocompatibility complex class I-related (MHC-I) molecule HFE (3) regulates iron homeostasis at the cellular level and systemically (4). Initial studies were consistent with HFE forming a complex with β2 microglobulin (β2m) and transferrin receptor 1 (TfR1) in the endoplasmic reticulum. Upon trafficking to the cell surface, HFE recycled in an endosomal compartment of intermediate pH and competed with transferrin for binding to TfR1 to limit cellular iron uptake (5–7). At physiological concentrations of iron-bound transferrin (10 μM holo-Tf), it was shown that HFE could not compete for binding to TfR1 because of its lower affinity, but that cellular iron levels were reduced by HFE expression. HFE therefore appeared to be regulating some aspect of iron import in the endosome, independent of TfR1 binding (8–10). HFE may also regulate iron export in some cell types (11, 12).
HFE regulates iron levels systemically by influencing expression of the peptide hormone hepcidin (13). This pathway is mediated by BMP6 and SMAD4 signaling in response to increased iron, proinflammatory stimuli, and hypoxia (14, 15). Hepcidin binds to ferroportin, an iron efflux channel expressed in intestinal epithelium, hepatocytes, and macrophages, thereby promoting ferroportin degradation, so that iron export is prevented (16, 17). Regulation of hepcidin transcription by hepatocytes required HFE to form a complex with transferrin receptor 2, not TfR1, a switch mediated by holo-Tf, which displaces HFE from TfR1 due to its greater affinity (18, 19).
Kaposi's sarcoma (KS)-associated herpesvirus (KSHV or HHV8) encodes two proteins, K3 and K5, which regulate MHC-I (20). Areas of endemic KS in Africa correlate with red–brown volcanic soils rich in iron, suggesting that iron represents an environmental variable in KS pathogenesis (21, 22). Iron is also associated with KS lesions by immunohistochemistry (23).
Given the homology of HFE to MHC-I, we asked whether K3 and K5 could regulate HFE. We show that K5, but not K3, down-regulated surface expression by targeting lysine-331 in the cytoplasmic tail of HFE for ubiquitination, leading to lysosomal degradation. In BC-3 cells, wild-type HFE was degraded rapidly upon viral reactivation, whereas an HFE K331R mutant line was unaffected. HFE was ubiquitinated on lysine-331 in unactivated BC-3 cells, providing evidence for an endogenous E3 ligase with characteristics similar to K5. The results are consistent with control of cellular iron homeostasis by ubiquitinated HFE, an endogenous pathway co-opted by KSHV to increase cytosolic iron levels required for lytic infection.
Results
Lysine 331 Is Required for K5-Mediated Regulation of HFE.
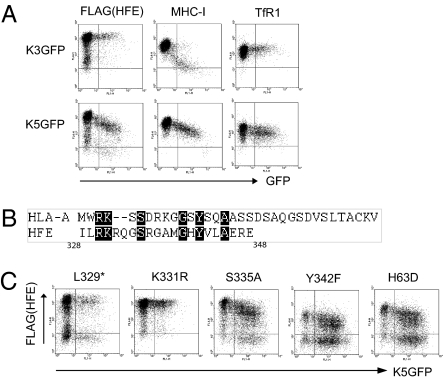
HeLa-M cells expressing FLAG-HFE were cotransfected with plasmids for K3 and K5 fused to GFP. Analysis by flow cytometry showed that both K3 and K5 down-regulated MHC-I, but that only K5 and not K3 regulated FLAG-HFE (Fig.1A).
Fig. 1.
HFE down-regulation by K5 requires lysine-331. (A) HeLa-M cells expressing FLAG-tagged HFE were transiently transfected with K3 and K5 as GFP fusions. After 48 h, cells were stained with anti-FLAG, MHC-I, and TfR1 primary and anti-mouse Alexa 633 secondary antibodies and analyzed by flow cytometry. (B) Comparison of the cytosolic tails of HLA-A2 and HFE identifies conserved amino acids. (C) Conserved residues were modified by in vitro mutagenesis. HFE variants were expressed in HeLa-M cells and cotransfected with K5GFP. Cells were stained with anti-FLAG antibody and analyzed by flow cytometry.
Amino acid alignment of the cytoplasmic tails of HLA-A2 and HFE highlighted six conserved residues (Fig.1B). We considered the conserved lysine K331 the likely target of K5. The more distal lysine in the HLA-A2 sequence, K340, shown to be the target for K3-mediated regulation, is not conserved (24). The other conserved residues S335 and Y342 may be targets for phosphorylation or trafficking motifs. We modified these amino acids and those associated with hemochromatosis, C282Y and H63D, by site-directed mutagenesis and tested regulation by K5. A stop codon was introduced to produce a truncated HFE molecule lacking the cytoplasmic tail, L329*. Anti-FLAG antibody staining in HFE variants cotransfected with K5GFP showed that S335A, H63D, and Y342F were sensitive to regulation by K5, but that K331R and L329* were resistant (Fig.1C). Therefore the membrane proximal lysine residue K331 was identified as the critical determinant of K5 regulation of HFE.
Increased Binding and Uptake of Transferrin in K5 Cells.
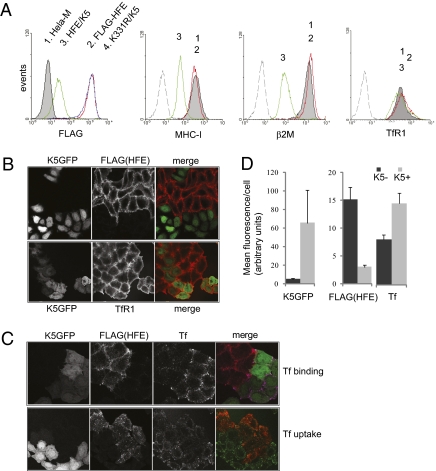
TfR1 expression was not significantly affected by transient transfection of K5 in HeLa-M cells expressing FLAG-HFE (Fig.1A). Using stable lines expressing FLAG-HFE and K5, HFE K331R/K5, and HFE wild-type alone, we compared staining of anti-FLAG, anti-MHC-I, anti-TfR1, and anti-β2m (Fig. 2A). Expression of untagged K5 in the lentiviral system was monitored using a second promoter driving GFP. Anti-FLAG staining on HFE/K5 cells was decreased by two orders of magnitude in comparison with cells without K5. Surface staining was rescued completely in K331R/K5 cells, confirming the potent effect of K5 on HFE and the critical dependence of lysine-331. MHC-I and β2m stainings were both decreased by an order of magnitude in K5+ cells. Flow cytometry (Fig. 2A) and confocal microscopy (Fig. 2B) confirmed that TfR1 was not affected significantly either by HFE expression or by K5 down-regulating HFE, as only slight variation in surface TfR1 was detected. Binding and uptake of Tf on the other hand were increased in K5+ cells (Fig. 2 C and D). Therefore, HFE competed effectively with Tf (at 60 nM concentration) for binding to TfR1, consistent with published reports (9). K5-mediated regulation of HFE relieved this competing interaction, allowing for increased Tf uptake.
Fig. 2.
K5 down-regulation of HFE but not TfR1. (A) Comparison of surface staining for anti-FLAG, MHC-I, β2m, and TfR1 antibodies by flow cytometry. Cell lines: 1, HeLa-M (shaded histogram); 2, HeLa FLAG-HFE plus empty lentiviral vector (red); 3, HFE/K5 (green); 4, K331R/K5 (blue). Dashed histogram, isotype control. (B) A mixed population of FLAG-HFE and HFE/K5 HeLa-M cells were seeded on coverslips. After 24 h, cells were fixed and stained with anti-FLAG (Upper) and anti-TfR1 (Lower) and anti-mouse Alexa 568 antibodies and then analyzed by confocal microscopy. Merged images: K5/GFP (green) and anti-FLAG and TfR1 (red). (C) FLAG-HFE and HFE/K5 HeLa-M cells growing on coverslips were processed for confocal microscopy as in B, except that Alexa 633 holo-transferrin (60 nM Tf) was used. (Upper) Anti-FLAG and transferrin binding to fixed cells, with K5GFP. (Lower) Anti-FLAG and transferrin uptake after 1 h at 37 °C. Cells were then fixed and permeabilized, and then secondary anti-mouse Alexa 568 antibody was applied. Merged images: (Upper) K5/GFP (green), anti-FLAG (red), and transferrin (purple); (Lower) anti-FLAG (red) and transferrin (green). (D) Images from C were analyzed using NIH Image software and relative fluorescence per cell was determined. Anti-FLAG staining in K5+ cells represented a decrease in relative fluorescence of 80% over K5-negative cells. An increase in Tf binding of 40% was detected in K5+ cells. Error bars represent SD by analysis of five fields per image.
K5 Targets HFE “High” for Degradation in a Post-Golgi Compartment.
No change in FLAG-HFE protein levels was detected by immunoblot (IB) in lysates from HFE cells ± K5 (Fig. S1A), although by microscopy, surface HFE was profoundly reduced (Fig. S1B). By this analysis, K5 removed specifically surface HFE without affecting total protein.
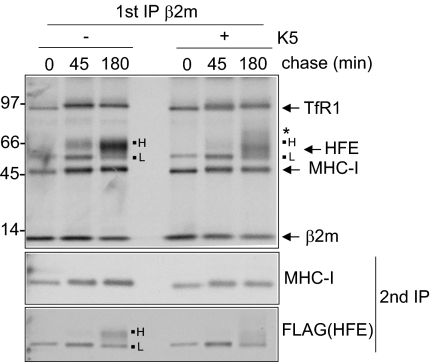
Maturation of HFE and MHC-I through the secretory pathway was examined by immunoprecipitation (IP) from metabolically labeled cells using antibody to β2m. TfR1, HFE, MHC-I, and β2m were recovered at the expected molecular weights (Fig. S1C). Two forms of HFE, low (L) and high (H), were detected, with conversion to the higher molecular weight (MW) form over time. HFE H was endoglycosidase H resistant (Fig. S1C), but PNGaseF sensitive (Fig. S1D), consistent with HFE glycosylation at the three predicted N-linked sites, in the endoplasmic reticulum, with further maturation occurring in the Golgi. Comparison of HFE and HFE/K5 cells by β2m IP showed that only HFE H was disrupted in K5+ cells (Fig. 3), particularly evident at the 45-min time point, where HFE H was absent. At 180 min the HFE H band was reduced in intensity, with slight smearing toward higher MW forms not seen in K5-negative cells. The data show that HFE/β2m/TfR1 complexes formed and trafficked normally in K5 cells, consistent with degradation of HFE high in a post-Golgi compartment.
Fig. 3.
K5 targets HFE “high” from a post-Golgi compartment. FLAG-HFE plus empty lentiviral vector and HFE/K5 cells were metabolically labeled and subjected to primary IP using anti-β2m at different chase time points and then secondary IP using anti-MHC-I and anti-FLAG antibodies and analyzed by SDS/PAGE. The appearance of the HFE high form was disrupted by K5 expression, with smearing toward higher molecular weight forms (indicated by *). Size markers are in kilodaltons.
HFE Is Ubiquitinated on K331 and Degraded in Lysosomes.
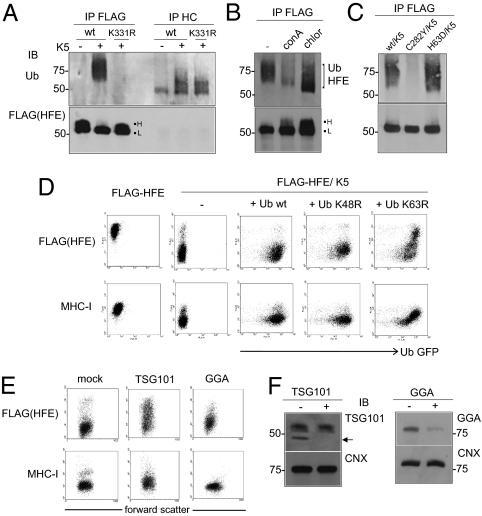
The smearing detected in HFE/K5 cells at the 180-min time point was consistent with HFE ubiquitination. We looked for ubiquitination of HFE in K5 cells by IP using anti-FLAG antibody and IB for ubiquitin (Fig. 4A). HC10 antibody was used similarly for the detection of ubiquitinated MHC-I. The ladder of higher MW bands indicative of polyubiquitination was detected for both MHC-I and FLAG-HFE in K5 cells. HFE ubiquitination was dependent on lysine-331 because the K331R variant was not ubiquitinated (Fig. 4A Upper). IB of anti-FLAG IPs showed that only HFE H was degraded (Fig. 4A Lower).
Fig. 4.
HFE is ubiquitinated on K331 and targeted to lysosomes. (A) IP from HeLa-M lysates of HFE, HFE/K5, and K331R/K5 cell lines with anti-FLAG and HC10 antibodies, followed by IB using anti-ubiquitin (Ub). (Upper) A smear characteristic of polyubiquitination was detected for both MHC-I and HFE in K5+ cells. The K331R variant was not ubiquitinated. Blots reprobed with rabbit anti-FLAG as a loading control showed that only HFE “high” was degraded (Lower). (B) Detection of ubiquitinated HFE in the presence of lysosomal inhibitors chloroquine (chlor) and concanamycin A (conA). HeLa-M cells expressing FLAG-tagged wild-type HFE and K5 were left untreated or incubated with appropriate inhibitors for 24 h. Cell lysates were prepared and analyzed by IP using anti-FLAG and then IB with anti-Ub HRP. (C) Ubiquitination by K5 of HFE variants associated with hemochromatosis. Lysates were prepared from HeLa-M cells expressing FLAG-tagged HFE wild type, C282Y, and H63D together with K5 and analyzed by IP using anti-FLAG antibody and IB using anti-Ub HRP. No modification of C282Y was detected, consistent with its trafficking defect and our proposed model of K5 action. H63D was ubiquitinated, with more ubiquitination detected compared with wild type. (D) Cell lines expressing HFE/K5 cotransfected with plasmids containing wild type, K48R, and K63R mutant ubiquitin (Ub) were stained with anti-FLAG and MHC-I and anti-mouse Alexa 633 secondary antibody and analyzed by flow cytometry. The Ub construct in the lentiviral vector was tagged with GFP at the C terminus, providing a measure of Ub expression. (E) Targeting of ubiquitinated HFE by ESCRT1/TSG101, but not by GGAs. HFE/K5 cells were transfected with TSG101 or GGA siRNA. After 48 h, cells were stained with anti-FLAG and MHC-I and then anti-mouse Alexa 633 antibodies and analyzed by flow cytometry. (F) Total cell protein (10 μg) of HFE/K5 cells ± TSG101 and GGA siRNA analyzed by IB, showing effective knockdown of TSG101 (expected 47 kDa) and GGA proteins. IB using anti-calnexin (CNX), used as a loading control, shows that CNX was unaffected by siRNA knockdown.
Ubiquitination of classical MHC-I by K3 leads to degradation in lysosomes (20, 25). To determine whether HFE shared this fate, HFE/K5 cells were treated with lysosomal inhibitors and the effect on HFE investigated (Fig. 4B). Treatment with chloroquine was effective at rescuing HFE H, correlating with accumulation of a lower MW band, consistent with monoubiquitinated HFE. Similar results were obtained using ammonium chloride, but the inhibitor of proteosomes MG132 was ineffective (Fig. S2). These results were consistent with the targeting of ubiquitinated HFE for degradation in lysosomes. Treatment with concanamycin A (conA), a V-type ATPase inhibitor, was less effective, with less ubiquitination detected in conA-treated cells (Fig. 4B). ConA acts by preventing the acidification of endosomes, implying that effective ubiquitination required endosome acidification.
Lysine-63 Linked Polyubiquitinated HFE Recruits TSG101.
K5 promotes complex lysine-63 linked polyubiquitin chains to down-regulate MHC-I (26). To see whether HFE was regulated similarly, we used mutant ubiquitin (Ub) molecules to outcompete endogenous Ub (Fig. 4D). Rescue of surface FLAG-HFE and MHC-I was detected with Ub K63R, but not with wild-type or K48R Ub variants. We concluded that HFE molecules were targeted by K5 for lysine-63 linked polyubiquitination.
Lysine-63 linked ubiquitination typically targets cell surface receptors to endolysosomes via the ESCRT pathway. The three protein complexes ESCRTI, -II, and -III are recruited sequentially to endosomal membranes by ubiquitinated cargo, driving internalization into multivesicular bodies (MVBs), which fuse with lysosomes (27). A key step is recruitment of ESCRT1/TSG101. To determine whether ubiquitinated HFE was targeted in this way, we used siRNA to TSG101 (24). FLAG-HFE and MHC-I molecules were restored to the cell surface by TSG101 knockdown, implicating the ESCRT pathway in the regulation of ubiquitinated HFE (Fig. 4 E and F). An alternative model invokes sorting of ubiquitinated cargo from the Golgi by GGAs (Golgi-localized, γ-ear-containing, Arf-binding proteins) (28). siRNA targeting GGAs did not significantly affect cell surface FLAG-HFE or MHC-I expression in K5 cells (Fig. 4 E and F).
Reactivated KSHV Targets K331 to Degrade HFE.
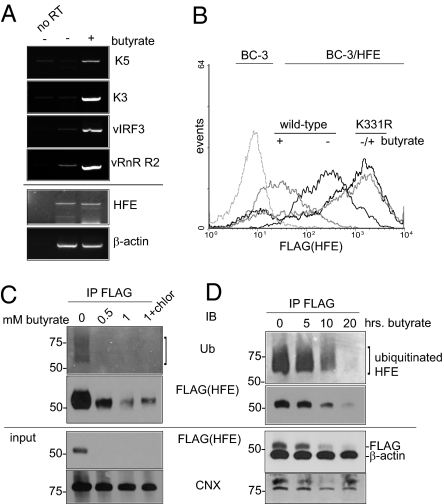
The effect of lytic KSHV on HFE was examined in the primary effusion lymphoma cell line BC-3, which is latently infected with KSHV. Sodium butyrate treatment of BC-3 cells was used to induce expression of lytic cycle genes, including K5 and K3 (Fig. 5A). KSHV transcripts vIRF3 and vRnR R2 subunit were also strongly induced, but endogenous transcripts β-actin and HFE were not. BC-3 cells expressed low levels of HFE mRNA, but no antibody is available to detect endogenous HFE protein. Therefore we created FLAG-HFE wild-type and K331R BC-3 cells. Butyrate treatment of BC-3/HFE wild-type cells showed down-regulation of anti-FLAG staining compared with untreated cells, whereas HFE K331R cells were unaffected (Fig. 5B). FLAG(HFE) staining was not decreased by butyrate treatment of cell lines that do not harbor KSHV (Fig. S3). We noted that surface FLAG staining was not comparable in untreated cells, as wild-type HFE exhibited intermediate expression compared with K331R. Expression of FLAG-HFE in this lentiviral system was monitored using a second promoter driving GFP, which showed no variation (Fig. S4).
Fig. 5.
Lytic KSHV degrades HFE by targeting K331. (A) Primary effusion lymphoma BC-3 cells untreated or treated with sodium butyrate (1 mM) to induce lytic phase KSHV. After 24 h, total RNA was prepared. Transcript expression was analyzed by RT-PCR and visualized by agarose gel. A no reverse transcriptase enzyme control (no RT) was included to show that transcript detection was dependent on first-strand cDNA synthesis. (B) BC-3/HFE wild-type and K331R cells, left untreated (−) or treated (+) with butyrate (1 mM) for 24 h, were analyzed for binding of anti-FLAG antibody by flow cytometry. Dashed line, binding of FLAG antibody to parental BC-3 cells. (C) Detection of ubiquitinated HFE in KSHV positive cells. Shown are BC-3/FLAG-HFE wild-type cells left untreated, treated with butyrate (0.5 and 1 mM), or treated with butyrate and chloroquine (1 mM and 10 μM, respectively). After 24 h, cell lysates were subjected to anti-FLAG antibody IP, followed by IB with anti-Ub. IB using rabbit anti-FLAG showed that HFE was degraded efficiently in butyrate-treated cells. IB for anti-CNX was used as a loading control. (D) BC-3/HFE wild-type cells treated with butyrate (1 mM) for 0, 5, 10, and 20 h, analyzed by anti-FLAG IP and anti-Ub IB as in C. IBs for β-actin and CNX were used as controls.
HFE Is Ubiquitinated in Unactivated BC-3 Cells.
We tested the effect of butyrate on HFE ubiquitination, by IP from BC-3/HFE wild-type cells, left untreated, treated with butyrate, or treated with butyrate and chloroquine. We detected ubiquitinated HFE in untreated BC-3 cells (Fig. 5C). This observation may indicate low K5 expression in unactivated cells, although K5 transcripts were not detected by RT-PCR (Fig. 5A). Alternatively, an endogenous E3 ligase could be ubiquitinating HFE. Such an activity would explain the intermediate expression detected by flow cytometry of wild-type HFE compared with K331R (Fig. 5B and Fig. S4). An endogenous E3 ligase could be acting upon K331 to control HFE expression, a characteristic distinct from K5, which targets HFE for ubiquitination and degradation. In contrast, no FLAG-HFE was detected by IB in butyrate-treated cells, showing that HFE was degraded efficiently by lytic KSHV. By IB of anti-FLAG IP, protein was detectable, with degradation of HFE by increasing butyrate concentration. Chloroquine rescued some protein as assessed by band intensity relative to control IB for calnexin (CNX).
In similar IP experiments, we examined the effect of 1 mM butyrate over a 20-h time course (Fig. 5D). We detected ubiquitinated HFE in untreated BC-3/HFE wild-type cells and degradation of HFE over time, compared with IB for β-actin and CNX. These data are consistent with an endogenous E3 ligase ubiquitinating HFE in untreated BC-3 cells, with K5 targeting HFE for degradation upon KSHV induction. Alternatively, ubiquitination in untreated cells is by K5 and other factors are induced by butyrate, which targets HFE molecules for degradation.
Regulation of TfR1 by Iron in HFE Wild-Type and K331R Cells.
KSHV presumably down-regulates HFE to affect iron homeostasis. We looked for evidence for this by monitoring changes in TfR1 in BC-3 cells. TfR1 expression is controlled at the posttranscriptional level by cytosolic iron acting to modulate the IRE-BP system (29). Low intracellular iron leads to stabilization of TfR1 mRNA and increased translation (8).
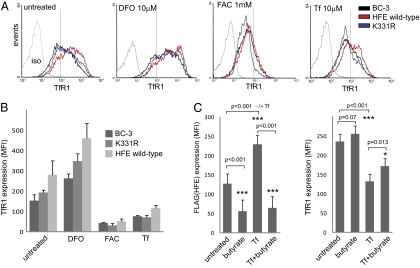
The regulation of TfR1 in BC-3, BC-3/HFE wild-type, and K331R cells, either left untreated or treated with the ferric iron chelator desferrioxamine (DFO), was compared with the response to iron delivery by ferric ammonium citrate (FAC) or holo-transferrin (Tf). Untreated HFE wild-type cells showed increased TfR1 surface expression compared with BC-3 and K331R cells (Fig. 6A Left). This increase in TfR1 was mimicked by DFO treatment (Fig. 6A Center Left) indicative of low cytosolic iron. In response to iron delivery by FAC, the cell lines responded by decreasing TfR1 (Fig. 6A Center Right). In response to Tf, TfR1 levels were again reduced, but the response was less profound in the HFE wild-type cells (Fig. 6A Right). These observations were consistent over multiple experiments (Fig. 6B) and show HFE acting to reduce cytosolic iron, causing TfR1 to be increased, an effect dependent upon HFE K331.
Fig. 6.
Regulation of TfR1 by iron in HFE wild-type and K331R cells. (A) Comparison of TfR1 expression on parental BC-3 and cells expressing wild-type and K331R HFE, left untreated (Left) and treated with desferrioxamine (DFO, Center Left), ferric ammonium citrate (FAC, Center Right), and holo-transferrin (Tf, Right). Cells were treated for 24 h, stained with anti-TfR1 and anti-mouse Alexa 633 antibodies, and analyzed by flow cytometry. Binding of isotype control (iso) is shown. (B) Chart shows mean fluorescence values for TfR1 expression from three experiments carried out as in A (except DFO treatment, which represents duplicate data). Error bars show SD. (C) Effect of lytic KSHV acting in the presence of iron on expression of FLAG(HFE) (Left) and TfR1 (Right). BC-3/HFE wild-type cells were left untreated or treated with butyrate, Tf, or butyrate and Tf together for 24 h; stained with anti-FLAG and anti-TfR1 antibodies; and then analyzed by flow cytometry. Charts show mean fluorescence from triplicate experiments. P values show significance tested by paired Student's t test. A P value <0.05 was considered significant.
Next we looked at the effect on HFE and TfR1 of lytic KSHV acting in the presence of iron. BC-3/HFE wild-type cells left untreated or treated with butyrate, Tf, or butyrate and Tf together were analyzed by flow cytometry using anti-FLAG and TfR1 antibodies (Fig. 6C). FLAG(HFE) expression was decreased by butyrate treatment and increased by Tf, consistent with previous results (Fig. 5 and Fig. S5), but decreased by butyrate acting in the presence of Tf (Fig. 6C Left and Fig. S6). TfR1 was decreased by Tf as expected, but increased slightly by butyrate treatment alone (P = 0.07, not significant) and more particularly by butyrate acting in the presence of Tf (*P = 0.013) (Fig. 6C Right). Similar results were obtained by butyrate acting in the presence of FAC (Fig. S6). TfR1 expression was not increased by butyrate treatment of cell lines that do not harbor KSHV (Fig. S3). These results indicated an iron requirement for lytic KSHV and with the virus targeting HFE to satisfy this demand.
Discussion
We characterized the molecular mechanism whereby K5, the E3 ubiquitin ligase encoded by KSHV, uses ubiquitination and lysosomal degradation to down-regulate HFE. The mechanism overlaps with that of classical MHC-I regulation by K3 and K5 (25, 26). Targeting specificity of these related E3 ligases is dependent upon the position of lysine residues in the cytoplasmic tail relative to the plasma membrane. K5 promotes complex, branched polyubiquitin chain formation to down-regulate MHC-I, which offers further scope for complexity in HFE regulation (26). We showed using butyrate-treated BC-3 cells that lytic KSHV also down-regulated HFE in a K331-dependent manner, which presumably involved K5, although other factors may be involved.
K5 targeted for degradation the fraction of the total cell pool of HFE molecules in the endosomal compartment. Targeting was efficient, because little HFE escaped to recycle to the cell surface. In the absence of K5, HFE inhibited binding and uptake of 60 nM Tf, showing that TfR1 was in complex with HFE at the plasma membrane. In K5 cells, binding of Tf increased, suggesting that K5 was targeting HFE molecules dissociated from TfR1. Structural studies show that binding of the two molecules via their extracellular domains is pH dependent (5). Concanamycin A, which inhibits endosome acidification, prevented effective K5-mediated ubiquitination of HFE (Fig. 4B). Our data therefore suggest a model whereby pH-dependent dissociation of the HFE/TfR1 complex occurs in early endosomes, allowing for K5-mediated ubiquitination on TfR1-free HFE. Instead of recycling to the cell surface, ubiquitinated HFE is sorted to MVBs via ESCRT machinery and degraded in lysosomes.
Down-regulation of classical MHC-I by K3/K5 serves to mask-virus infected cells from surveillance by circulating cytotoxic T and NK cells (30). K5 down-regulation of HFE could be acting similarly to prevent engagement of another receptor. Alternatively, KSHV may manipulate iron levels to aid its replication. Induction of lytic KSHV required iron, as evidenced by the increase in TfR1 on viral induction (Fig. 6C). By inspection of the KSHV genome (NC_009333), the only molecule with an obvious iron requirement is orf60 ribonucleotide reductase R2 subunit (vRnR R2), the enzyme that provides nucleotide substrates for DNA biosynthesis. Transcripts for this gene were induced in lytic phase (Fig. 5A). It is possible that KSHV down-regulates HFE to increase iron availability for DNA biosynthesis.
By comparing the response to iron delivery of wild-type HFE and K331R BC-3 cells, we detected differences in the regulation of TfR1, which was used to monitor changes in cytosolic iron (Fig. 6). The increase in TfR1 by expression of wild-type HFE alone was indicative of low cytosolic iron, presumably by increased iron export. The response of wild-type HFE cells to 10 μM holo-Tf, but not to FAC, also indicated a low iron phenotype compared with parental BC-3 and K331R cells, possibly as a result of controlled iron uptake. Taken together, the results suggest that HFE acts to balance iron import and export, effects dependent on lysine-331 and iron delivery from Tf. The mechanism(s) did not rely on inhibition of Tf binding, because TfR1 was increased by HFE expression in untreated cells. Physiological concentrations of Tf were used, as these have been shown to outcompete HFE for TfR1 binding (6) and K331R cells did not inhibit iron delivery by Tf, despite high cell surface expression. These observations are more consistent with regulation of iron import/export in the endosome, with the transporter DMT1/Nramp2 or ferric reductase being potential targets, as proposed by other workers (6).
Ubiquitination of HFE lysine-331 leading to intermediate HFE expression was detected in unactivated BC-3 cells. Expression of K5 at low levels or of an endogenous E3 ligase with characteristics similar to K5 would account for this observation. More work will be required to resolve these alternatives. HFE protein has been detected in few tissues (31–33) and it will also be important to assess whether HFE ubiquitination represents a general mechanism controlling HFE expression and iron homeostasis in these sites.
Materials and Methods
Antibodies.
Monoclonal M2 and rabbit polyclonal anti-FLAG (Sigma), anti-HLA class I W6/32 and HC10 (lab stocks), anti-β2 microglobulin (Dako), anti-EEA1 (Molecular Probes), anti-TfR1 (Pharmingen), anti-calnexin (Stressgen), anti-TSG101 (Abcam), and anti-ubiquitin HRP (P4D1; Santa Cruz) were used. Secondary antibodies were goat anti-mouse Alexa 488, 568, and 647 and goat anti-rabbit Alexa 488, 568, and 647 (Molecular Probes) and goat anti-rabbit and anti-mouse as HRP conjugates (Dako).
DNA Constructs.
HFE cDNA was cloned into the pFLAG vector (Sigma) using BglII/BamHI to produce FLAG-HFE. Primers were HFE.F agatctgcgttcacactctctgcac and HFE.R ggatcctcactcacgttcagctaagac. HFE variants were produced by site-directed mutagenesis. K3 and K5 as GFP fusion constructs (24) and the pHRsin lentivirus system were used (34). FLAG-HFE was cloned into the pHRsin vector. Internal NotI and BamHI sites were removed by rounds of restriction digestion followed by T4 DNA polymerase treatment to create blunt ends, which were ligated. Resulting inserts were resequenced and then subcloned using BamHI/NotI into pHRsin.
Tissue Culture, Transfection, and Flow Cytometry.
HeLa-M and BC-3 cells were maintained in RPMI1640 medium plus 10% FCS, pen/strep (100 units), and l-glutamine (200 mM). HeLa cells growing in six-well plates were transfected using Fugene (Roche). Stable lines were produced by cell sorting of anti-FLAG positive cells maintained in medium supplemented with G418 (1 mg/mL). FACScalibur was used for flow cytometry. For lentiviral transduction, 293T cells were cotranfected with packaging vectors pCMVR8.91 and pMDG together with pHRsin UbEm lentivirus plasmids. After 48 h, culture supernatants were harvested, filtered (0.2-μm filter), and applied to cells. Reagents for depletion of TSG101 by siRNA were described previously (24). HFE/K5 cells growing in six-well plates in complete medium lacking pen/strep were transfected with 200 pmol TSG101 siRNA (Dharmacon), using Dharmafect 1. After 48 h cells were analyzed by flow cytometry and by IB to confirm TSG101 depletion. Depletion of GGAs was achieved similarly except using Smartpool siRNA (Dharmacon) targeted to GGAs (kindly provided by J. Hirst and M. S. Robinson, Department of Clinical Biochemistry, Cambridge University, Cambridge, UK). For induction of KSHV, BC-3 cells were treated with sodium butyrate. Human holo-transferrin and ferric ammonium chloride were used. Desferrioxamine was a gift from T. M. Cox (Cambridge University, UK).
Metabolic Labeling, Immunoprecipitation, and Immunoblot.
HeLa-M cells were starved for 1 h in methionine/cysteine-free medium and then labeled with [35S]methionine/cysteine for 20 min. Cells were removed to unlabeled complete medium for the indicated times to chase. Cells were lysed in IP buffer [50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 0.5% Triton-X, 2 mM PMSF, 5 mM iodoacetamide, EDTA-free protease inhibitor] for 10 min at 4 °C and precleared using protein A-Sepharose, followed by IP using the indicated antibodies and protein A. For re-IP, sample beads from primary IP were resuspended in 1% SDS, incubated at 70 °C for 5 min, and diluted to 0.1% SDS using IP buffer, before adding secondary antibodies and protein A. Samples were treated with endoglycosidase H or PNGaseF, before analysis on 10% SDS/PAGE. Gels were fixed (10% acetic acid, 20% methanol) and then dried and exposed to X-ray film (Kodak Biomax MR). For IBs, proteins were transferred to PVDF membrane, blocked with dried milk (5% Marvel/PBS, 0.1% Tween20) and incubated for 1 h with primary and HRP-conjugated secondary antibodies. Blots were visualized with ECL reagent. The following inhibitors were used: ammonium chloride (50 mM), chloroquine (100 μM), MG132 (10 μM), and concanamycin A (50 nM). For ubiquitination assays, 2 × 106 cells were lysed in 100 μl IP buffer, containing 1% SDS and 100 units of benzonase, and incubated at 70 °C for 10 min. Lysates were diluted to 1 ml with IP buffer and precleared, followed by IP at 4 °C using anti-FLAG and protein A.
Antibody and Transferrin Binding and Internalization Assay.
HeLa-M cells expressing FLAG-HFE with/without K5GFP were grown on coverslips. Cells were fixed for 10 min (3% parafomaldehyde/1× PBS) and treated for 5 min in 0.1% Triton-X100 before staining with primary and secondary antibodies. Coverslips were mounted (Fluoromount G) and cells were visualized using confocal microscopy (Zeiss LSM510 META), using a 63× objective, and analyzed using LSM Image Browser software (Zeiss) before saving in Adobe Photoshop. For internalization, cells growing on coverslips were stained with anti-FLAG M2 antibody and directly labeled holo-transferrin (Tf Alexa633; Molecular Probes) for 30 min at 4 °C, washed twice with cold 1× PBS, and then incubated for 1 h at 37 °C. Samples were removed into 3% paraformaldehye/1× PBS to fix and washed twice (1× PBS) before staining with appropriate secondary antibody.
RT-PCR.
Total RNA was purified from cell pellets using TriReagent. Transcripts were amplified using the One-Tube RT-PCR System (Stratagene). Primers were K5F tccaaggacgtagaagaggg K5R caccggcttttttgtgggcgc, K3F atggaagatgaggatgttcc K3R ggagacactataagccccatcg, vIRF3F atggcgggacgcaggcttacc vIRF3R gtcatcacatgtaactgaacgc, VRNRF ctgtatacaagcgatcacgacgg VRNRR caaatcgtcagtcacacacgtgg, and HFEF atgggcccgcgagccaggccgg HFER ctcacgttcagctaagacgtag.
Supplementary Material
Acknowledgments
We thank Simon McCallum and Anna Petrunkina–Harrison (MoFlow), Matthew Gratian and Mark Bowen (microscopy), and Dr. M. R. Wills (Department of Medicine, Cambridge University) for help with BC-3 experiments. We also thank Drs. Adrian Kelly and Howard Davidson for helpful discussion. This work was supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003421107/-/DCSupplemental.
References
- 1.Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 2.Drakesmith H, Prentice A. Viral infection and iron metabolism. Nat Rev Microbiol. 2008;6:541–552. doi: 10.1038/nrmicro1930. [DOI] [PubMed] [Google Scholar]
- 3.Feder JN, et al. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 4.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: Molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 5.Lebrón JA, et al. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111–123. doi: 10.1016/s0092-8674(00)81151-4. [DOI] [PubMed] [Google Scholar]
- 6.Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem. 1999;274:9022–9028. doi: 10.1074/jbc.274.13.9022. [DOI] [PubMed] [Google Scholar]
- 7.Giannetti AM, Björkman PJ. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem. 2004;279:25866–25875. doi: 10.1074/jbc.M401467200. [DOI] [PubMed] [Google Scholar]
- 8.Riedel HD, et al. HFE downregulates iron uptake from transferrin and induces iron-regulatory protein activity in stably transfected cells. Blood. 1999;94:3915–3921. [PubMed] [Google Scholar]
- 9.Zhang AS, Davies PS, Carlson HL, Enns CA. Mechanisms of HFE-induced regulation of iron homeostasis: Insights from the W81A HFE mutation. Proc Natl Acad Sci USA. 2003;100:9500–9505. doi: 10.1073/pnas.1233675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson H, Zhang AS, Fleming WH, Enns CA. The hereditary hemochromatosis protein, HFE, lowers intracellular iron levels independently of transferrin receptor 1 in TRVb cells. Blood. 2005;105:2564–2570. doi: 10.1182/blood-2004-03-1204. [DOI] [PubMed] [Google Scholar]
- 11.Drakesmith H, et al. The hemochromatosis protein HFE inhibits iron export from macrophages. Proc Natl Acad Sci USA. 2002;99:15602–15607. doi: 10.1073/pnas.242614699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies PS, Enns CA. Expression of the hereditary hemochromatosis protein HFE increases ferritin levels by inhibiting iron export in HT29 cells. J Biol Chem. 2004;279:25085–25092. doi: 10.1074/jbc.M400537200. [DOI] [PubMed] [Google Scholar]
- 13.Bridle KR, et al. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 14.Wang RH, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Andriopoulos B, Jr, et al. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 17.Nemeth E, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, et al. Interaction of the hereditary hemochromatosis protein HFE with transferrin receptor 2 is required for transferrin-induced hepcidin expression. Cell Metab. 2009;9:217–227. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–214. doi: 10.1016/j.cmet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coscoy L, Ganem D. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA. 2000;97:8051–8056. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonart T. Iron: A target for the management of Kaposi's sarcoma? BMC Cancer. 2004;4:1–8. doi: 10.1186/1471-2407-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler JL. Endemic Kaposi's sarcoma in Africa and local volcanic soils. Lancet. 1993;342:1348–1351. doi: 10.1016/0140-6736(93)92252-o. [DOI] [PubMed] [Google Scholar]
- 23.Wada DA, Perkins SL, Tripp S, Coffin CM, Florell SR. Human herpesvirus 8 and iron staining are useful in differentiating Kaposi sarcoma from interstitial granuloma annulare. Am J Clin Pathol. 2007;127:263–270. doi: 10.1309/GMH9CENH4909AWVB. [DOI] [PubMed] [Google Scholar]
- 24.Hewitt EW, et al. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duncan LM, et al. Lysine-63-linked ubiquitination is required for endolysosomal degradation of class I molecules. EMBO J. 2006;25:1635–1645. doi: 10.1038/sj.emboj.7601056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boname JM, et al. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11:210–220. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr Opin Cell Biol. 2008;20:4–11. doi: 10.1016/j.ceb.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelham HR. Membrane traffic: GGAs sort ubiquitin. Curr Biol. 2004;14:R357–R359. doi: 10.1016/j.cub.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 30.Coscoy L. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat Rev Immunol. 2007;7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 31.Waheed A, et al. Association of HFE protein with transferrin receptor in crypt enterocytes of human duodenum. Proc Natl Acad Sci USA. 1999;96:1579–1584. doi: 10.1073/pnas.96.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkkila S, et al. Association of the transferrin receptor in human placenta with HFE, the protein defective in hereditary hemochromatosis. Proc Natl Acad Sci USA. 1997;94:13198–13202. doi: 10.1073/pnas.94.24.13198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bastin J, Drakesmith H, Rees M, Sargent I, Townsend A. Localisation of proteins of iron metabolism in the human placenta and liver. Br J Haematol. 2006;134:532–543. doi: 10.1111/j.1365-2141.2006.06216.x. [DOI] [PubMed] [Google Scholar]
- 34.Demaison C, et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of immunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.