Abstract
With malaria parasites (Plasmodium spp.), Toxoplasma, and many other species of medical and veterinary importance its iconic representatives, the protistan phylum Apicomplexa has long been defined as a group composed entirely of parasites and pathogens. We present here a report of a beneficial apicomplexan: the mutualistic marine endosymbiont Nephromyces. For more than a century, the peculiar structural and developmental features of Nephromyces, and its unusual habitat, have thwarted characterization of the phylogenetic affinities of this eukaryotic microbe. Using short-subunit ribosomal DNA (SSU rDNA) sequences as key evidence, with sequence identity confirmed by fluorescence in situ hybridization (FISH), we show that Nephromyces, originally classified as a chytrid fungus, is actually an apicomplexan. Inferences from rDNA data are further supported by the several apicomplexan-like structural features in Nephromyces, including especially the strong resemblance of Nephromyces infective stages to apicomplexan sporozoites. The striking emergence of the mutualistic Nephromyces from a quintessentially parasitic clade accentuates the promise of this organism, and the three-partner symbiosis of which it is a part, as a model for probing the factors underlying the evolution of mutualism, pathogenicity, and infectious disease.
Keywords: symbiosis, mutualism, parasitism, protist phylogeny, molgulid tunicate
With their evolutionary relationships buried in ancient lineages and entangled in extensive morphological and genomic diversity, protists continue to pose phylogenetic challenges to biologists. Endosymbiotic† protists have commonly proved recalcitrant material for phylogenetic analysis, as their profound and often fast-evolving adaptations to life inside other organisms have contributed additional layers of disguise to their evolutionary origins (2, 3).
One long-standing enigma has been the endosymbiotic marine protist Nephromyces. The phylogenetic affinities of this organism have been in question since the 19th-century zoologist Lacaze–Duthiers first described multiple “parasitic elements” of uncertain taxonomic identity in an unexpected habitat: the lumen of a ductless, urate- and calcium oxalate-rich organ of uncertain function, the so-called renal sac, in molgulid ascidian tunicates (Urochordata, phylum Chordata) (4–6). Giard (7) later classified these microbial “elements” as chytrid fungi, and named them Nephromyces.
Although Nephromyces was first described as a parasite, its ubiquity in molgulids argues strongly for Nephromyces infection as a net benefit to its molgulid hosts. Studies to date have documented Nephromyces in every adult individual of every Molgula species surveyed, regardless of population, geographical location, environmental conditions, season, or year of collection; Nephromyces was also found in all adult hosts examined in at least one population of another molgulid genus, Bostrichobranchus (6, 8, 9). The 100% prevalence (percentage of host individuals infected) of Nephromyces among adult molgulids is all the more impressive given the fact that Nephromyces is a nonhereditary symbiont, transmitted horizontally to new hosts, via ambient seawater, each host generation (10). This ubiquitous pattern of infection contrasts with typical patterns of parasitism; even though prevalence in parasitic associations can also sometimes be locally high in particular host populations or environmental conditions, overall prevalence of a parasite within a given host species nevertheless varies over space and time.
Mirroring the consistent infection of adult molgulids with Nephromyces, the obligately symbiotic Nephromyces has itself been found only in molgulids, with all but a few stages of its morphologically eclectic life history (Fig. 1) limited to the renal sac lumen (6, 11). The apparently universal, mutually exclusive association of these two clades in nature thus suggests that the biology and evolutionary histories of Nephromyces and molgulid tunicates are closely, and mutualistically, intertwined.
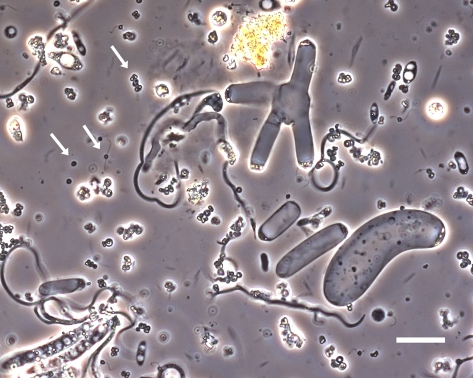
Fig. 1.
Nephromyces cells from renal sac fluid in M. retortiformis. Arrows indicate spores and flagellated cells. Larger filamentous cells are trophic stages. (Scale bar: 20 μm.)
An understanding of the evolutionary ancestry of Nephromyces could offer clues to the origins of this surprising symbiosis, but phenotypic information alone has been unable to clarify the phylogenetic relationships of this organism. Though Nephromyces does resemble fungi in its chitinous walls, hyphal-like trophic stages, and the absence of chloroplasts, none of these characteristics is unique to fungi (6, 12). Furthermore, several traits of Nephromyces, including tubular mitochondrial cristae and a posteriorly biflagellate cell stage (6, 12, 13), are atypical for fungi. Finally, the life cycle of Nephromyces does not resemble closely that of any fungal or protistan taxa (11).
From these diverse features, biologists have drawn diverse taxonomic conclusions. Several 20th-century investigators concurred with Giard that Nephromyces was a chytrid or other “lower fungus” (9, 14, 15); others questioned not only the chytrid affinities of Nephromyces, but even its very existence (refs. 16, pp 80–81 and 355–356, and 17). More recently, Nephromyces has been bounced from group to group among protistan phyla, placed at the base of the animal/fungal divergence (18), or grouped in various clades with other biflagellate protists of uncertain affinities (19).
Results and Discussion
To resolve the taxonomic ambiguities of Nephromyces, we sequenced short-subunit ribosomal DNA (SSU rDNA) from Nephromyces cells isolated from four Molgula species: M. occidentalis, M. citrina, M. manhattensis, and M. retortiformis. Analysis of these sequences indicates that Nephromyces is an apicomplexan (Fig. 2).
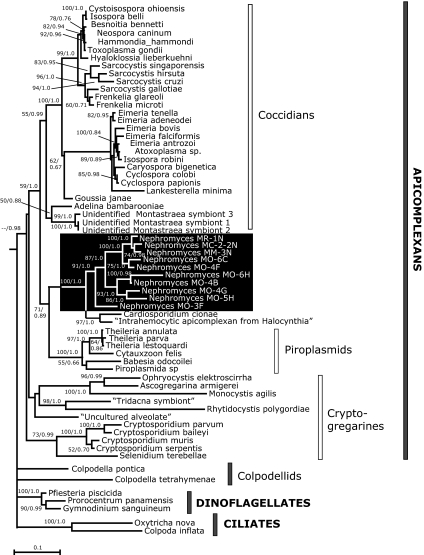
Fig. 2.
Phylogenetic tree of Apicomplexa, based on 18S ribosomal RNA genes, with representatives of known apicomplexans, and Nephromyces sequences from four Molgula species, including seven cloned sequences of Nephromyces isolated from M. occidentalis (sequences from 18 additional M. occidentalis-Nephromyces clones, not included in the analysis, were identical or nearly identical to clones 4F or 6C). Colpodellids, dinoflagellates, and ciliates were used as outgroups. The topology was constructed using maximum likelihood (PhyML); support values are expressed as percent bootstrap values (greater than 50%) and Bayesian posterior probabilities (>0.5). (Scale bar: 0.1 substitutions per site.) See Table S1 for accession numbers of taxa used in analysis.
We confirmed the identity of these sequences with fluorescence in situ hybridization (FISH), testing apicomplexan-specific and Nephromyces-specific SSU rRNA oligonucleotide probes on Nephromyces from M. manhattensis and M. occidentalis, and on Toxoplasma gondii (as a non-Nephromyces apicomplexan control). Of these samples, Toxoplasma and Nephromyces bound to the apicomplexan-specific probes, but only Nephromyces, from both M. manhattensis and M. occidentalis, bound to the Nephromyces-specific probes (Fig. 3); host tissue did not bind to any of these probes. The 25 cloned SSU rDNA sequences of Nephromyces isolated from M. occidentalis (Nephromyces MO; Fig. 2) showed notable diversity; although 20 of these sequences (represented by 6C and 4F in Fig. 2) were nearly identical, the other five sequences were fairly divergent from the majority (4B, 6H, 5H, 4G, and 3F in Fig. 2). Sample Nephromyces sequences from other host species (M. manhattensis: Nephromyces MM; M. citrina: Nephromyces MC; M. retortiformis: Nephromyces MR; Fig. 2) varied both from Nephromyces of M. occidentalis and from each other. Despite this diversity, all these sequences clustered together as a single, strongly supported clade.
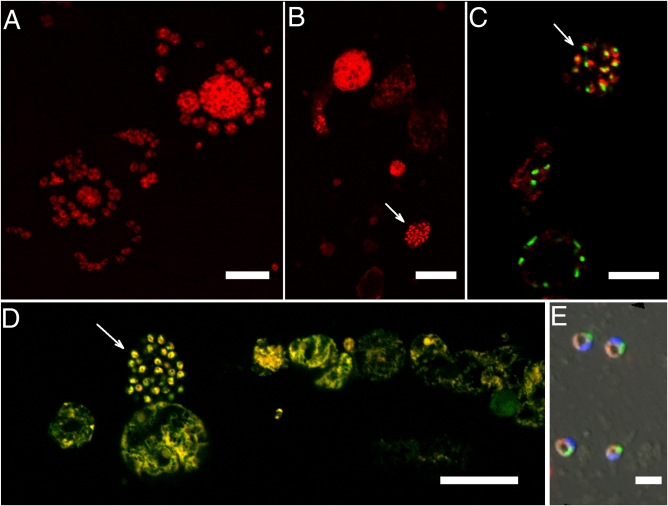
Fig. 3.
FISH of Nephromyces and Toxoplasma using apicomplexan-specific (Api-L) and Nephromyces-specific (Neph-1) SSU rRNA probes. Both probes were bound to fluor Cy3 (red). (Scale bars: A, B, D, 20 μm; C, 10 μm; E, 3 μm.) (A) Toxoplasma, Api-L; (B) Nephromyces from M. occidentalis, Api-L. Arrow indicates spores or flagellated cells. All other cells are uncleaved sporangia or trophic stages. (C) Nephromyces from M. manhattensis, Neph-1 (with eubacterial probe, EUB 338, linked to the fluor BODIPY FL (green). Toxoplasma did not bind to the Neph-1 probe. Arrow indicates spores or flagellated cells. All other cells are trophic stages. (D) Nephromyces from M. manhattensis, Api-L [merged with general eukaryote probe, EUK-516, linked to green (Fluos) fluor. Yellow represents binding to both Api-L and EUK-516]. Arrow indicates spores or flagellated cells. All other cells are uncleaved sporangia or trophic stages. (E) Nephromyces spores, Api-L, with EUB 338-BODIPY FL (green), and DAPI (blue).
Several phenotypic features of Nephromyces support the phylogenetic conclusions of SSU rDNA sequence analysis. Most compellingly, two structural features show provocative similarities to key apicomplexan characters. The nonflagellate, motile infective stage—seen transiently in Molgula blood after inoculation of M. manhattensis and M. occidentalis (11), and, significantly, the only stage of Nephromyces known to cross epithelial boundaries in Molgula—strongly resembles infective stages (sporozoites) of Plasmodium, Toxoplasma, Cryptosporidium, and other apicomplexans (Fig. 4A); sporozoite-like cells are also seen in the renal sac in at least one molgulid species, M. retortiformis (Fig. 4B). In addition, Nephromyces spores (the host-transfer stages, which give rise to the infective cells) contain inclusions reminiscent of rhoptries of the apical complex, a key structure in host-cell invasion among apicomplexa, and the ultrastructural hallmark of the apicomplexa and sister lineages (11, 13, 20–22) (Fig. 4 C and D).
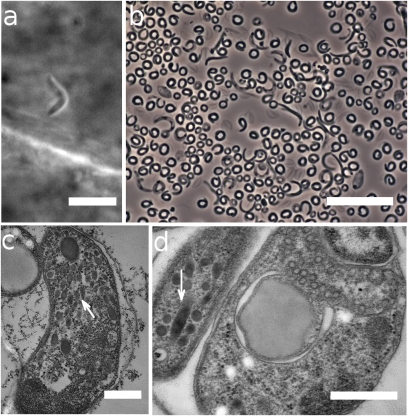
Fig. 4.
Apicomplexan-like cells in Nephromyces. (A) Infective stage of Nephromyces, in heart of laboratory-raised M. occidentalis, shortly after inoculation with Nephromyces spores. Photo clip from video (Zeiss Axiocam HS camera). (Scale bar: 5 μm.) (B) Sporozoite-like cells in the renal sac of M. retortiformis. (Scale bar: 20 μm.) (C and D) Transmission electron micrographs of two Nephromyces spores from M. occidentalis, with arrows showing rhoptry-like inclusions. (Scale bar: 0.5 μm.)
Further, Nephromyces shares with other apicomplexa a number of features not unique to apicomplexa, but nonetheless consistent with apicomplexan affinities. Like Nephromyces, all apicomplexans are horizontally transmitted, obligate symbionts (10, 20). The filamentous, hypha-like cells of Nephromyces possess membrane indentations suggestive of micropores, a feature characteristic of alveolate protists (apicomplexans, dinoflagellates, and ciliates; figure 1A in ref. 13) (22, 23). The high urate oxidase activity of Nephromyces, along with its urate-rich host habitat, suggests that Nephromyces may share with other apicomplexans a metabolic dependence on host purines, although with urate catabolism the more likely route of purine utilization, rather than the hypoxanthine- or adenosine-based purine salvage pathways seen in Toxoplasma and other apicomplexans (6, 24, 25). Finally, like other apicomplexa (but paradoxically so, given the fact that peroxisomes are the typical locus of urate oxidase in eukaryotes), there is thus far no ultrastructural evidence for peroxisomes in Nephromyces cytoplasm (13, 26).
A number of evolutionary issues have yet to be clarified. The significance of SSU rDNA sequence diversity seen in Nephromyces, both within and between host species, is still unknown. Further genomic, morphological, and developmental studies will help address the possibility of multiple infections and sexual recombination of Nephromyces within a single host, the degree of Nephromyces host specificity, and general patterns of coevolution between Nephromyces and its molgulid hosts.
Our sequence analysis shows substantial support for an affiliation of Nephromyces with piroplasmid apicomplexans (Fig. 2); however, definitive resolution of the fine-scale relationships of Nephromyces to piroplasmids and other apicomplexans must await additional genomic and ultrastructural data.
But even with phylogenetic details still to be resolved, the broader affinities of Nephromyces are nevertheless unequivocal. Bayesian and maximum-likelihood SSU rDNA sequence analyses, in concert with the strong morphological resemblance of Nephromyces spores and infective stages to apicomplexan sporozoites and other apicomplexan cells, indicate clearly that Nephromyces is not merely a sister taxon to the apicomplexa but a member of a distinctive, but bona fide, apicomplexan clade.
Several features nevertheless distinguish Nephromyces from other apicomplexans. Apart from its apicomplexan-like reproductive and infective stages, much of the Nephromyces life cycle bears little resemblance to typical apicomplexan life histories. The presence of conspicuously flagellated stages is unusual among apicomplexa, although biflagellate microgametes, resembling somewhat the biflagellate stages of Nephromyces, are found among some coccidians (6, 11, 21, 27–29). In contrast to coccidia and piroplasmida (but similar in some respects to gregarines), almost the entire life cycle of Nephromyces takes place in an extracellular environment (6, 11). Like the apicomplexans Cryptosporidium and Gregarina, but unlike Plasmodium and Toxoplasma, there is thus far no ultrastructural evidence for apicoplasts in Nephromyces (13, 30).
The microhabitat of Nephromyces is also unusual, compared with that of other tissues colonized by apicomplexans. Molgulids are considered a highly derived group of ascidians, a perspective supported not only by sequence analysis but also by their distinctive ecological, developmental, and morphological features, including the renal sac itself and the presence of Nephromyces (6, 31). Accumulation of urate and/or calcium deposits is widespread among other ascidian families either in blood cells or, in a few families, in multiple, small vesicles that may be analogous and/or homologous to the renal sac (6). But of these various “storage” tissues, the renal sac is the only one known to be colonized regularly by symbiotic microbes; it is also by far the largest such structure, and the only one abutting the heart. The particular features of its closed, extracellular, calcium-, oxalate-, and nitrogen-rich renal sac habitat, coupled with the high urate oxidase activity of this organism (6, 24), suggest a distinctive physiological niche for Nephromyces, compared with that of other apicomplexa.
Finally, Nephromyces contains hereditarily transmitted intracellular bacteria (13). These bacterial symbionts seem certain to have important metabolic effects on Nephromyces, including the possibility (among other possible metabolic contributions) that the bacteria are the source of urate oxidase activity found in their peroxisome-free apicomplexan hosts.
Only two other apicomplexans with bacterial endosymbionts have been described, both of them also from marine invertebrate hosts (32, 33). Our sequence analysis indicates that at least one of these, Cardiosporidium cionae, is a relative of Nephromyces. C. cionae is a parasite (of varying prevalence) found in pericardial bodies of Ciona intestinalis, an ascidian only distantly related to the Molgulidae (31, 33). Though the morphology of C. cionae is not identical to that of Nephromyces, it does share several features with Nephromyces, including biflagellate cells and cells reminiscent of Nephromyces spores and sporangia, as well as intracellular bacteria. Paralleling morphological data, SSU rDNA sequences confirm the distinctness of C. cionae from Nephromyces (among other differences, SSU rDNA of Nephromyces lacks a 66-bp insert found in C. cionae), as well as the relatedness of these two taxa. Another similar SSU rDNA sequence (GenBank accession no. EF558768), from an intrahemocytic apicomplexan parasite isolated from another ascidian, Halocynthia roretzi, also groups with C. cionae and Nephromyces in our analysis. However, despite the fact that H. roretzi, a pyurid ascidian, is considered a closer relative to molgulids than to Ciona, its apicomplexan parasite shows a closer relationship to C. cionae (Fig. 2) than to Nephromyces, with Nephromyces as a distinct subclade. These data suggest that Nephromyces may have a coevolutionary history possibly shaped both by bacterial endosymbionts and by life in ascidian hosts, with significant modifications correlated with colonization of molgulids; further sampling seems likely to reveal additional features and further complexity in this lineage.
Despite its several key similarities to other apicomplexa, however, Nephromyces nevertheless remains unique among the Apicomplexa in the beneficial outcome of its interactions. Phylogenetic relatedness between pathogenic and mutualistic endosymbionts has been documented in several microbial clades. Among eukaryotic examples, ascomycete and basidiomycete fungi and dinoflagellates contain both mutualistic and parasitic symbionts, as well as free-living species (34–37). Bacteria provide several well-documented examples of especially close relatedness of mutualists and pathogens, including mutualistic and pathogenic members in Rhizobiaceae (38, 39) and epsilonproteobacteria (40), and pathogenic and mutualistic species of the genera Burkholderia and Vibrio (41, 42). But the singular emergence of the mutualistic Nephromyces from an otherwise exclusively parasitic clade (20, 43, 44) offers a particularly striking example of the evolutionary malleability of interaction outcomes in many symbiotic associations and a reminder of the many commonalities of endosymbiotic life, whatever the fitness outcome for hosts. Probing the metabolic and ecological foundations of the bacterial-Nephromyces-molgulid symbiosis, and the possible correlation of its benign outcome with the several distinctive features of both the microbial and animal partners, should yield fresh perspectives on the factors determining the evolution of both mutualism and of infectious disease.
Materials and Methods
Collection of Nephromyces Cells and DNA; Cloning and Sequencing.
We isolated Nephromyces from the renal sac of M. manhattensis (Cape Cod and Gloucester, MA), M. citrina (Cape Cod, MA), M. retortiformis (Passamaquoddy Bay, New Brunswick, Canada), and M. occidentalis (supplied by Gulf Specimen Marine Lab, Panacea, FL). DNA was extracted from Nephromyces (from M. manhattensis and M. occidentalis) using guanidium isothiocyanate (GITC) buffer (45, 46).
Nephromyces DNA was amplified with a general eukaryote forward primer (3F: 5′ GTT CGA TTC CGG AGA GGG A) and an apicomplexan-specific reverse primer (Api1R: 5′ TAA TCT ATC CCC ATC ACG ATG C–3′). PCR products were ligated into a T-tailed vector, then transformed into chemically competent Escheria coli. Cloned products were recovered by PCR amplification with vector primers M13R and T7, then sequenced in both directions.
Phylogenetic Analysis.
Seven SSU rDNA Nephromyces sequences from M. occidentalis (chosen from a SSU rDNA dataset of 25 cloned sequences to represent the range of sequence diversity) and one cloned Nephromyces sequence apiece from M. retortiformis, M. manhattensis, and M. citrina, respectively, were added to a SSU rDNA dataset of representative apicomplexans and other alveolates. Final datasets of 65 sequences were aligned using ClustalW. The alignments were manually edited using McClade to exclude regions containing missing data as well as unalignable positions. Phylogenetic trees were constructed using maximum-likelihood (ML) and Bayesian methods with PhyML (47) and MrBayes (48), respectively, with the graphical interface TOPALi v2 (49). Because deep SSU phylogenies are known to be sensitive to highly diverged sequences, we ran preliminary analyses in each case to identify and exclude sequences resulting in significantly long branches that could cause tree distortion. For example, taxa such as Plasmodium and Hepatozoon were excluded from final trees because of their high sequence divergence.
Analyses were performed following a general time-reversible (GTR) model of sequence evolution using a gamma correction for site-to-site rate variation. We used eight categories of rates plus invariable sites and the shape parameter alpha estimated from the data. For ML, we assessed node robustness by bootstrap analysis with 100 replicates. For Bayesian analysis, we set MrBayes to operate four Markov chain Monte Carlo (MCMC) computations (default temperature = 0.2). In each case, a total of 1 million generations was calculated with trees sampled every 10 generations and with a prior burn-in of 25%. Posterior probabilities correspond to the frequency at which a given node is found in the post–burn-in trees.
Fluorescence in Situ Hybridization.
For design of custom FISH probes, SSU rDNA sequences of Nephromyces from M. occidentalis and M. manhattensis were compared with those of six coccidian species (Isospora suis, I. felis, Eimeria catrone, E. pilarensis, Sarcocystis capracanis, and T. gondii) and three piroplasmids (Babesia felis, Cytauxzoon felis, and Theileria ovis). Custom probes were conjugated with the red fluor Cy3 (Thermo Scientific or Invitrogen) or with the green fluor BODIPY FL (Invitrogen). The Api-L probe (5′ ATC TCT AGT CGG CAT AGT TTA TGG T), designed to bind to many apicomplexa, has exact matches to SSU rRNA of all of the above coccidians, some piroplasmids (B. felis and T. ovis), C. cionae, and Nephromyces from at least four host (Molgula) species. A second probe, Neph-1 (5′ CTC TTA AGT TTC TGA AAG AG), designed to bind specifically to Nephromyces rRNA, showed an exact match to the most common Nephromyces-cloned SSU rDNA sequences from M. occidentalis in this study, and a single mismatch to the Nephromyces SSU rDNA sequence from M. manhattensis used in our phylogenetic analysis. Other apicomplexan sequences showed mismatches at 4 (C. cionae, Halocynthia apicomplexan, B. felis, and T. ovis), 7 (T. gondii), or more of the 20 bases. Complementary probes included a general eukaryote probe (EUK 516) and a general eubacterial probe (EUB-338 I-II-III) (50, 51), conjugated to the green fluors Fluos (Thermo Scientific) or BODIPY FL and DAPI.
For in situ hybridization, symbiont and host tissue samples from Nephromyces (from M. manhattensis and M. occidentalis) and Toxoplasma gondii (in tissue cultures of mouse kidney cells) were fixed in 4% paraformaldehyde in sea water or phosphate buffer and stored in ethanol buffer or glycerol buffer until use. Standard hybridization protocols with 20% (Fig. 3E) or 35% (Fig. 3 A–D) formamide hybridization buffers were used. Some samples were embedded in polyacrylamide before hybridization (52).
Supplementary Material
Acknowledgments
We thank K. B. Seah for sequencing rDNA from M. manhattensis, M. retortiformis, and M. citrina and for calling our attention to the Halocynthia-apicomplexan rDNA sequence. We thank P. Keeling for laboratory support and advice; N. Pierce, S. Palumbi, R. Woollacott, J. Wernegreen, D. Patterson, and the Huntsman Marine Science Centre (St. Andrews, NB, Canada) for use of laboratory facilities; B. Striepen and L. Sharling (University of Georgia, Athens, GA) for providing Toxoplasma cultures for FISH; S. Lücker and the International FISH course (University of Vienna) for advice on FISH; and A. T. Newberry, J. T. Carlton, M. Sogin, and J. Wernegreen for review of the manuscript. This work was supported by the Baker Foundation, the Eppley Foundation, National Science Foundation Grant EF-0340538, National Institutes of Health/National Library of Medicine Grant 5G 13LM009396-02, Arizona State University West and the Radcliffe Institute for Advanced Study (Harvard University) (to M.B.S.). Funding was also provided by a Libbie Hyman Memorial Scholarship (Society for Integrative and Comparative Biology) and an R. C. Frazee Scholarship (Huntsman Marine Science Centre) (to C.R.). C.H.S. is a Scholar of the Canadian Institute for Advanced Research, supported by grants from the Tula Foundation to the University of British Columbia Centre for Microbial Diversity and Evolution and Dalhousie University Centre for Comparative Genomics and Evolutionary Bioinformatics.
Footnotes
The authors declare no conflict of interest.
Data deposition: SSU rDNA sequences have been deposited in the GenBank database (accession nos. HM469375–HM469384).
*This Direct Submission article had a prearranged editor.
†We define symbiosis in the original broad sense of Anton de Bary as the intimate association (living together) between two or more species, encompassing parasitic, commensal, and mutualistic (mutually beneficial) associations (1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002335107/-/DCSupplemental.
References
- 1.Saffo MB. Coming to terms with a field: Words and concepts in symbiosis. Symbiosis. 1992;14:17–31. [Google Scholar]
- 2.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Edman JC, et al. Ribosomal RNA sequence shows Pneumocystis carinii to be a member of the fungi. Nature. 1988;334:519–522. doi: 10.1038/334519a0. [DOI] [PubMed] [Google Scholar]
- 4.de Lacaze-Duthiers H. Account of solitary ascidians from the coasts of France (Translated from French) Archiv Zool Exp Gen. 1874;3:309–311. [Google Scholar]
- 5.Saffo MB, Lowenstam HA. Calcareous deposits in the renal sac of a molgulid tunicate. Science. 1978;200:1166–1168. doi: 10.1126/science.200.4346.1166. [DOI] [PubMed] [Google Scholar]
- 6.Saffo MB. Symbiogenesis and the evolution of mutualism: Lessons from the Nephromyces-bacterial-molgulid symbiosis. In: Margulis L, Fester R, editors. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. Cambridge, MA: MIT Press; 1991. pp. 410–429. [Google Scholar]
- 7.Giard A. On Nephromyces, a new genus of parasitic fungi from the kidney of Molgulidae (Translated from French) C R Acad Sci Paris. 1888;106:1180–1182. [Google Scholar]
- 8.Saffo MB. Distribution of the endosymbiont Nephromyces Giard within the ascidian family Molgulidae. Biol Bull. 1982;162:95–104. [Google Scholar]
- 9.Buchner P. In: Endosymbiosis of Animals with Plant Microorganisms, Mueller B, Foeckler FH, translators. New York: Interscience; 1965. [Google Scholar]
- 10.Saffo MB, Davis W. Modes of infection of the ascidian Molgula manhattensis by its endosymbiont Nephromyces Giard. Biol Bull. 1982;162:105–112. [Google Scholar]
- 11.Saffo MB, Nelson R. The cells of Nephromyces: Developmental stages of a single life cycle. Can J Bot. 1983;61:3230–3239. [Google Scholar]
- 12.Saffo MB, Fultz S. Chitin in the symbiotic protist Nephromyces. Can J Bot. 1986;64:1306–1310. [Google Scholar]
- 13.Saffo MB. Symbiosis within a symbiosis: Intracellular bacteria in the endosymbiotic protist Nephromyces. Mar Biol. 1990;107:291–296. [Google Scholar]
- 14.Harant H. A contribution to the natural history of ascidians and their parasites. 1. Chytridiomycetes (Translated from French) Ann Inst Oceanogr Monaco. 1931;8:349–352. [Google Scholar]
- 15.Azéma M. Studies on the blood and excretion in the Ascidicea (Translated from French) Ann Inst Oceanogr Monaco. 1937;17:1–150. [Google Scholar]
- 16.Johnson TW, Jr., Sparrow FK., Jr . Fungi in Oceans and Estuaries. Germany: Cramer, Weinheim; 1961. [Google Scholar]
- 17.Alderman D. Fungal diseases of marine animals. In: Gareth-Jones ED, editor. Recent Advances in Aquatic Mycology. New York: Wiley; 1976. pp. 223–260. [Google Scholar]
- 18.Beakes GW. Relationships between lower fungi and protozoa. In: Coombs GH, Vickerman K, Sleigh MA, Warren M, editors. Evolutionary Relationships Among Protozoa. London: Chapman & Hall; 1998. pp. 351–373. [Google Scholar]
- 19.Cavalier-Smith T. Protist phylogeny and the high-level classification of protozoa. Eur J Protistol. 2003;39:338–348. [Google Scholar]
- 20.Perkins FO, Barta JO, Clopton RE, Peirce MA, Upton SJ. Apicomplexa. In: Lee JJ, Leedale GF, Bradbury P, editors. Illustrated Guide to the Protozoa. 2nd Ed. Lawrence, KS: Society of Protozoologists; 2000. pp. 190–369. [Google Scholar]
- 21.Leander BS, Keeling PJ. Early evolutionary history of dinoflagellates and apicomplexans (alveolata) as inferred from hsp90 and actin phylogenies. J Phycol. 2004;40:341–350. [Google Scholar]
- 22.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 23.Appleton PL, Vickerman K. Presence of apicomplexan-type micropores in a parasitic dinoflagellate, Hematodinium sp. Parasitol Res. 1996;82:279–282. doi: 10.1007/s004360050113. [DOI] [PubMed] [Google Scholar]
- 24.Saffo MB. Nitrogen waste or nitrogen source? Urate degradation in the renal sac of molgulid tunicates. Biol Bull. 1988;175:403–409. [Google Scholar]
- 25.Chaudhary K, et al. Purine salvage pathways in the apicomplexan parasite Toxoplasma gondii. J Biol Chem. 2004;279:31221–31227. doi: 10.1074/jbc.M404232200. [DOI] [PubMed] [Google Scholar]
- 26.Schlüter A, et al. The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol Biol Evol. 2006;23:838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 27.Lainson R. Intestinal coccidia (Apicomplexa: Eimeriidae) of Brazilian lizards. Eimeria carmelinoi n.sp., from Kentropyx calcarata and Acroeimeria paraensis n.sp. from Cnemidophorus lemniscatus lemniscatus (Lacertilia: Teiidae) Mem Inst Oswaldo Cruz. 2002;97:227–237. doi: 10.1590/s0074-02762002000200016. [DOI] [PubMed] [Google Scholar]
- 28.Vieira LS, Lima JD, Ribeiro MFB, Bozzi IA, Camargos ERS. Ultrastructure of endogenous stages of Eimeria ninakohlyakimovae Yakimoff & Rastegaieff, 1930 Emend. Levine, 1961 in experimentally infected goat. Mem Inst Oswaldo Cruz. 1997;92:533–538. doi: 10.1590/s0074-02761997000400017. [DOI] [PubMed] [Google Scholar]
- 29.Kopečná J, et al. Phylogenetic analysis of coccidian parasites from invertebrates: Search for missing links. Protist. 2006;157:173–183. doi: 10.1016/j.protis.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Toso MA, Omoto CK. Gregarina niphandrodes may lack both a plastid genome and organelle. J Eukaryot Microbiol. 2007;54:66–72. doi: 10.1111/j.1550-7408.2006.00229.x. [DOI] [PubMed] [Google Scholar]
- 31.Zeng L, Jacobs MW, Swalla BJ. Coloniality has evolved once in Stolidobranch Ascidians. Integr Comp Biol. 2006;46:255–268. doi: 10.1093/icb/icj035. [DOI] [PubMed] [Google Scholar]
- 32.Azevedo C, Padovan I, Corral L, Padovan P. Ultrastructural description of an unidentified apicomplexan oocyst containing bacteria-like hyperparasites in the gill of Crassostrea rizophorae. Dis Aquat Organ. 2005;65:153–157. doi: 10.3354/dao065153. [DOI] [PubMed] [Google Scholar]
- 33.Ciancio A, et al. Redescription of Cardiosporidium cionae (Van Gaver and Stephan, 1907) (Apicomplexa: Piroplasmida), a plasmodial parasite of ascidian haemocytes. Eur J Protistol. 2008;44:181–196. doi: 10.1016/j.ejop.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 34.Berbee M. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol Mol Plant Pathol. 2001;59:165–187. [Google Scholar]
- 35.Lutzoni F, Pagel M, Reeb V. Major fungal lineages are derived from lichen symbiotic ancestors. Nature. 2001;411:937–940. doi: 10.1038/35082053. [DOI] [PubMed] [Google Scholar]
- 36.Gast RJ. Molecular phylogeny of a potentially parasitic dinoflagellate isolated from the solitary radiolarian, Thalassicolla nucleata. J Eukaryot Microbiol. 2006;53:43–45. doi: 10.1111/j.1550-7408.2005.00071.x. [DOI] [PubMed] [Google Scholar]
- 37.Konovalova GV. Parasitic dinoflagellates and ellobiopsids (Ellobiopsidae) of the coastal waters of the Sea of Japan. Russ J Mar Biol. 2008;34:28–37. [Google Scholar]
- 38.Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Evol Microbiol. 2001;51:89–103. doi: 10.1099/00207713-51-1-89. [DOI] [PubMed] [Google Scholar]
- 39.Duan Y, et al. Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact. 2009;22:1011–1020. doi: 10.1094/MPMI-22-8-1011. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa S, et al. Deep-sea vent epsilon-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partida-Martinez LP, et al. Burkholderia rhizoxinica sp. nov. and Burkholderia endofungorum sp. nov., bacterial endosymbionts of the plant-pathogenic fungus Rhizopus microsporus. Int J Syst Evol Microbiol. 2007;57:2583–2590. doi: 10.1099/ijs.0.64660-0. [DOI] [PubMed] [Google Scholar]
- 42.Ruby EG. The Euprymna scolopes-Vibrio fischeri symbiosis: A biomedical model for the study of bacterial colonization of animal tissue. J Mol Microbiol Biotechnol. 1999;1:13–21. [PubMed] [Google Scholar]
- 43.Huang J, Kissinger JC. Horizontal and intracellular gene transfer in the apicomplexa: The scope and functional consequences. In: Katz LA, Bhattacharya D, editors. Genomics and Evolution of Microbial Eukaryotes. New York: Oxford Univ Press; 2006. pp. 123–136. [Google Scholar]
- 44.Morrison DA. Evolution of the Apicomplexa: Where are we now? Trends Parasitol. 2009;25:375–382. doi: 10.1016/j.pt.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Tkach V, Pawlowski J. A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol. 1999;44:147–148. [Google Scholar]
- 46.Tillet D, Neilan BA. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J Phycol. 2000;36:251–258. [Google Scholar]
- 47.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 48.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 49.Milne I, et al. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: Development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 51.Loy A, Maixner F, Wagner M, Horn M. probeBase—an online resource for rRNA-targeted oligonucleotide probes: New features 2007. Nucleic Acids Res. 2007;35(Database issue):D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daims H, Lücker S, Wagner M. daime, a novel image analysis program for microbial ecology and biofilm research. Environ Microbiol. 2006;8:200–213. doi: 10.1111/j.1462-2920.2005.00880.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.