Abstract
Intracellular detection of virus infections is a critical component of innate immunity carried out by molecules known as pathogen recognition receptors (PRRs). Activation of PRRs by their respective pathogen-associated molecular patterns (PAMPs) leads to production of proinflamatory cytokines, including type I IFN, and the establishment of an antiviral state in the host. Out of all PRRs found to date, retinoic acid inducible gene I (RIG-I) has been shown to play a key role in recognition of RNA viruses. On the basis of in vitro and transfection studies, 5′ppp RNA produced during virus replication is thought to bind and activate this important sensor. However, the nature of RNA molecules that interact with endogenous RIG-I during the course of viral infection has not been determined. In this work we use next-generation RNA sequencing to show that RIG-I preferentially associates with shorter, 5′ppp containing viral RNA molecules in infected cells. We found that during Sendai infection RIG-I specifically bound the genome of the defective interfering (DI) particle and did not bind the full-length virus genome or any other viral RNAs. In influenza-infected cells RIG-I preferentially associated with shorter genomic segments as well as subgenomic DI particles. Our analysis for the first time identifies RIG-I PAMPs under natural infection conditions and implies that full-length genomes of single segmented RNA virus families are not bound by RIG-I during infection.
Keywords: Sendai, influenza, interferon, pathogen-associated molecular pattern, defective-interfering particle
The retinoic acid inducible gene I (RIG-I)–like receptor (RLR) family of viral sensors contains three members that include the retinoic acid inducible gene (RIG-I), melanoma differentiation factor 5 (MDA5), and Laboratory of Genetics and Physiology gene 2 (LGP2) (1–4). Both RIG-I and MDA5 have been shown to play an important role in recognition of RNA viruses. For most RNA viruses both receptors contribute to IFN induction, although the relative contribution may be cell type specific (5–7). Some viruses, such as picornaviruses and influenza virus, appear to be recognized by only one of the sensors, with picornaviruses being sensed by MDA5 and influenza viruses by RIG-I (1, 8, 9). The substrate specificities of RIG-I and MDA5 have not been clearly established, although from RNA transfection experiments in knockout cells it appears that RIG-I recognizes RNA of various lengths with 5′-triphosphates and some partial double-stranded characteristics, whereas MDA5 senses only very long dsRNA molecules (>2,000 nt) in a phosphate-independent manner (10–14). All RLRs are members of the DExD/H family of RNA helicases and contain an ATP-dependent helicase domain and a C-terminal regulatory domain (RD). The N termini of RIG-I and MDA5 contain two tandem CARD domains required for downstream signaling through their adaptor, MAVS (15–18). The RD domain of RIG-I is responsible for recognition and binding to its RNA substrates in a 5′-phosphate–dependent manner, whereas the helicase domain has affinity for dsRNA (19–21). In uninfected cells RIG-I is thought to exist in an inactive state; the C-terminal RD domain is proposed to interact with the N-terminal CARD domain and block it from association with MAVS. RNA binding to the RD of RIG-I likely induces a conformational change in the protein, resulting in CARD exposure and association with the CARD domain of MAVS.
Because both RIG-I and MDA5 are localized in the cytoplasm, it is imperative for these receptors to be able to distinguish self RNA from viral RNA to prevent IFN production in the absence of infection. The characteristics of RNA molecules capable of activating RIG-I have been well established through numerous biochemical and knockout studies. The signature features of RIG-I agonists are a 5′-triphosphate group at the end of an RNA molecule longer than at least 19 nt and some dsRNA regions (10, 11). Additionally, 5′ppp containing RNAs rich in U residues have been found to act as more potent inducers of RIG-I, indicating that sequence composition might play a role in activation (22). It is yet unclear whether ssRNA, even in the presence of a 5′-triphosphate group, is capable of inducing RIG-I activity, and at least in the case of shorter RNA molecules it appears that some double-stranded characteristics are required for its activation (12, 13).
Although RNA molecules capable of inducing RIG-I have been well characterized, it remains to be seen which if any of these RNAs are actually interacting with RIG-I in virus-infected cells. A recent study using overexpressed RIG-I has shown that this protein associates with negative stranded viral RNA in Sendai virus-infected (SeV) cells and concluded that genomic RNA serves as a inducer of RIG-I signaling (3). In our study we examine SeV-infected cells and analyze RNA molecules that interact with endogenous RIG-I protein both early and late in viral infection. By applying deep sequencing analysis to examine the isolated RNA species we were able to identify the exact nature of RIG-I–associated viral RNA in an unbiased manner. Through this approach we determined that in SeV-infected cells, RIG-I specifically associates with the defective interfering RNA genomes and not with the full-length genomes, mRNA, and leader or trailer RNAs. The immunostimulatory effects of RIG-I–associated RNA in SeV-infected cells were abolished upon removal of all three or two 5′-terminal phosphates. In influenza PR8 ΔNS1 virus-infected cells we observed that RIG-I associates with all genomic segments, but preferentially associates with shorter RNA molecules, such as the NS and M segments, and the internal deletion defective interfering (DI) particles generated by PB1 and PA segments. On the basis of our work we conclude that under natural infection conditions RIG-I preferentially associates with shorter viral RNAs that contain 5′ triphosphates and some dsRNA regions. This study represents a unique analysis of endogenous RIG-I/pathogen-associated molecular pattern (PAMP) complexes present during viral infections.
Results
Isolation of RIG-I/RNA Complexes from Virus-Infected Cells.
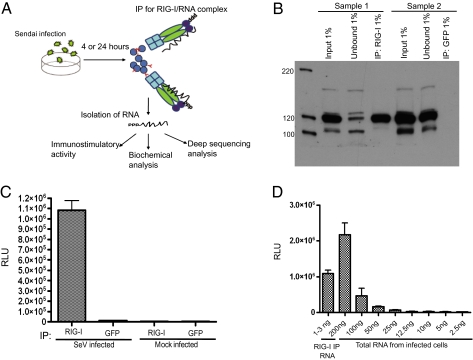
To analyze endogenous RNA substrates of RIG-I, A549 human lung carcinoma cells were infected with Sendai virus Cantel (Sev-C) at a high multiplicity of infection (MOI) and infection was allowed to proceed for 24 h. At this time the cells were lysed and RIG-I/RNA complexes were immunoprecipitated with a monoclonal antibody against RIG-I. Fig. 1A outlines the overall schematic for the experimental procedure. To make sure that we could distinguish between RIG-I–associated RNA and RNA precipitated in a nonspecific way, an anti-GFP antibody was used in parallel. As can be seen from Fig. 1B, precipitation of endogenous RIG-I was very efficient and the vast majority of RIG-I protein was isolated from cell lysates. Following immunoprecipitation (IP), coimmunoprecipitated RNA was isolated by phenol/chlorophorm extraction and transfected into a 293T ISRE-FF (containing a firefly luciferase gene under the control of an IFN-stimulated promoter) reporter cell line to analyze the immunostimulatory activity of the isolated RNA species. Transfection of RIG-I–associated RNA into the reporter cells resulted in a 90-fold increase in reporter activity compared with the GFP control, indicating that the immunostimulatory RNA was specifically pulled down in association with RIG-I (Fig. 1C). Comparison of ISRE-FF activity between RIG-I–associated RNA and decreasing amounts of total RNA from SeV-C–infected A549 cells revealed that RIG-I immunoprecipitation greatly concentrates the immunostimulatory activity of the RNA generated in virus-infected cells, demonstrating that RIG-I pulldown is specifically enriching for immunogenic RNA (Fig. 1D).
Fig. 1.
Immunoprecipitation of RIG-I/RNA complexes from 24-h SeV infections and enzymatic analysis of RIG-I–associated RNA. (A) A schematic representation of the experimental procedure for characterization of RNA from RIG-I IPs. (B) Western blot analysis from RIG-I pulldowns shows the efficiency of RIG-I immunoprecipitations. (C) Immunostimulatory activity of RNA from RIG-I and control (GFP) IPs upon transfection into 293T ISRE-FF cells. (D) Enrichment of immunostimulatory RNA with RIG-I IP compared with total RNA from SeV-infected cells.
Biochemical Analysis of RIG-I–Associated RNA from SeV-C–Infected Cells.
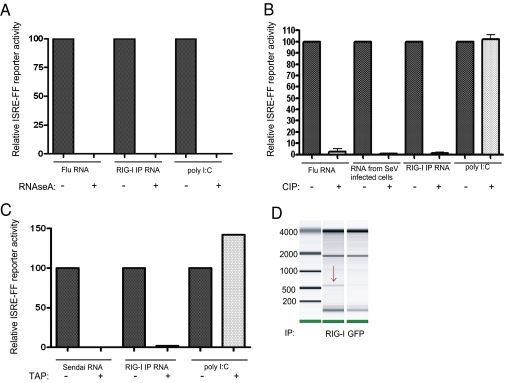
Previous studies have shown that multiple types of RNA molecules are capable of activating RIG-I. These RNAs have been described to contain 5′-triphosphates, diphosphates and monophosphates, and 3′-monophosphates; to be either single- or double-stranded; and to span the length from 19 to thousands of nucleotides long (3, 10–14, 20, 21, 23–25). Enzymatic analysis of immunoprecipitated RNA bound to RIG-I allowed us to characterize the biochemical nature of endogenous RIG-I inducers in SeV-infected cells. Treatment of isolated RNA with RNAseA led to complete loss of immunostimulatory activity in both RIG-I–associated RNA and control RNA, confirming that the PAMPs associated with RIG-I in SeV-infected cells are indeed RNA molecules (Fig. 2A). To address the phosphate composition of RIG-I–associated RNA we treated the isolated RNA with calf alkaline phosphatase (CIP), an enzyme that removes all 5′ and 3′ phosphates. Treatment of RIG-I IP RNA with CIP resulted in complete loss of its immunostimulatory activity similarly to control influenza virus genomic RNA, and in contrast to poly(I:C), a synthetic dsRNA molecule with a 5′ monophosphate that does not rely on phosphate composition for its immunostimulatory activity (Fig. 2B). Treatment of RIG-I–associated RNA with tobacco acid pyrophosphotase (TAP), which removes the first two terminal phosphates and leaves on a monophosphate group, also led to a complete loss of immunostimulatory activity, demonstrating that RNA species associated with RIG-I during SeV infection require an intact 5′ triphosphate for immogenicity (Fig. 2C).
Fig. 2.
(A) RNAseA treatment of RIG-I bound RNA as well as the control RNAs completely abolishes their immunostimulatory activity. (B) CIP treatment of RIG-I bound RNA as well as flu RNA and RNA from SeV-infected cells but not poly(I:C) leads to loss of all immunostimulatory activity. (C) TAP treatment of RIG-I bound RNA and purified SeV virus RNA unlike poly(I:C) leads to a complete loss in immunostimulatory activity. (D) Agilent mRNA chip of RIG-I–associated RNA reveals a distinct 550-nt band not present in the control IP.
Deep Sequencing of RIG-I–Associated RNA from SeV-C–Infected Cells.
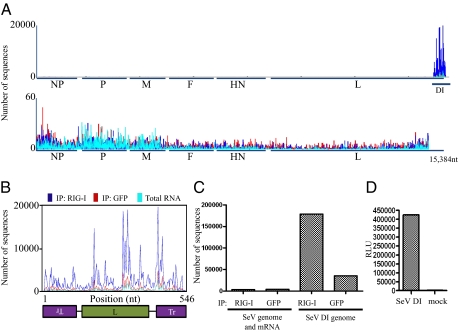
To identify the exact nature of RIG-I–associated RNA in an unbiased manner, deep sequencing analysis was performed on the isolated RNA species. The RNA was prepared for sequencing according to Illumina mRNA-seq protocol and sequenced on the Illumina Genome Analyzer. The Illumina platform provides deep coverage, on the order of 10–25 million reads per sequencing sample with relatively short length reads of 29 nt. On the basis of sample preparation methodology both negative and positive sense RNAs are amplified in an identical way and the two forms cannot be distinguished when mapped to their genomic location. Sequences were mapped to the SeV genome and relative abundances of these sequences between RIG-I pulldown and GFP control pulldown, as well as total RNA from SeV-infected cells, were compared. Fig. 3A shows the graphical representation of sequences mapped to the SeV genome. Individual peaks on the graph correspond to a sequencing read that starts at that particular position and extends in either direction. The x axis corresponds to all possible 15,384 positions in the SeV genome, and the y axis shows the number of reads that begin at that position. Analysis of obtained reads revealed significant variations in peak intensities between close positions on the genome; this variation is clearly observed in Fig. 3B. These peaks were similarly distributed between the RIG-I IP sample and the GFP IP sample, indicating to us that they were most likely due to sequencing biases and not to real differences in RNA abundances that could be due to cellular processing of viral RNA. To make sure that this is the case we sequenced viral RNA isolated from purified Sendai virus. Analysis of peak distributions across the same region of the genome (i.e., positions 14,900 and 15,384) revealed that the same nucleotide positions were overrepresented in the purified virus RNA as in our IP samples. On the basis of this evidence we conclude that the variation in peak intensities observed between adjacent genomic positions is due to sequencing biases introduced by the Illumina platform. Examination of SeV sequences from total cellular RNA clearly illustrated that the vast majority of viral RNA mapped to the 5′ end of the SeV genome [Fig. 3A (teal color) and Fig. S1]. Specifically, RNA mapping to a region of the genome between positions 14,932 and 15,384 was much more abundant in infected cells than RNA mapping to the rest of the SeV genome. Because it is known from previous studies that the SeV-C copy-back DI particle genome maps to precisely those positions, we concluded that the majority (≈95%) of viral RNA species present in infected cells at 24 h postinfection (hpi) are of a copy-back DI nature (26). Comparison of RIG-I–associated RNA with that of the control IP revealed that the RIG-I pulldown was specifically enriched in DI RNA, with RIG-I samples containing approximately seven times more DI RNA than control samples (Fig. 3 A–C). None of the other SeV RNAs, including genomic RNA, mRNAs, and leader or trailer were overrepresented in the RIG-I pulldown (Fig. 3A Lower). SeV DI copy-back genomes consist of the SeV trailer sequence at the 5′ end, followed by the partial sequence of the L gene and a sequence that is the exact complement to the trailer (antitrailer) at the 3′ end of the molecule (Fig. 3B). This unique DI genome structure results in a 546-nt RNA molecule with a relatively long perfect dsRNA portion (92 nt) (very different from typical RNA virus genomes, which contain only short regions of perfect dsRNA). Visualization of a 550-nt band on the Agilent bioanalyzer RNA chip in the RIG-I IP sample but not in the control IP supports the conclusion that the DI genome is preferentially interacting with RIG-I (Fig. 2D). Identification of copy-back DIs as a RIG-I PAMP agrees with previous characterization of these molecules as exceptionally good inducers of the IFN response (26).
Fig. 3.
Deep sequencing analysis of RIG-I–associated and control IP RNA from SeV virus-infected cells. (A) RNA from RIG-I IP and control (GFP) IP and total RNA from SeV-infected cells (blue, red, and teal, respectively) were subjected to Illumina deep sequencing. Obtained sequencing reads are mapped to their starting position on the virus genome; the y axis shows the number of sequences mapped to a particular position. (Upper) All sequences mapped to the entire genome are shown. (Lower) The last 484 nt are removed to allow better visualization of the rest of the genome (note the difference in the y scale between Upper and Lower). (B) Sequencing reads mapped to the genome of the DI particle show enrichment for RIG-I–associated sequences throughout the entire DI molecule. (C) Comparison of numbers of sequences that map to the DI genome or the rest of the SeV genome in the RIG-I and control (GFP) IPs. (D) Induction of ISRE-FF reporter by transfection of T7 SeV DI RNA compared with mock transfected cells.
Analysis of RIG-I–Associated RNA at Early Times of SeV Infection.
We next determined whether the same or different viral RNA species are associated with RIG-I relatively early in infection. We followed the same approach as described for the 24 h infection with the exception of lysing cells 4 h postinfection. As can be seen from Fig. S2A, we were able to isolate immunostimulatory RNA in a RIG-I–specific manner from these cells, and this RNA was again subjected to deep sequencing analysis. The relative amount of DI genomes in these cells was lower than at the 24-h time point with ≈34% of SeV RNA molecules mapping to the DI genome. Despite DI's lower abundance in these cells it was again found to be the only Sendai RNA that was specifically associating with RIG-I and we did not see any RIG-I–specific binding of the full-length genome (Fig. S2 B and C). Therefore it appears that RIG-I interacts with the same SeV-derived RNA molecule both early and late in infection, namely DI RNA.
Confirmation of Deep Sequencing Data with Quantitative PCR.
To validate our deep sequencing analysis with an independent method we chose to perform TaqMan Q-PCR RNA quantification. On the basis of the unique structure of the SeV copy-back DI, with the 3′ end of the molecule containing an antitrailer (Fig. 3B), it is possible to design PCR primers that will detect only the DI RNA and not the full-length genome, L mRNA, or trailer RNA. Comparison of relative abundances of DI RNA and genome RNA/L mRNA between total RNA from infected cells, RIG-I IP and GFP IP at 24 hpi confirmed that only DI-specific sequences were enriched in RIG-I IPs (Fig. S3A), validating conclusions from our deep sequencing data. To obtain sense-specific information about ratios of DI genome to full-length genome (excluding L mRNA sequences) we performed the same Q-PCR analysis except with sense-specific RT amplification. Again we saw that only DI genomic RNA and not the full-length genome was preferentially interacting with RIG-I (Fig. S3C). We also attempted to analyze RIG-I–associated RNA very early on in SeV-infected cells by allowing the infection to progress for only 30 min or 1 h. Immunoprecipitation of RIG-I at these early time points did not produce any immunostimulatory RNA (Fig. S3B) nor could we detect any significant differences in either full-length genome or DI genome abundance in RIG-I–associated RNA (Fig. S3C). This failure to detect immunostimulatory RNA very early in infection could possibly be due to limited sensitivity of our methodology or requirement for higher levels of virus replication to induce the antiviral response.
Isolation and Deep Sequencing of RIG-I–Associated RNA from Influenza PR8ΔNS1 Virus Infections.
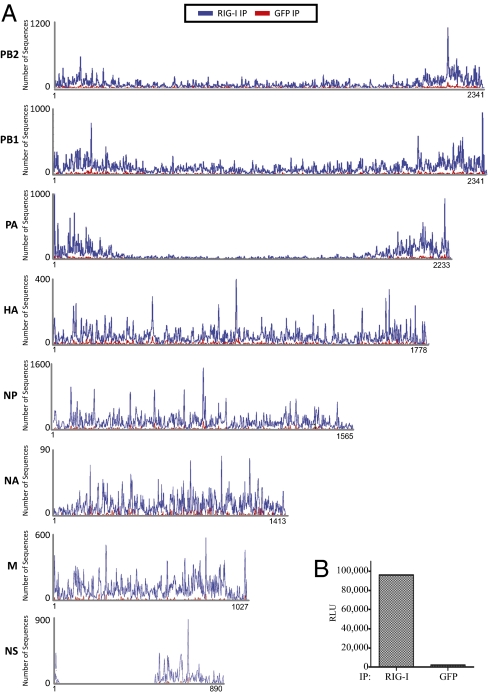
We next attempted to characterize RNA molecules associated with RIG-I during influenza virus infection, as it possesses a very different genome organization and replication cycle compared with Sendai virus. Because wild-type influenza virus is very efficient at blocking IFN induction through the action of its well-characterized IFN antagonist NS1 (27), we decided to infect A549 cells with PR8ΔNS1 virus. This mutant virus is lacking the RNA sequence that codes for the NS1 protein and therefore it is unable to block IFN production and RIG-I up-regulation in infected cells. Isolation of RIG-I/RNA complexes from A549 cells infected with a high MOI of PR8ΔNS1 virus produced RNA that was specifically immunostimulatory upon transfection into the 293T ISRE-FF reporter cell line compared with the RNA isolated from the control anti-GFP pulldown (Fig. 4B). Again to identify which viral RNA species were specifically interacting with RIG-I in infected cells we performed deep sequencing analysis of all isolated RNA. The obtained sequences were mapped to the influenza virus PR8 genomes and analyzed for abundances between the RIG-I IP sample and the control IP sample. Fig. 4A shows the obtained sequencing reads mapped to each segment of the influenza virus genome. For all genomic segments we saw a higher abundance of RNA in RIG-I IP samples than in the control samples, with the average ratio between RIG-I IP and control of 2.6. To establish that this difference represents a significant change in abundance between the two samples we compared the relative abundances of eight randomly picked cellular mRNAs from the same sequencing dataset. The average ratio between RIG-I IP and the control IP for these eight mRNAs was 1.1 (Table S1). Therefore, we conclude that we have identified RNAs that specifically interact with RIG-I in the course of influenza virus infection.
Fig. 4.
Immunoprecipitation and deep sequencing analysis of RIG-I–associated RNA from 24-h influenza virus infections. (A) Sequencing reads from RIG-I and control (GFP) IPs were mapped to the individual PR8 influenza virus genomic segments. Each peak corresponds to the beginning position of that particular read and the number of sequences starting at that position is represented on the y axis. RIG-I enrichment (blue) can be observed on all genomic segments. (B) Transfection of RIG-I–associated RNA into the ISRE-FF reporter cells shows high immunostimulatory activity of this RNA compared with the control (GFP) IP.
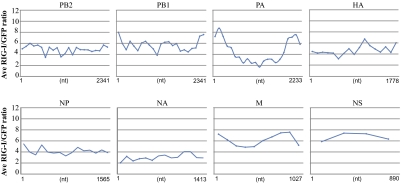
To see whether specific regions within the individual genome segments were more enriched in the RIG-I pulldown we calculated the RIG-I IP/control IP ratio at each nucleotide position on each segment. These ratios were then averaged over 100-nt intervals and allowed us to visualize the relative enrichment ratios over the length of each genomic segment (Fig. 5). This analysis revealed that the 5′ and 3′ regions in the PB1 and PA segments were more overrepresented in RIG-I pulldowns than the rest of those segments. We hypothesized that these regions might represent internal deletion DI particles that have previously been shown to be associated with influenza virus replication (28). RT-PCR of the PA gene with primers corresponding to the ends of the segment indeed produced a 650-nt product that was not observed in the same RT-PCR performed with purified RNA from PR8 influenza virus virions; this RNA was sequenced and confirmed to map to the ends of the PA segment. Comparison of RIG-I IP/control IP RNA ratios between the segments identifies the two DI RNAs from PA and PB1 segments as well as the NS and M segments as being the most enriched RNA molecules in the RIG-I pulldown (Fig. 5). On the basis of these observations we propose that RIG-I binds to all segments of the flu genome but preferentially associates with shorter RNA molecules such as the shorter influenza virus segments and short DI particles generated from the larger segments.
Fig. 5.
Analysis of average local enrichment of influenza virus RNA in RIG-I versus control (GFP) immunoprecipitations. For each genomic segment ratios of RIG-I/GFP sequencing reads were calculated at each nucleotide position. The average of these ratios for every 100 nt was calculated and is shown.
Immunostimulatory Activity of Individual PR8 RNA Genomic Segments.
To check that RIG-I–associated viral RNA molecules identified in our pulldowns could act as PAMPs and induce an antiviral response, we generated six of these RNAs by T7 promoter-driven in vitro transcription. The size and purity of produced RNAs were checked by denaturing agarose gel electrophoresis (Fig. S4A). Transfection of equimolar ratios of these RNAs into the ISRE-FF reporter cells resulted in induction of the FF reporter, indicating that all of these RNAs were indeed immunogenic (Fig. S4B). With the exception of NA RNA, which gave the lowest amount of induction, we did not see any significant differences in reporter stimulation between the different flu RNAs, and thus immunogenicity of these RNAs was not proportional to RNA length. Currently we do not understand the molecular basis or significance of the observed differences in RIG-I association between various influenza RNAs. These differences could potentially reflect variability in RNA segment accessibility to RIG-I during the course of infection, either structurally or spatially. Also, because T7 transcribed RNAs are naked RNAs transfected into cells, their localization, stability, and interactions with RIG-I might be fundamentally different from those that take place during viral infection.
Discussion
By subjecting the RNA to both biochemical and deep sequencing analysis we have been able to pinpoint the exact nature of RNA molecules that interact with RIG-I during the course of SeV and influenza virus infections and induce the antiviral response through the RIG-I signaling pathway. The application of deep sequencing techniques allowed us to examine the RNA molecules without any a priori assumptions about their identity and eliminated biases commonly introduced by more traditional methods such as Northerns and quantitative PCR where the selection of primers and probes can greatly influence the observed result. We conclude that viral replicating RNA constitutes the majority of the viral immunostimulatory RNA associated with RIG-I. In infections with both viruses we observed that shorter molecules preferentially associate with RIG-I over longer ones even though all of these RNAs should contain identical 5′-triphosphorylated ends. The preference of RIG-I for shorter substrates might explain why influenza virus has been shown to be exclusively recognized by RIG-I unlike most other RNA viruses that normally rely on both RIG-I and MDA5 recognition. All of the molecules we have identified as specifically interacting with RIG-I also contain some dsRNA regions directly adjacent to the triphosphate, supporting conclusions from previous work that this panhandle-type architecture of the RNA molecule may be preferentially recognized by RIG-I (12, 13). The identification of the influenza virus NS segment and SeV DI RNA as preferred substrates of RIG-I also agrees with their predicted roles as RIG-I PAMPs on the basis of the U-rich composition of this RNA (22). It will be very interesting to see what types of RNAs associate with RIG-I in infections with other viruses and whether generation of DI particles by Mononegavirales members is a prerequisite for RIG-I activation. The extent of secondary structure characteristics in viral RNA would be expected to vary highly between various viruses and the exact role these differences might play in RIG-I activation is an interesting and complex question. The fact that CIP and TAP treatment led to complete loss of DI's ability to induce an immunostimulatory response is interesting and illustrates that the 5′ triphosphate (although a diphosphate has not been experimentally ruled out in this study) is required even in the context of a relatively long dsRNA region, such as SeV DI RNA. Our finding that DI subgenomic particles preferentially associate with RIG-I provides an explanation for the historical observation that viruses containing these particles act as superior IFN inducers (26, 29). The approach we used in our work provides a powerful tool for analysis of RIG-I–associated RNA from various viral infections in multiple cell types as well as other protein/RNA complexes.
Materials and Methods
RIG-I/RNA Complex Immunoprecipitation.
A549 human lung carcinoma cells were infected with a high MOI of SeV-C or PR8ΔNS1 viruses. Infections were allowed to proceed for 24 or 4 h and the cells were washed five times with cold PBS and lysed in a 0.5% Nonidet P-40 buffer. Lysates were frozen at −80 °C before being subjected to immunoprecipitation with either an anti-RIG-I antibody or an anti-GFP antibody (Abcam ab1218). Fractions from IPs were obtained during the process and frozen for future protein analysis. RNA was isolated from agarose beads by proteinase K treatment in SDS buffer and phenol/chlorophorm extraction followed by ethanol precipitation. Immunostimulatory activity of isolated RNA was analyzed by transfecting a small fraction of RNA into a 293T ISRE-FF reporter cell line and measurement of FF activity.
Biochemical Analysis of RNA.
Isolated RNA was subjected to treatment with RNAseA (Qiagen), CIP (Promega), and TAP (Epicentre) and transfected into the 293T ISRE-FF reporter cell line following each treatment to assay potential loss of immunostimulatory activity. As controls, purified influenza PR8 virus RNA, purified SeV-C RNA, total RNA from SeV-C–infected cells, and poly(I:C) were used in various experiments. Viral RNA was isolated from sucrose cushion purified virus with phenol/chlorophorm or TRIzol (Invitrogen) extraction, and total cellular RNA was isolated with TRIzol extraction. Agilent RNA chip analysis was performed at the microarray facility at Mount Sinai School of Medicine using the mRNA chip.
Deep Sequencing Analysis of RNA.
Total RNA isolated from immunoprecipitations or from cell lysates was prepared for Illumina sequencing using the mRNA-Seq (Illumina) sample preparation kit according to manufacturer's instruction. To analyze all RNA species present, the initial poly(A) RNA isolation step was omitted. Because ribosomal RNA presented an overwhelming portion of all RNA in either immunoprecipitations or total cellular RNA, a RiboMinus Eukaryote Kit for RNA-Seq (Invitrogen) was used before deep sequencing to remove a large portion of ribosomal species. The RNA was checked following ribosomal RNA removal for its ability to induce the ISRE-FF reporter, thereby excluding the removed ribosomal sequences as possible inducers of RIG-I. Sequencing was performed on the Illumina Genome Analyzer in the Mount Sinai sequencing facility. Obtained sequences were mapped to human and viral genomes and relative abundances were analyzed between RIG-I pulldown and control samples. Average ratios for influenza virus genomic segments between RIG-I pulldown and control pulldowns were calculated by determining the relative sequence abundance at each position on the genomic segment and calculating the average of those ratios over every 100 nt.
TaqMan Quantitative PCR Analysis.
Q-PCR analysis was performed with utilization of Roche LightCycler 480 technology. All Q-PCR reactions incorporated multiplexed human actin β internal controls and relative abundance of each RNA was calculated with reference to this control.
T7 RNA Transcription.
Templates for T7 RNA transcription were synthesized from PR8 pDZ plasmids coding for individual RNA segments of influenza PR8 virus (30). T7 SeV-C DI particle template was created by RT amplification of SeV DI RNA from infected cells. A truncated T7 promoter was added to each DNA segment by PCR. T7 transcription reactions were carried out with a T7 MEGAscript kit (Ambion). RNA was purified with RNAeasy columns (Qiagen) and analyzed on denaturing agarose gels for correct size and purity.
Supplementary Material
Acknowledgments
We thank Dr. Estanislao Nistal-Villan (Mount Sinai School of Medicine, New York, NY) for the RIG-I antibody, Dr. Adam Vigil (University of California, Irvine, CA) for assistance with manuscript preparation, Dr. Luis Martinez-Sorbido (University of Rochester, Rochester, NY) for the 293T ISRE-FF cell line, Richard Cadagan and Osman Lizardo for technical assistance, and Mount Sinai sequencing facility for assistance with deep sequencing processing and analysis. This research was partly funded by National Institutes of Health Grants R01AI46954, U19AI83025, U01AI082970, and U54AI57168 and by the National Institute of Allergy and Infectious Diseases-funded Center for Research in Influenza Pathogenesis (HHSN266200700010C). A.B. is supported by National Institute of Allergy and Infectious Diseases Training Program in Mechanisms of Virus–Host Interactions (2T32AI007647-11).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005077107/-/DCSupplemental.
References
- 1.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 2.Foy E, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140:397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 5.Yount JS, Gitlin L, Moran TM, López CB. MDA5 participates in the detection of paramyxovirus infection and is essential for the early activation of dendritic cells in response to Sendai virus defective interfering particles. J Immunol. 2008;180:4910–4918. doi: 10.4049/jimmunol.180.7.4910. [DOI] [PubMed] [Google Scholar]
- 6.Melchjorsen J, et al. Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J Virol. 2005;79:12944–12951. doi: 10.1128/JVI.79.20.12944-12951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato H, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Gitlin L, et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 10.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 11.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawai T, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 16.Meylan E, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 17.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Xu LG, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Takahasi K, et al. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol Cell. 2008;29:428–440. doi: 10.1016/j.molcel.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 22.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 24.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ranjith-Kumar CT, et al. Agonist and antagonist recognition by RIG-I, a cytoplasmic innate immunity receptor. J Biol Chem. 2009;284:1155–1165. doi: 10.1074/jbc.M806219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strahle L, Garcin D, Kolakofsky D. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology. 2006;351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 27.García-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 28.Odagiri T, Tashiro M. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol. 1997;71:2138–2145. doi: 10.1128/jvi.71.3.2138-2145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marcus PI, Sekellick MJ. Defective interfering particles with covalently linked []RNA induce interferon. Nature. 1977;266:815–819. doi: 10.1038/266815a0. [DOI] [PubMed] [Google Scholar]
- 30.Quinlivan M, et al. Attenuation of equine influenza viruses through truncations of the NS1 protein. J Virol. 2005;79:8431–8439. doi: 10.1128/JVI.79.13.8431-8439.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.