Abstract
Protein modification by conjugation of small ubiquitin-related modifier (SUMO) is involved in diverse biological functions, such as transcription regulation, subcellular partitioning, stress response, DNA damage repair, and chromatin remodeling. Here, we show that the serine/arginine-rich protein SF2/ASF, a factor involved in splicing regulation and other RNA metabolism-related processes, is a regulator of the sumoylation pathway. The overexpression of this protein stimulates, but its knockdown inhibits SUMO conjugation. SF2/ASF interacts with Ubc9 and enhances sumoylation of specific substrates, sharing characteristics with already described SUMO E3 ligases. In addition, SF2/ASF interacts with the SUMO E3 ligase PIAS1 (protein inhibitor of activated STAT-1), regulating PIAS1-induced overall protein sumoylation. The RNA recognition motif 2 of SF2/ASF is necessary and sufficient for sumoylation enhancement. Moreover, SF2/ASF has a role in heat shock-induced sumoylation and promotes SUMO conjugation to RNA processing factors. These results add a component to the sumoylation pathway and a previously unexplored role for the multifunctional SR protein SF2/ASF.
Keywords: posttranslational modification, splicing factor, RNA processing, E3 ligase
Ser/Arg-rich (SR) proteins were first described as regulators of both constitutive and alternative splicing (1, 2). They are characterized by a modular structure consisting of a C terminal domain-rich in arginine and serine dipeptides (RS domain) and one or two N-terminal RNA-recognition motifs (RRMs) (1). Although RS domains were first identified as protein-protein interaction platforms, it has been shown that they contact the RNA directly at the splicing branch point and the 5′ splice site (3, 4). In addition, RRMs, originally reported to contact the RNA, were shown to mediate protein-protein interactions (5). The function of SR proteins exceeds splicing regulation (2, 6). They regulate transcription (7), mRNA export (8), mRNA stability (9, 10), translation (5, 11), and genome stability (12, 13). Splicing factor 2/alternative splicing factor [SF2/ASF, recently renamed SRSF1 (14)] is a prototypical member of the SR protein family (15, 16). Its second RRM (RRM2) is required for translation regulation via mammalian target of rapamycin binding (5) and for SF2/ASF recruitment to nuclear stress bodies (nSBs) upon heat shock (17, 18).
Small ubiquitin-related modifier (SUMO) is a transient and reversible posttranslational protein modifier (19–21). SUMO proteins (SUMO1 to -4) are expressed in an immature proform that carries a C-terminal stretch of variable length. Removal of this C-terminal extension by SUMO-specific proteases (SENPs) leaves an invariant Gly-Gly motif that marks the C terminus of the mature protein (22). The steps involved in the SUMO pathway resemble those of the ubiquitin pathway (23). The first step is the ATP-dependent activation of a mature SUMO protein by the SUMO-specific E1 activating enzyme heterodimer (SAE I/SAE II in mammals). Next, SUMO is transferred from SAE II to the E2 conjugating enzyme Ubc9, forming a thioester linkage between the catalytic Cys residue of Ubc9 and the C-terminal carboxy group of SUMO (24). Finally, Ubc9 transfers SUMO to the substrate: an isopeptide bond is formed between the C-terminal Gly residue of SUMO and a Lys side chain of the target (25). Although most targets can be sumoylated in the presence of E1 and E2 in vitro, the process is usually facilitated by SUMO E3 ligases in vivo (26–28). The E3 ligases best characterized to date include the protein inhibitor of activated STAT (PIAS) (29), the polycomb protein Pc2 (27), and the nuclear pore complex protein RanBP2 (28). It is difficult to conceive, however, that the scarce number of components identified to date account for the regulation of this complex pathway (19). Thus, it is expected that additional proteins (i.e., cofactors) function to regulate the sumoylation pathway, as it is the case for ubiquitin E3 ligase complexes (30, 31). Reversibility of the sumoylation process is achieved by the action of SENPs that deconjugate SUMO from its target proteins.
We report here that the SR protein SF2/ASF is a regulator of protein sumoylation. SF2/ASF greatly stimulates sumoylation both in vivo and in vitro, and its depletion inhibits overall SUMO conjugation. SF2/ASF interacts with the SUMO E2 conjugating enzyme Ubc9 and with specific substrates, facilitating the transfer of SUMO from the thioester intermediate to the substrate. SF2/ASF also interacts with the SUMO E3 ligase PIAS1 both physically and functionally. These effects are dependent on the RRM2 of SF2/ASF, which is also sufficient to stimulate sumoylation. SF2/ASF has a role in hyperthermic stress-induced SUMO conjugation and stimulates the sumoylation of RNA processing factors. Taken together, these results show that SF2/ASF acts as a cofactor stimulating SUMO conjugation: it displays some characteristics of an E3 ligase and also regulates the function of the well-characterized E3 ligase PIAS1.
Results and Discussion
SF2/ASF Regulates Protein Sumoylation in Cultured Cells.
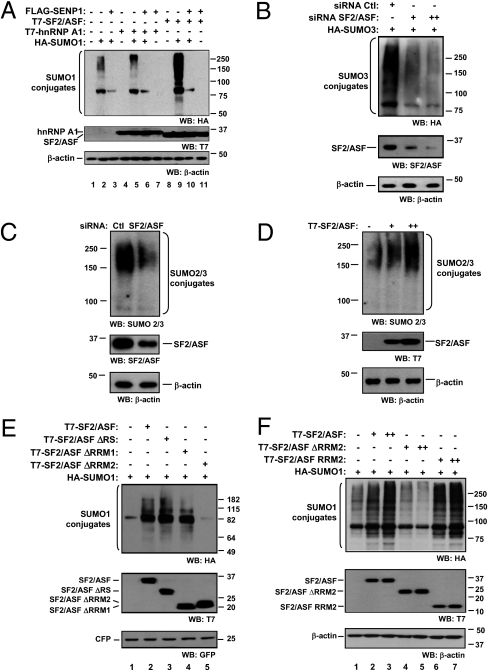
Considering that (i) splicing regulators and ubiquitin/Ubl regulators are found together in protein complexes and share a localization pattern within the cell (32–35), and (ii) sumoylation regulates different processes that are interconnected with splicing, we decided to gain insight into the role of SR proteins in the SUMO pathway. We chose the best-characterized SR protein, SF2/ASF, whose involvement in signal-induced splicing and translation regulation was previously studied in our group (36, 37). Coexpression of SF2/ASF with mature SUMO1 in HEK 293T cells strongly stimulated global protein sumoylation. Another splicing regulator, heterogeneous nuclear ribonucleoprotein (hnRNP) A1, has little or no effect on protein sumoylation (Fig. 1A). The effect of SF2/ASF on sumoylation is dose-dependent and comparable to the one exerted by the SUMO E3 ligase PIAS1 (Fig. S1A). In every case, SUMO1 conjugates are lost when SENP1 is coexpressed (Fig. 1A and Fig. S1A). SF2/ASF also enhances SUMO3 conjugation and both effects are observed in different cell lines (Fig. S2). We then addressed the role of endogenous SF2/ASF in this process. SUMO3 and SUMO1 conjugation was drastically impaired in SF2/ASF-depleted cells (Fig. 1B and Fig. S1B, respectively).
Fig. 1.
SF2/ASF promotes SUMO conjugation in living cells in a RRM2-dependent manner. In every case, HEK 293T cells were transfected with the indicated plasmids and siRNA. Unless clearly stated, cells were lysed 48 h posttransfection in Laemmli sample buffer. Proteins were separated by SDS/PAGE and subject to Western blot, as indicated at the bottom of each panel. (A) Cells were transfected with HA-SUMO1, T7-SF2/ASF, T7-hnRNP A1, and FLAG-SENP1, as indicated (500 ng each). (B) Cells were transfected with a control siRNA (siRNA Ctl) or with an SF2/ASF-specific siRNA [12 nM (+) or 25 nM (++)]. After 24 h, cells were retransfected with HA-SUMO3. (C) Cells were transfected with a control siRNA (Ctl) or with siRNA against SF2/ASF (25 nM), and after 72 h cells were lysed in Laemmli buffer. (D) Cells were transfected with T7-SF2/ASF [500 ng (+) or 1 μg (++)]. (E) Cells were transfected with 500 ng HA-SUMO1, 500 ng of T7-SF2/ASF or its deletion mutants, and 50 ng of pECFP for transfection efficiency and loading control. (F) Cells were transfected with 500 ng HA-SUMO1 and 100 (+) or 500 ng (++) of T7-SF2/ASF, T7-SF2/ASF ΔRRM2 or T7-SF2/ASF RRM2.
We assessed the impact of SF2/ASF expression levels on endogenous SUMO2/3 conjugation. Diminishing SF2/ASF expression by siRNA reduces (Fig. 1C), but its overexpression stimulates SUMO2/3 conjugation in a dose-dependent manner (Fig. 1D).
These results demonstrate that SF2/ASF is a hitherto unexplored regulator of the sumoylation pathway. Its physiological expression is required for maintaining normal overall sumoylation levels, as shown by the RNAi strategy, and reminiscent of the effect of the yeast E3 ligases Siz1 and Siz2 gene disruption that abolishes modification of most targets (38).
RRM2 Is Necessary and Sufficient for SF2/ASF-Stimulated Sumoylation.
We used deletion mutants (Fig. S3) to analyze the role of each SF2/ASF domain on SUMO conjugation. SF2/ASF mutants lacking the RRM1 (ΔRRM1) or the RS domain (ΔRS) show similar sumoylation-enhancing activity to wild-type SF2/ASF. In contrast, the mutant lacking the RRM2 (ΔRRM2) is unable to stimulate sumoylation (Fig. 1 E and F and Fig. S2). Remarkably, expression of the RRM2 by itself is sufficient to stimulate sumoylation (Fig. 1F).
We took advantage of the fact that the ΔRRM1 and RRM2 constructs are siRNA-resistant (Fig. S4 A and B) and the effect of SF2/ASF knockdown on SUMO conjugation was rescued by expression of either one of these constructs, ruling out any nonspecific effect of this siRNA (Fig. S4C).
The effect of other members of the SR protein family was analyzed (Fig. S5). SRp20 [recently renamed SRSF3 (14)], which does not contain an RRM2, failed to stimulate sumoylation. We then tested two SR proteins that harbor RRM2s differing in their degree of identity with SF2/ASF RRM2, SRp30c [recently renamed SRSF9 (14)], and SRp40 [recently renamed SRSF5 (14)] (72 and 37% identity, respectively) (Fig. S5B). SRp30c but not SRp40 is able to stimulate protein sumoylation (Fig. S5C).
These results point to the RRM2 as the major determinant of SF2/ASF sumoylation-stimulatory function, and also indicate that the mere presence of any RRM2 is not sufficient to confer sumoylation-enhancing activity. A structural and mutational analysis of different RRM2 domains should provide insightful information in this respect.
To compare SF2/ASF ability to regulate sumoylation with its activity as an alternative splicing regulator, we analyzed three well-characterized SF2/ASF-responsive splicing eventes (fibronectin EDI exon, CFTR exon 9, and adenovirus E1a) (Fig. S6 A–C) upon overexpression of SF2/ASF or its deletion mutants. The RRM2 domain alone, although sufficient to enhance sumoylation, either fails to alter splicing patterns of FN EDI exon and CFTR exon 9, or alters the splicing pattern in a different fashion than SF2/ASF, as shown for adenovirus E1a transcripts (Fig. S6 D–I). This lack of correlation between SF2/ASF effects on sumoylation and splicing suggest that the role for SF2/ASF in the sumoylation process cannot be explain exclusively by its splicing activity, and point to a splicing-independent function for SF2/ASF in the sumoylation process.
SF2/ASF Interacts with Ubc9 and Stimulates Sumoylation of Specific Substrates.
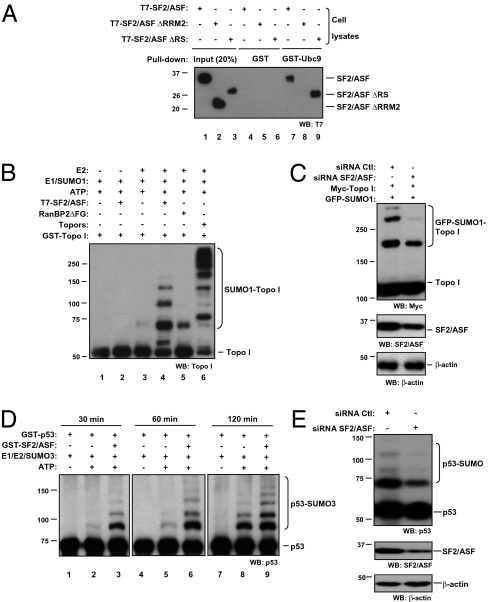
To test whether SF2/ASF directly interacts with the sumoylation machinery, we performed GST pull-down assays by incubating GST-Ubc9 with cell extracts expressing full-length SF2/ASF or its deletion mutants. T7-SF2/ASF is pulled down by GST-Ubc9 but not by GST alone. The ΔRRM2 mutant is unable to bind Ubc9, but the ΔRS mutant that is as efficient as wild-type SF2/ASF in stimulating sumoylation does interact with Ubc9 (Fig. 2A). SF2/ASF-Ubc9 interaction is independent from RNA or DNA, as it is not disturbed by the addition of RNase/DNase to the reaction mixture (Fig. S7A), and appears to be direct because T7-SF2/ASF purified from HEK 293T cells (Fig. S7B) could be pulled down by GST-Ubc9 (Fig. S7C). GST-Ubc9 also pulled down endogenous SF2/ASF (Fig. S7D).
Fig. 2.
SF2/ASF interacts with Ubc9 and promotes Topo I and p53 sumoylation in vitro and in living cells. (A) HEK 293T cells were transfected either with T7-SF2/ASF wt, ΔRRM2 or ΔRS and lysates were prepared as described in Materials and Methods. Cleared lysates were incubated with 2 μg GST or GST-Ubc9 and pulled down with glutathione Sepharose beads. After SDS/PAGE, SF2/ASF binding was analyzed by Western blotting with an anti-T7 antibody. (B) In vitro sumoylation reactions were performed using GST-Topoisomerase I (residues 1–200, “Topo I”) as a substrate. Topo I (1 μg, ∼1 μM) was incubated with 150 ng E1 (∼65 nM), 30 ng Ubc9 (∼85 nM), and 1 μg SUMO1 (∼4.5 μM), either with or without purified T7-SF2/ASF (200 ng, ∼360 nM), GST-RanBP2ΔFG (10 ng, ∼8 nM), or GST-Topors (268/644) (400 ng, ∼300 nM) for 30 min. Reactions were stopped by addition of 1 vol of Laemmli sample buffer. One-fourth of the reaction was run in SDS/PAGE and analyzed by Western blot as indicated at the bottom of each panel. (C) HEK 293T cells were transfected either with control (Ctl) or SF2/ASF-specific siRNA (15 nM) and, 24 h later, transfected with full-length Myc-tagged Topo I (500 ng) and GFP-SUMO1 (500 ng). Cells were lysed 48 h later in Laemmli sample buffer and subject to Western blot, as indicated at the bottom of each panel. (D) SF2/ASF stimulates p53 sumoylation in vitro. GST-p53 (250 ng, ∼167 nM) was incubated with E1 (150 ng, ∼65 nM), E2 (30 ng, ∼85 nM), and SUMO3 (1 μg, ∼4.5 μM), either with or without GST-SF2/ASF (200 ng, ∼190 nM), in sumoylation assay buffer (66). Aliquots were taken from the reaction at the indicated time points and analyzed by SDS/PAGE, followed by Western blotting with an anti-p53 antibody. (E) Depletion of SF2/ASF affects p53 sumoylation in vivo. HEK 293T cells were transfected either with control (Ctl) or SF2/ASF-specific siRNA (15 nM). Cells were lysed 72 h later in Laemmli sample buffer and subject to Western blot, as indicated at the bottom of each panel.
To address whether the sumoylation-stimulatory function of SF2/ASF can take place independently of its other known regulatory activities, we tested the effect of SF2/ASF in cell-free in vitro sumoylation reactions with well-known sumoylation substrates. Sumoylation of Topoisomerase I (hereafter Topo I) is weakly seen upon incubation with E1, E2, and SUMO1 (Fig. 2B). Addition of purified T7-SF2/ASF increased sumoylation of Topo I. However, although the active fragment of RanBP2, RanBP2ΔFG, displays marginal stimulation of Topo I sumoylation compared with SF2/ASF, the SUMO E3 ligase Topors exerts a stronger effect, enhancing the formation of high molecular-weight SUMO1-Topo I conjugates, as previously reported (39). SF2/ASF is unable to conjugate SUMO in the absence of Ubc9, ruling out a role as an E2-conjugating enzyme (Fig. 2B). GST-SF2/ASF purified from bacteria also enhances in vitro sumoylation of Topo I (Fig. S8A). In agreement with these results, knockdown of endogenous SF2/ASF by siRNA inhibits Topo I sumoylation in living cells (Fig. 2C).
In vitro sumoylation reactions were also performed with p53 as a substrate (40, 41). Limiting amounts of Ubc9 were used and SUMO3 conjugation to p53 was hardly detectable at early time-points. Addition of purified GST-SF2/ASF increased the amount of SUMO-conjugated p53 in a time-dependent manner (Fig. 2D). Similar results were obtained with SUMO1 (Fig. S8B). Consistently, depletion of SF2/ASF by siRNA diminishes SUMO conjugation to p53 in living cells (Fig. 2E).
SF2/ASF interacts with Topo I, as previously described (42, 43), and with p53 (Fig. S8 C and D). The nature and biological relevance of this latter interaction await further investigation. Furthermore, SF2/ASF has no effect on sumoylation of Sp100, a substrate of the E3 ligase RanBP2 (Fig. S8E) (28).
Thus, SF2/ASF directly affects the sumoylation machinery by interacting with the E2 conjugating enzyme and promoting the sumoylation of specific substrates, Topo I and p53, both in vitro and in vivo.
Our initial finding that depletion of SF2/ASF drastically impairs sumoylation in living cells could be due in part to the reported effects of SF2/ASF depletion on cell cycle and apoptosis (13). However, a direct action of SF2/ASF on the SUMO pathway is evident from the results obtained with in vitro sumoylation assays.
SF2/ASF Stimulates SUMO Transfer from Ubc9 to the Substrate.
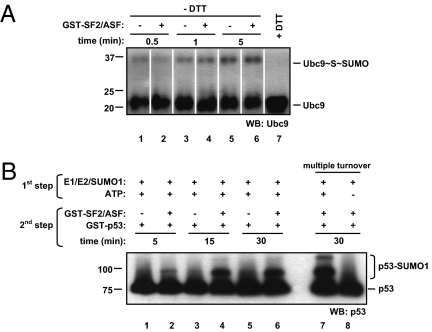
Based on the results described above, SF2/ASF could be acting at two nonmutually exclusive levels: promotion of Ubc9-SUMO thioester linkage formation or stimulation of SUMO transfer from Ubc9 to the substrate. Although Ubc9 loading increased with time, SF2/ASF failed to enhance the rate of SUMO-Ubc9 thioester bond formation (Fig. 3A). SUMO transfer from Ubc9 to p53 was then measured in single turnover reactions, as previously described (44). SF2/ASF clearly stimulates this step in the SUMO conjugation pathway (Fig. 3B), indicating that SF2/ASF action takes place downstream of Ubc9.
Fig. 3.
SF2/ASF promotes the transfer of SUMO from Ubc9 to p53. (A) SF2/ASF does not stimulate Ubc9-SUMO1 thioester bond formation. Recombinant E1 (150 ng), E2 (100 ng), and SUMO1 (200 ng) were incubated as described in Material and Methods for the indicated time points, either with or without GST-SF2/ASF (200 ng). Reactions were stopped by addition of an equal volume of 2× nonreducing SDS-sample buffer (lanes 1–6) or 2× Laemmli sample buffer containing 100 mM DTT (lane 7), and analyzed by Western blotting with an anti-Ubc9 antibody. (B) SF2/ASF stimulates SUMO1 transfer from Ubc9 to p53. Recombinant E1 (150 ng), E2 (300 ng), and SUMO1 (200 ng) were incubated as described in Materials and Methods. After diluting the reaction in EDTA-containing buffer, the SUMO1-loaded E2 was incubated with GST-p53 (200 ng), either with or without GST-SF2/ASF (200 ng) for the indicated time points. In lanes 7 and 8, EDTA was omitted, enabling further rounds of Ubc9 loading and transfer (“multiple turnover”). Reactions were stopped by addition of an equal volume of 2× Laemmli sample buffer and analyzed by SDS/PAGE, followed by Western blotting with an anti-p53 antibody.
E3 ligases are known to facilitate the transfer of ubiquitin or Ubl proteins from an E2 enzyme to a substrate protein and to display substrate specificity. Except for ubiquitin HECT E3 ligases, all other known E3 ligases form complexes with the SUMO-charged E2 and the target (19). Thus, SF2/ASF shares some characteristics with E3 ligases. Further biochemical and structural characterization is needed to determine the precise mechanism of action of SF2/ASF at the SUMO transfer step.
SF2/ASF Interacts with PIAS1 and Regulates its E3 Activity.
Given that PIAS1 copurifies with the spliceosome (34) and resembles scaffold-attachment factors known to interact with SR proteins (33), we tested if SF2/ASF could interact with PIAS1. GST pull-down assays demonstrate this interaction, which is dependent on SF2/ASF RRM2 (Fig. S9A) and seems to be direct, as purified T7-SF2/ASF could be pulled down by GST-PIAS1 (Fig. S9B). SF2/ASF-PIAS1 interaction takes place in a DNA/RNA-independent manner (Fig. S9C). Coimmunoprecipitation assays show that PIAS1 interacts with wild-type SF2/ASF but not with the mutant lacking the RRM2 in whole-cell lysates (Fig. S9D).
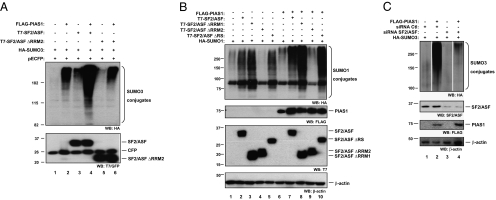
As expected, PIAS1 enhances SUMO conjugation. Coexpression of PIAS1 and SF2/ASF leads to a synergistic effect on overall protein sumoylation (Fig. 4A). The mutant SF2/ASF lacking the RRM2 was unable to enhance PIAS-mediated sumoylation (Fig. 4A). When a suboptimal amount of PIAS1 was transfected, it still enhanced the ability of wild-type SF2/ASF, ΔRRM1, or ΔRS mutants to stimulate sumoylation (Fig. 4B). Furthermore, PIAS-enhanced SUMO conjugation is impaired when SF2/ASF is depleted by siRNA (Fig. 4C), indicating that the SUMO E3 ligase activity of PIAS1 depends on the presence of SF2/ASF. These experiments show an RRM2-dependent functional synergism between PIAS1 and SF2/ASF that stimulate protein sumoylation in living cells. It is worth noting that the functional interaction between PIAS1 and SF2/ASF appears to involve yet unidentified cellular proteins because it could not be recapitulated with in vitro sumoylation assays (Fig. S9E). The results presented here are not enough to rule out an effect of SF2/ASF on PIAS1 translation, as suggested by Fig. 4B (lanes 6–10). However, under this hypothesis, depletion of SF2/ASF by siRNA should reduce PIAS protein levels, which does not seem to be the case. Keeping in mind the already described activity of SF2/ASF in the translation process (5, 11, 36, 37), this potential additional level of control remains to be explored. Considering that SF2/ASF has no SUMO E2 activity, its role as a coregulator of a SUMO E3 ligase adds a further level of regulation to the sumoylation pathway, resembling the case of ubiquitin E3 ligase complexes (31).
Fig. 4.
PIAS1 activity is regulated by SF2/ASF. (A) SF2/ASF enhances PIAS1 activity on SUMO conjugation. HEK 293T cells were transfected with HA-SUMO3 (500 ng) and FLAG-PIAS1, T7-SF2/ASF or T7-SF2/ASF ΔRRM2 (500 ng each) as indicated, together with 50 ng of pECFP for transfection efficiency and loading control (total amount of DNA, 2 μg). Cells were lysed in Laemmli sample buffer after 48 h. Western blotting was performed with the antibodies indicated at the bottom of each panel. (B) HEK 293T cells were transfected with HA-SUMO1 (500 ng), FLAG-PIAS1 (100 ng), T7-SF2/ASF, or its deletion mutants (500 ng each) as indicated (total amount of DNA, 2 μg). Cells were lysed in Laemmli sample buffer after 48 h. Western blotting was performed with the antibodies indicated at the bottom of each panel. (C) SF2/ASF is critical for PIAS1-mediated SUMO conjugation. HEK 293T cells were transfected either with a control siRNA (siRNA Ctl) or an SF2/ASF specific siRNA. After 24 h, cells were retransfected with the plasmids expressing HA-SUMO3 and FLAG-PIAS1 (500 ng). Forty-eight hours later, cells were lysed in Laemmli sample buffer and subject to Western blot, as indicated at the bottom of each panel.
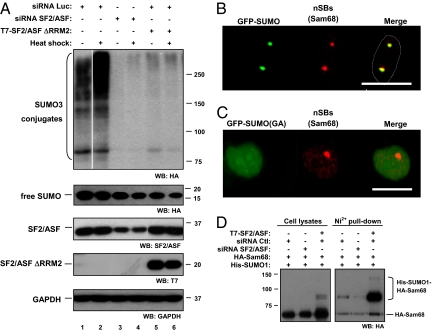
SF2/ASF Has a Role in Heat Shock-Stimulated Sumoylation.
We decided to study SF2/ASF role in the heat-shock response because this treatment enhances SUMO conjugation (45) (Fig. 5A). Furthermore, SUMO2 and SUMO3 are required for cells to survive to hyperthermic stress (46). Depletion of SF2/ASF by siRNA greatly inhibits heat shock-induced sumoylation, pointing to SF2/ASF as a key factor in this regulatory phenomenon. Moreover, overexpression of SF2/ASF ΔRRM2 exerts similar effects to SF2/ASF depletion (Fig. 5A), suggesting this mutant could be acting in a dominant-negative manner.
Fig. 5.
SF2/ASF participates in stress-induced sumoylation and regulates Sam68 sumoylation in living cells. (A) HEK 293T cells were transfected with the indicated siRNAs. After 24 h, cells were retransfected in every case with an HA-SUMO3 plasmid (500 ng) and with a T7-SF2/ASF ΔRRM2 plasmid (1 μg), when indicated. Forty-eight hours later, cells were exposed to heat shock (42 °C for 15 min) or left untreated, lysed in Laemmli sample buffer, and subject to Western blot as indicated at the bottom of each panel. (B and C) HEK 293T cells were transfected with GFP-SUMO1 or the GFP-SUMO1(GA) mutant 24 h after plating. The next day, cells were heat-treated as indicated in Materials and Methods. Cells were then fixed, permeabilized, and incubated with a Sam68 antibody as a nSB marker. Alexa 637-conjugated secondary antibody was used. Sam68 is shown in red and GFP-SUMO in green. (Scale bars, 20 μm.) (D) HEK 293T cells were transfected with the indicated siRNAs and 24 h later with the indicated plasmids. After 48 h, cells were harvested and lysates were subject to Ni-NTA agarose purification under denaturing conditions. His-tagged sumoylated proteins were subject to Western blot using an anti-HA antibody. A fraction of each cell lysate (3%) was run in parallel as input control.
Considering that (i) SF2/ASF shifts its subnuclear localization in response to heat shock from splicing speckles to nSBs in an RRM2-dependent manner (17, 18), and (ii) nSBs are known to recruit heat-shock transcription factors, as well as a subset of pre-mRNA processing factors (47) leading to changes in splicing patterns (48), we asked whether nSBs are sites of SUMO-conjugated proteins. Upon heat shock, Sam68, an RNA processing factor and a hallmark of nSBs (47–49), colocalized with wild-type GFP-SUMO1 but not with a mutant that is unable to conjugate to target proteins [GFP-SUMO1(GA)] (50) (Fig. 5 B and C). These results indicate that nSBs colocalized with SUMO-conjugated proteins but not with the free SUMO pool. Thus, upon heat shock, sumoylated proteins localize in nSBs, where SF2/ASF as well as other pre-mRNA processing factors reside. It is tempting to speculate that SF2/ASF could be part of a regulatory network accounting not only for splicing regulation but also for the regulation of sumoylation-dependent protein activity required for cell recovery upon hyperthermic stress.
SF2/ASF and Sumoylation of RNA Processing Factors.
Taking into account that (i) many RNA binding proteins have been reported to be sumoylated (51–54) and (ii) recent studies have shown an enrichment in the fraction of RNA processing- and RNA binding-proteins upon purification of SUMO substrates in vivo (55, 56), we wondered whether SF2/ASF could enhance sumoylation of RNA processing factors.
Cell extracts from HEK 293T transfected with His-SUMO1 were subject to Ni-NTA purification to enrich His-tagged sumoylated proteins. Pulled-down proteins were analyzed by Western blot with specific antibodies against RNA metabolism-related factors known to be sumoylation substrates (57–59). SF2/ASF overexpression stimulates, but its knockdown diminishes SUMO conjugation to the nucleolar protein Nop58 (Fig. S8 F and G) and to Sam68 (Fig. 5D).
An effect of sumoylation on the activity of different RNA-binding proteins has been reported (57, 60, 61), and sumoylation has been shown to regulate mRNA 3′-end processing (61, 62). SUMO modification of nucleolar proteins controls their subnuclear distribution and members of the SENP family are concentrated within the nucleolus. SUMO conjugation to Topo I regulates its nucleolar delocalization upon cell treatment with anticancer drugs (63). SENP3-dependent sumoylation status of nucleophosmin (NPM1) is involved in the regulation of rRNA processing and ribosome synthesis (64). Our finding that SF2/ASF expression levels control the sumoylation status of RNA metabolism-related proteins lead us to propose SF2/ASF as a putative molecular and functional link between the RNA processing and sumoylation machineries, which of course awaits further investigation.
In conclusion, we have described a previously unknown role for the SR protein SF2/ASF in the sumoylation pathway. SF2/ASF exerts its effect at least at two different levels: it interacts with Ubc9 promoting the sumoylation of specific substrates and it regulates the SUMO E3 ligase activity of PIAS1. We have identified specific targets of this activity and deciphered SF2/ASF mechanism of action. Finding additional targets for SF2/ASF sumoylation-regulatory task and deepening into its physiological relevance is our immediate future challenge.
Materials and Methods
Cell Lines.
HEK 293T cells were grown in DMEM supplemented with 10% FBS, 4.5 g/L glucose, and 110 mg/L sodium pyruvate.
Plasmids, siRNAs, and Transfection.
HEK 293T cells were transfected with Lipofectamine 2000 (Invitrogen). The list of expression vectors used is available in SI Materials and Methods. For knockdown experiments, siRNA duplexes were transfected at the concentration indicated in each figure. Small interfering RNA targeting luciferase mRNA was used as a control.
Immunofluorescence of Heat-Shocked Cells.
HEK 293T cells were transfected with GFP-SUMO1 or the GFP-SUMO1(GA) mutant 24 h after plating. The next day, cells were transferred to a 42 °C water bath for 1 h and then allowed to recover for 1 h at 37 °C. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100 in PBS. More details of the protocol are available in SI Materials and Methods. Sam68 was used as an nSBs marker, as described (47–49).
Recombinant Proteins.
GST, GST-Ubc9, and GST-SUMO3 were expressed in Escherichia coli M15(pREP4) cells and GST-SF2/ASF, GST-Topors (268/644), GST-PIAS1 in E. coli BL21(DE3) Rosetta strain by induction with 1 mM IPTG and purified with glutathione Sepharose beads (GE Healthcare). T7-SF2/ASF and T7-SF2/ASF ΔRRM2 were purified from transfected HEK 293T lysates exactly as described (65). Proteins were analyzed by SDS/PAGE and Coomassie staining for quantification and purity. Recombinant GST-Topo I, SUMO1, and SUMO3 were purchased from LAE Biotech. SUMO E1, SUMO E2, GST-p53, GST-RanBP2ΔFG, and GST-Sp100 were from BIOMOL/Enzo Life Sciences.
Immunoprecipitation.
HEK 293T cells were lysed in 1 mL of lysis buffer [20 mM Tris-HCl pH 7.5, 150 mM KCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 1 mM β-glycerophosphate, 10% glycerol, 1× Complete Protease Inhibitor (Roche)] and incubated for 30 min at 4 °C. After centrifugation for 20 min at 4 °C, supernatants were used immediately for coimmunoprecipitation or kept at −80 °C. Details of this protocol are available in SI Materials and Methods.
GST Pull-Down Assays.
Lysates from HEK 293T cells expressing the indicated proteins or purified recombinant proteins were used for pull-down experiments, as described (30).
Ni2+ Pull Down.
HEK 293T cells were transfected in 6-cm dishes with the indicated siRNAs (10 nM) and 24 h later with the indicated plasmids. After 48 h, His-SUMO1 conjugated proteins were purified under denaturing conditions using Ni-NTA agarose beads according to the manufacturer's instructions (QIAGEN).
In Vitro Sumoylation Reactions.
In vitro sumoylation reactions were performed as described (66). Recombinant proteins were added at the concentrations indicated in each figure legend.
Thioester Formation Assay.
Assays were carried out essentially as described (30), with 100 ng E1, 100 ng E2, 200 ng SUMO1, and 200 ng recombinant SF2/ASF when indicated.
SUMO Transfer Reactions.
Recombinant E1 (150 ng), E2 (300 ng), SUMO1 (200 ng), GST-p53 (200 ng), and GST-SF2/ASF (200 ng) were used. A detailed protocol is available in SI Materials and Methods.
Western Blot and Antibodies.
Protein samples were resolved by SDS/PAGE and transferred to Hybond-P membranes (GE Healthcare). Membranes were blocked and incubated with the primary antibody. After washing, membranes were incubated with HRP-conjugated secondary antibodies (Biorad). Membranes were developed using ECL plus reagent (GE Healthcare). The list of antibodies used is available in SI Materials and Methods.
The experiments were performed at least three times, and representative images are shown in each case.
Supplementary Material
Acknowledgments
An extended list of people and institutions that helped with this work is available in SI Acknowledgments. This work was supported by grants from Agencia Nacional de Promoción Científica y Tecnológica of Argentina; Universidad de Buenos Aires; Consejo Nacional de Investigaciones Científicas y Técnicas of Argentina; and European Alternative Splicing Network.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004653107/-/DCSupplemental.
References
- 1.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen H, Green MR. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol Cell. 2004;16:363–373. doi: 10.1016/j.molcel.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 4.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 5.Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Long JC, Caceres JF. The SR protein family of splicing factors: Master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 7.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemaire R, et al. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 2002;16:594–607. doi: 10.1101/gad.939502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 11.Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Wang J, Manley JL. Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation. Genes Dev. 2005;19:2705–2714. doi: 10.1101/gad.1359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge H, Manley JL. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 16.Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 17.Chiodi I, et al. RNA recognition motif 2 directs the recruitment of SF2/ASF to nuclear stress bodies. Nucleic Acids Res. 2004;32:4127–4136. doi: 10.1093/nar/gkh759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metz A, Soret J, Vourc'h C, Tazi J, Jolly C. A key role for stress-induced satellite III transcripts in the relocalization of splicing factors into nuclear stress granules. J Cell Sci. 2004;117:4551–4558. doi: 10.1242/jcs.01329. [DOI] [PubMed] [Google Scholar]
- 19.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 20.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 22.Melchior F, Schergaut M, Pichler A. SUMO: Ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 24.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin D, et al. Identification of a substrate recognition site on Ubc9. J Biol Chem. 2002;277:21740–21748. doi: 10.1074/jbc.M108418200. [DOI] [PubMed] [Google Scholar]
- 26.Hochstrasser M. SP-RING for SUMO: New functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 27.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 28.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 29.Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- 30.Carbia-Nagashima A, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 31.Joazeiro CA, Weissman AM. RING finger proteins: Mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 32.Ihara M, Stein P, Schultz RM. UBE2I (UBC9), a SUMO-conjugating enzyme, localizes to nuclear speckles and stimulates transcription in mouse oocytes. Biol Reprod. 2008;79:906–913. doi: 10.1095/biolreprod.108.070474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nayler O, et al. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26:3542–3549. doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan JA, et al. Protein inhibitors of activated STAT resemble scaffold attachment factors and function as interacting nuclear receptor coregulators. J Biol Chem. 2002;277:16993–17001. doi: 10.1074/jbc.M109217200. [DOI] [PubMed] [Google Scholar]
- 36.Blaustein M, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 37.Blaustein M, et al. SF2/ASF regulates proteomic diversity by affecting the balance between translation initiation mechanisms. J Cell Biochem. 2009;107:826–833. doi: 10.1002/jcb.22181. [DOI] [PubMed] [Google Scholar]
- 38.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 39.Hammer E, Heilbronn R, Weger S. The E3 ligase Topors induces the accumulation of polysumoylated forms of DNA topoisomerase I in vitro and in vivo. FEBS Lett. 2007;581:5418–5424. doi: 10.1016/j.febslet.2007.10.040. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez MS, et al. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weger S, Hammer E, Heilbronn R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005;579:5007–5012. doi: 10.1016/j.febslet.2005.07.088. [DOI] [PubMed] [Google Scholar]
- 42.Labourier E, et al. Interaction between the N-terminal domain of human DNA topoisomerase I and the arginine-serine domain of its substrate determines phosphorylation of SF2/ASF splicing factor. Nucleic Acids Res. 1998;26:2955–2962. doi: 10.1093/nar/26.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kowalska-Loth B, Girstun A, Trzcińska AM, Piekiełko-Witkowska A, Staroń K. SF2/ASF protein binds to the cap region of human topoisomerase I through two RRM domains. Biochem Biophys Res Commun. 2005;331:398–403. doi: 10.1016/j.bbrc.2005.03.180. [DOI] [PubMed] [Google Scholar]
- 44.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 45.Tempé D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 46.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 47.Denegri M, et al. Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell. 2001;12:3502–3514. doi: 10.1091/mbc.12.11.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biamonti G. Nuclear stress bodies: A heterochromatin affair? Nat Rev Mol Cell Biol. 2004;5:493–498. doi: 10.1038/nrm1405. [DOI] [PubMed] [Google Scholar]
- 49.Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: A proteomic analysis. Proc Natl Acad Sci USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navascues J, et al. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Struct Biol. 2008;163:137–146. doi: 10.1016/j.jsb.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 53.Vassileva MT, Matunis MJ. SUMO modification of heterogeneous nuclear ribonucleoproteins. Mol Cell Biol. 2004;24:3623–3632. doi: 10.1128/MCB.24.9.3623-3632.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vertegaal AC, et al. A proteomic study of SUMO-2 target proteins. J Biol Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 55.Blomster HA, et al. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pungaliya P, et al. TOPORS functions as a SUMO-1 E3 ligase for chromatin-modifying proteins. J Proteome Res. 2007;6:3918–3923. doi: 10.1021/pr0703674. [DOI] [PubMed] [Google Scholar]
- 57.Babic I, Cherry E, Fujita DJ. SUMO modification of Sam68 enhances its ability to repress cyclin D1 expression and inhibits its ability to induce apoptosis. Oncogene. 2006;25:4955–4964. doi: 10.1038/sj.onc.1209504. [DOI] [PubMed] [Google Scholar]
- 58.Lyman SK, Gerace L, Baserga SJ. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA. 1999;5:1597–1604. doi: 10.1017/s1355838299991288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajan P, et al. Regulation of gene expression by the RNA-binding protein Sam68 in cancer. Biochem Soc Trans. 2008;36:505–507. doi: 10.1042/BST0360505. [DOI] [PubMed] [Google Scholar]
- 60.Desterro JM, et al. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell. 2005;16:5115–5126. doi: 10.1091/mbc.E05-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vethantham V, Rao N, Manley JL. Sumoylation regulates multiple aspects of mammalian poly(A) polymerase function. Genes Dev. 2008;22:499–511. doi: 10.1101/gad.1628208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mo YY, Yu Y, Shen Z, Beck WT. Nucleolar delocalization of human topoisomerase I in response to topotecan correlates with sumoylation of the protein. J Biol Chem. 2002;277:2958–2964. doi: 10.1074/jbc.M108263200. [DOI] [PubMed] [Google Scholar]
- 64.Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008;9:273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cazalla D, Sanford JR, Cáceres JF. A rapid and efficient protocol to purify biologically active recombinant proteins from mammalian cells. Protein Expr Purif. 2005;42:54–58. doi: 10.1016/j.pep.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 66.Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.