Abstract
New Zealand's extinct flightless moa radiated rapidly into a large number of morphologically diverse species, which produced an equally large range of egg morphologies. The exact number of moa species, as well as the characteristics of the eggs they laid, remains contentious. Moreover, like most extinct species, we understand little about their nesting and incubation habits. We used a modified ancient DNA extraction procedure to recover exogenous mitochondrial and nuclear DNA from the inside and outside surfaces of moa eggs. We used sequences from the inside of 69 eggshells to directly assign these remains to seven of the 10 currently recognized moa species. In addition we were able to assign, to the species level, six of the rare reconstructed “whole” eggs. These molecular results enabled us to identify two distinct lineages within the genus Euryapteryx. Members of these lineages differed in eggshell thickness, with one lineage being characterized by a relatively thin eggshell. Unexpectedly, several thin-shelled eggs were also shown to belong to the heaviest moa of the genera Dinornis, Euryapteryx and Emeus, making these, to our knowledge, the most fragile of all avian eggs measured to date. Moreover, sex-specific DNA recovered from the outer surfaces of eggshells belonging to species of Dinornis and Euryapteryx suggest that these very thin eggs were likely to have been incubated by the lighter males. The thin nature of the eggshells of these larger species of moa, even if incubated by the male, suggests that egg breakage in these species would have been common if the typical contact method of avian egg incubation was used.
Keywords: avian eggs, exogenous DNA, endogenous DNA, ratite, evolution
New Zealand's extinct moa (Aves: Dinornithiformes) were ratite birds. Living species of ratites include the emu and cassowary of Australia and New Guinea, the kiwi of New Zealand, and the rhea of South America. Extinct members include the elephant bird of Madagascar as well as approximately 10 species of moa (1–4). Moa were flightless and were particularly diverse in both size and in morphology (1, 2, 4–8). For example, members of the genus Dinornis were characterized by large body size, weighing as much as 250 kg, with extreme sexual dimorphism in which females were approximately twice the size of males (2, 5). In contrast, individuals of the coastal moa Euryapteryx curtus weighed as little as 9 kg (4) and females were approximately 20% larger than males. As a group, moa represented one of the most dramatic examples of morphological radiation in the history of vertebrates (1, 2, 6) and have been of international interest since originally described by Richard Owen in 1839 (9). This remarkable ratite group vanished soon after the settlement of New Zealand by Polynesians late in the 13th century (8, 10).
Many questions about moa biology and evolution remain, despite more than 100 y of scientific investigation. Moa eggs ranged from 10 times the volume of a standard chicken egg (approximately 120 mm × 95 mm; ∼0.06 kg) to more than 80 times the volume of a standard chicken egg (approximately 240 mm × 178 mm; ∼4.5 kg) (4, 8, 11, 12). They also varied in color from bright olive green to various shades of off-white, and the thicknesses ranged from 0.50 to 1.89 mm (4, 11, 12). Although moa eggshell fragments are abundant in New Zealand's many natural and archaeological sites, “whole” moa eggs are rare, with only 36 being known to date (11, 12). Of these, only three have been associated with their respective species as a result of their fortuitous association with identifiable skeletal material, and in one instance by the retrieval of an almost complete embryonic skeleton from within the egg (13).
The methods moa used to incubate their eggs, and which sex incubated, is unknown. In extant ratites the incubation of eggs and the rearing of young is often the role of the male, who may simultaneously incubate one to approximately 30 eggs from a number of hens (14, 15). To determine the specific identity of a range of whole eggs and egg fragments and to investigate how moa nested, we aimed to recover both endogenous and exogenous ancient DNA from the inside and outside surfaces respectively of moa eggs. Our aims were first to identify these eggs to their respective species, and second to reveal more about moa egg morphology and incubation behavior.
Results and Discussion
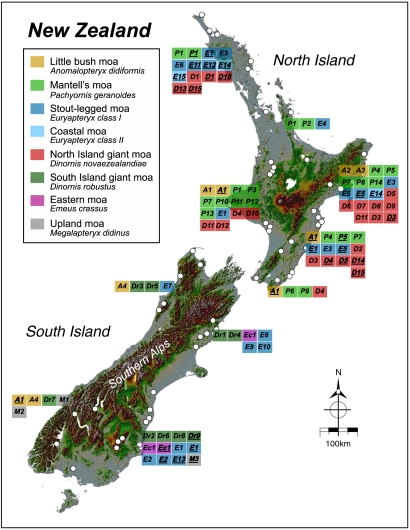
By using DNA sequences previously published by the present group (1–3) and others available on GenBank (Table S1), PCR primers were designed to amplify a short fragment of the moa mitochondrial Hypervariable Region I (HVRI). Using these primers, a fragment of about 70 bp was amplified, which contained a highly variable signature region of approximately 30 bp. We sequenced an additional 26 subfossil moa samples from a wide range of localities across New Zealand (Fig. 1 and Table 1; Table S2 provides more details). Together with the GenBank sequences, this database comprised a total of 82 reference samples.
Fig. 1.
Moa HVRI haplotype locations. Haplotypes are shown in blocks adjacent to the coastal area where they were found. White circles represent actual sample locations (for location and sample names, refer to Fig. S1 and Table S2). Haplotypes obtained from moa eggshells are underlined.
Table 1.
HVRI haplotypes recovered from subfossil moa bones and egg shells
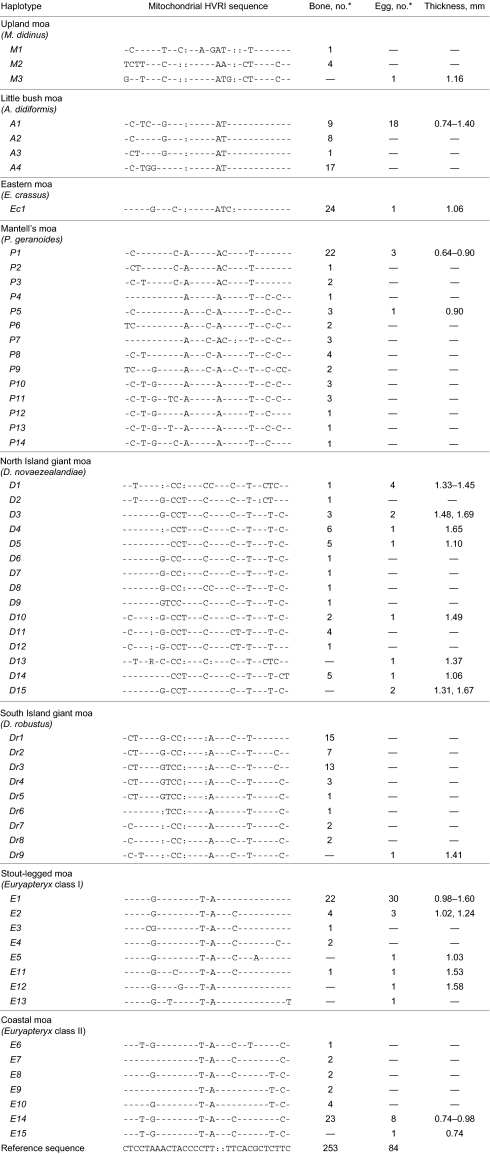 |
*Number of moa bones and eggshells measured that provided clear species and haplotype identification.
Haplotypes detected are given with each haplotype representing a unique sequence. All sequences are compared with a reference sequence given at the bottom.
By targeting the 70-bp HVRI fragment, we were able to recover DNA from 83 of 117 eggshell samples analyzed (71%). These samples were recovered from a range of sites including archaeological middens, rock-shelters and caves, dunes, as well as alluvial deposits. Eggs from archaeological sites have been shown to range in age from 400 to 700 y BP (16). For example, moa midden material from large sites at Rakaia Mouth, Wakanui, Waitaki Mouth (Oamaru), and Wairau Bar dated from 481 ± 51 y BP to 638 ± 44 y BP. Moa samples from natural sites typically ranged in age from approximately 1,300 to 19,000 y BP. For example, moa bones recovered from Makirikiri and Pyramid Valley swamps and Honeycomb Hill cave (Oparara) dated from 1,296 ± 28 y BP to 18,901 ± 192 y BP (2, 6, 8, 16).
We obtained 69 sequences from the inside surfaces of the eggs, and 41 of these also yielded DNA from the outer surface. An additional 15 sequences were obtained from eggshell outer surfaces only (Table S2). A total of 53 unique haplotypes were detected among the moa subfossil remains, of which 14 were also detected in eggshell DNA. A further eight haplotypes were found in eggshell only (Table 1). In all cases the haplotypes found in eggshells were the same or similar to the haplotypes found in moa bone recovered from the same geographic region (Fig. 1).
Amplification of a longer HVRI fragment of 156 bp resulted in a 40% reduction in amplification success. Similarly, purification of DNA extracts using silica binding and elution (DNeasy Blood and Tissue Kit; Qiagen) also resulted in a reduction of amplification success for the 70 bp fragment. This is possibly a result of the inability of silica to efficiently retain DNA fragments of less than 100 bp with standard extraction procedures (17). For this reason, all DNA extractions were precipitated in the presence of linear polyacrylamide (18), which we found ideal for the recovery of small amounts of highly fragmented DNA such as that from ancient material.
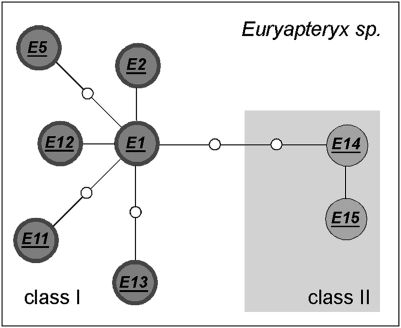
Sequences from inside egg surfaces could be directly assigned to seven of the 10 currently recognized species of moa by using the constructed database (Table 1). We established that olive green eggshells (sample AIM LB12037) belonged to the South Island's upland moa Megalapteryx didinus (Table 1 and Table S2) (11). We also identified the first known eggshell fragments of the small Mantell's moa Pachyornis geranoides (19). The eggshell of this species is relatively thin (0.64–0.90 mm) and is characterized by 40 to 70 pore depressions per cm2 on the outer surface, with a maximum pore-depression length of 0.7 mm. A single eggshell fragment was attributed to the large eastern moa Emeus crassus, and at 1.06 mm thick was very similar to the one known eggshell sample of this species that was recovered from the pelvis of an E. crassus individual identified by bone morphology (20). Several eggshells of varying thicknesses (0.74–1.40 mm) could be attributed to the little bush moa Anomalopteryx didiformis, common to both New Zealand's North and South Islands (4, 6). For the genus Euryapteryx, difficult to group into species despite extensive studies of skeletal material and large-scale mitochondrial analysis including the COI barcoding region (1, 3), we detected two classes of eggshell thickness: one thick class (class I; 0.98–1.60 mm) associated with haplotypes E1, E2, E5, E11, E12, and E13, and one thin class (class II; 0.74–0.98 mm) associated with the haplotypes E14 and E15, suggesting individuals of two distinct sizes within this genus (Table 1 and Fig. 2). These are likely to represent two species.
Fig. 2.
Spanning network analysis of Euryapteryx haplotypes detected in eggshell. Haplotypes are grouped as class I (those derived from eggshells 0.98–1.60 mm thick) and class II (those derived from eggshells 0.74–0.98 mm thick). White circles depict intermediate, undetected haplotypes.
DNA sequences retrieved from four of the whole moa eggs (Fig. 3 and Table S2) showed that a large egg from Central Otago (sample AIM LB4016) belonged to the form of Euryapteryx (class I) associated with the thicker eggshells. The other three eggs (egg 6 from Waitomo, HBM 26043 from Hukanui, and HBM 5466 from Havelock North; Fig. 3 and Table S2) all belonged to the little bush moa A. didiformis. These identifications matched assumptions based on moa species known to have been present in these areas (11). However, sampling of fragments remaining after the partial reconstruction of an egg thought to belong to the North Island giant moa D. novaezealandiae (Egg 5, Table S2), and another reported to be from the heavy-footed moa Pachyornis elephantopus (OU PLR355; Fig. 3 and Table S2) (11), suggested that these eggs are actually from, respectively, the relatively small A. didiformis and the South Island giant moa D. robustus. Surprisingly, eggshell identified by DNA as D. robustus was thinner than expected (1.41 mm; Table 1). Similarly, we showed that eggshell fragments of the North Island giant moa D. novaezealandiae were as thin as 1.06 mm (Table 1). These shells are very thin for giant birds whose females are estimated to weigh as much as 250 kg and raises questions as to how these eggs may have been incubated (2, 4, 5).
Fig. 3.
Whole eggs sampled. (A) AIM LB4016; (B) HBM 5466; (C) HBM 26043; (D) HBM Hk7b (partial egg only); (E) OU PLR355; (F) WCM cmm collection [WO 227; egg 5 (large) and egg 6 (small)] (11). For E and F, only shell fragments associated with these eggs were sampled. AIM, Auckland Museum; HBM, Hawke's Bay Museum; OU, University of Otago; WCM, Waitomo Caves Museum.
However, for the thickest of class I Euryapteryx shells (boldface in Table 2) and all those belonging to the giant moa species D. novaezealandiae and D. robustus, the outside shell surfaces yielded DNA from the males only (Table 2). Furthermore, in the two cases in which the outer mitochondrial haplotype differed from that of the inner shell surface, the sex recovered from the outer shell surface DNA was male (Table 2). This indicates that, in these cases, the DNA was sloughed from the male, and most likely this occurred during the incubation process. We suggest that, for P. geranoides, the larger (class I) form of Euryapteryx, and both species of Dinornis, males were likely to be the incubators of eggs (Table 2). As expected, the haplotype of the female DNA detected on the outside surface of eggshell samples always matched that detected on the inside surface, suggesting that the outer shell DNA in these cases is most likely that remaining from the female during the egg laying process. Although our sample size was relatively small, no evidence for interspecific brood parasitism was detected.
Table 2.
Sexes recovered from the inside and outside of moa eggs
| Inside | Outside | ||
| Haplotype | Sex | Haplotype | Sex |
| A. didiformis (M/F, 30–50 kg) | |||
| — | — | A1 | F |
| A1 | — | A1 | F |
| A1 | — | A1 | F |
| A1 | — | A1 | F |
| A1 | F | A1 | F |
| A1 | M | A1 | F |
| A1 | M | — | — |
| A1 | M | A1 | M |
| A1 | F | A1 | F |
| A1 | M | A1 | F |
| E. crassus (M, 36–49 kg; F, 55–75 kg) | |||
| Ec1 | M | Ec1 | F |
| P. geranoides (M, 15–20 kg; F, 21–32 kg) | |||
| P5 | M | P5 | M |
| D. novaezealandiae (M, 61–90 kg; F = 98–170 kg) | |||
| — | — | D16 | M |
| — | — | D5 | M |
| D1 | F | — | — |
| — | — | D1 | M |
| D1 | F | D1 | — |
| D10 | F | - | — |
| D3 | F | D3 | — |
| D3 | M | D3 | M |
| D. robustus (M, 57–114 kg; F, 92–249 kg) | |||
| Dr1 | M | Dr1 | M |
| Euryapteryx sp. Class I (M, 55–80 kg; F, 67–105 kg) | |||
| E5 | F | E1/2 | M |
| E1 | F | E1 | — |
| E1 | M | E1 | F |
| E1 | F | — | — |
| E1 | M | — | — |
| E1 | M | — | — |
| — | — | E1 | F |
| E1 | F | E1 | F |
| E1 | M | E1 | F |
| E1 | F | E1 | M |
| E1 | — | E1 | F |
| E1 | F | — | — |
| E1 | F | — | — |
| E1 | F | E1 | — |
| — | — | E12 | M |
| E1 | F | E1 | — |
| — | — | E1 | F |
| E11 | M | E1 | M |
| Euryapteryx sp. class II (M, 15–20 kg; F, 20–30 kg) | |||
| E14 | F | E15 | — |
Haplotypes and sexes are presented for DNA extracted from the inside and outside surfaces of moa eggshells. Male DNA detected on outside shell surfaces are shown in bold. Weights of males and females of the different species are shown in kg (Table S3).
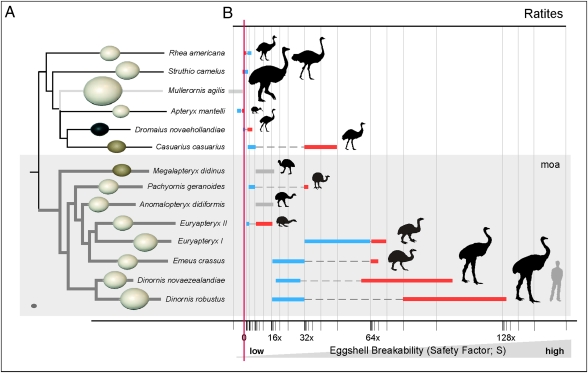
The relatively large size of ratites generally means their eggs are more vulnerable to breakage during incubation than those of any other bird (21–23). Hence, perhaps it is not surprising that, in all living ratites, the lighter sex (nearly always the male) incubates the eggs (Fig. 4B) (24). Experimentally, it has been shown that the likelihood of eggshell breakage (expressed as safety factor; S) can be calculated using “yield point force” (F; the force required for egg breakage when applied to the egg poles). This is commonly derived from eggshell thickness (L) as F = (0.01469L2.0424/m) − 1, where m is the mass of the incubating animal (21). Negative S values indicate a significant risk of egg breakage during nesting (21), and every S value decrease of 0.1 unit doubles the likelihood of egg breakage. By using these measures, we show that the eggs of Dinornis were far more susceptible to breakage than any of the 3,434 avian species measured to date (21). This is the case, even if the lighter male (approximately 75 kg) incubated, with these egg thicknesses being more characteristic of a bird approximately one third of this weight (Fig. 4B).
Fig. 4.
Ratite egg phylogeny and eggshell strength. (A) Phylogeny of ratites based on extensive mitochondrial DNA sequencing (1, 2, 6, 32). The light grey line indicates the uncertain position of Mullerornis within the tree (32). Relative egg sizes are shown based on volumes calculated from shape dimensions or shell thickness (4, 22, 23). A dark grey chicken egg is shown in the light grey box (Bottom Left) for size comparison. (B) Egg breakability factors (S) calculated using mass of the incubating bird and egg thickness (Table S3). Blue indicates males, red indicates females, grey indicates no (or unknown) sexual dimorphism.
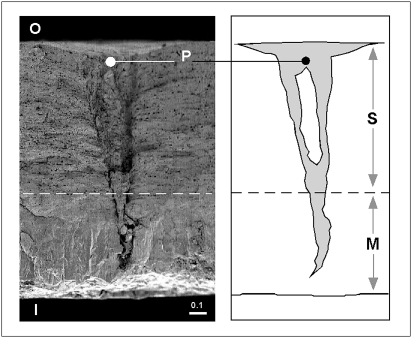
Our success in retrieving exogenous DNA from egg surfaces is most likely the result of a refinement of current ancient DNA extraction procedures, in addition to the targeting of short (<100 bp) DNA fragments. The chemical composition of the shell and its pore-rich structure (25, 26) may also fortuitously enhance the collection and storage of exogenous DNA (Fig. 5). Attempts to extract DNA from several smooth interpore regions on the outer surface consistently failed to yield amplifiable DNA, in direct contrast to extractions that included pore-rich regions. We suggest that cells sloughed from the laying or incubating bird are collected in the pores and are effectively held there as a result of the known tight interaction of calcium from the eggshell with the DNA backbone (27).
Fig. 5.
Radial section of “thick” moa eggshell (AIM LB4015, 1.3 mm thick). I, inside eggshell surface; O, outside surface. A single branched pore is marked P. The outer thick spongy layer (S) and inner mammillary layer (M; 25) are indicated by the arrows and separated by a dotted line. SEM courtesy of D. K. Zelenitsky (26). Scale bar in millimeters. Outline of pore is shown (Right).
The detection of male DNA on the outer shell surfaces from the three largest moa species—D. robustus, D. novaezealandiae, and the larger morphological form of Euryapteryx (class I)—suggests that, for these species at least, male incubation was likely. However, considering the weight of even the smaller males of these species, the possibility of egg breakage is still many times greater than that of any other bird and raises questions how these eggs were successfully incubated (Fig. 4). One possibility is that eggs were incubated in a nest specifically constructed to support the weight of the incubating animal. However, the exact structure of moa nests remains unclear, and what little evidence there is suggests that moa nests were unsupported, being very similar to those of emus and cassowaries (24, 28) in comprising a simple scrape in the ground surrounded by a thin layer of coarse, woody vegetation and stripped bark (29, 30). In the absence of an adequately supported nest, we suggest it was unlikely that the larger moa—Dinornis, Emeus, and Euryapteryx—would have been able to incubate their eggs using the contact incubation method practiced by nearly all birds, including the other ratites (24).
The recovery of DNA from the outer surfaces of moa eggs indicates that DNA deposited, by contact only, on exogenous materials can survive for hundreds to thousands of years. This work has allowed both the direct association of moa species to eggs and, considering the fragile nature of the eggs, has raised some questions about how these extinct animals nested.
Materials and Methods
Materials.
Whole moa eggs and shell fragments (Fig. 3 and Table S2) were made available by Auckland Museum, Hawke's Bay Museum, Waitomo Caves Museum, and the University of Otago (Table S2). Sequences from these samples were compared with moa of known species and known provenance (Fig. 1, Fig S1, and Table S2). All sampling was carried out on-site and samples were stored in Eppendorf safe-lock tubes and transferred directly to a −80 °C freezer in the ancient DNA facility at the Institute of Natural Resources, Massey University, Auckland, New Zealand.
DNA Extraction from Eggshell.
Approximately 3 mm3 of eggshell dust was collected by careful slicing (using a scalpel) of eggshell surfaces onto disposable Whatman Benchkote squares. The scalpel blade was wiped with 10% bleach between samples. The eggshell dust was incubated at 55 °C with rotation for approximately 30 min in 200 μL of 0.5 M EDTA/0.01% Triton X-100 with approximately 50 μg of proteinase K. The solution was extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1) followed by chloroform, and the DNA was precipitated from the aqueous layer by the addition of 0.5 volumes (100 μL) of 7.5 M ammonium acetate, 5 μL of linear polyacrylamide carrier (18) and 2.5 volumes (500 μL) of ethanol. The mix was incubated at –20 °C for 20 min and then centrifuged at 15,000 × g for 15 min. The resulting white pellet was washed with 0.8 mL of 80% ethanol and resuspended in 40 μL of water.
DNA Amplification.
A small, highly variable region of the mitochondrial HVR1 region was amplified by PCR using the moa-specific primers: mcrshFF, AGTCGACGCTTCTAGCTTATTCCTATGTAGTGCTYGTAAGGTCTAA; and mcrshRR, CATGCTACCTGCTACTGTCATCTGGTACYAGGGATCATATCACCGTG. These primers harbor generic primer sequences (underlined) at their 5′ termini to allow direct sequencing of very short products (approximately 120 bp; Table S1) (31). A second set of HVR primers, mCR11f (CTGGTATCAGATGGATTTCTT) and mCR4 (1), was used to determine the size of DNA extracted from eggshell. These primers amplified a 156-bp fragment. Approximately 1 μL (up to 5 ng) of DNA was amplified by PCR in 10 μL volumes containing 50 mM Tris-Cl pH 8.8, 20 mM (NH4)2SO4, 2.5 mM MgCl2, 1 mg/mL BSA, 200 μM of each dNTP, 40 ng of each primer, and approximately 0.3 U of platinum Taq (Invitrogen). The reaction mix was overlaid with mineral oil and subjected to amplification in a Hybaid OmniGene thermal cycler using the following parameters: 94 °C for 2 min (×1) and then 94 °C for 20 s and 54 °C to 60 °C for 40 s (×50). Amplified DNAs were detected by agarose gel electrophoresis in 0.5× Tris-borate-EDTA buffer, stained with 50 ng/mL ethidium bromide in 0.5× Tris-borate-EDTA buffer, and then visualized over UV light.
DNA Purification and Sequencing.
Silica bed DNA purification was carried out using a DNeasy blood and tissue kit as outlined by the manufacturer (Qiagen). Positive amplifications were purified by centrifugation through approximately 50 μL of dry Sephacryl S200HR and then sequenced at the Allan Wilson Centre Genome Sequencing Service (Auckland, New Zealand) using Applied Biosystems BigDye Terminator v3.1 chemistry and an ABI3730 Genetic Analyzer (Applied Biosystems). Direct sequencing was carried out using the primer F+4 (CCTCAGTCGACGCTTCTAGCTT) and/or R (CATGCTACCTGCTACTGT). The F+4 primer contains an additional 4 bp at the 5′ terminus to allow single pass sequencing. All sequences were obtained from at least two independent amplifications with a subset of samples being extracted at least twice and sequenced in the reverse direction, using primer R. All sequences obtained were compared with moa sequences generated specifically for this work and those retrieved from GenBank of known species and provenance (Fig. 1 and Table S2).
Sexing by DNA.
Genetic sexing was carried out as described elsewhere (2). Results were considered positive only when multiple successful amplifications of the sex-specific locus (for females) and the autosomal control locus were obtained. For females the sex-specific locus was amplified twice, with as many as eight repeats being performed on males (33).
Ancient DNA.
In accordance with procedures for ancient DNA analysis, all DNA extractions were carried out in a physically separate, dedicated ancient DNA facility at the Institute for Natural Sciences at Massey University, Auckland, New Zealand. All sequences were obtained from at least one extraction and multiple amplifications. In addition, for sequence verification, repeat extractions and amplifications were carried out on 10 randomly selected samples, and several samples were extracted and amplified at Griffith University Ancient DNA Laboratories (Nathan, Australia).
Morphometrics and Allometry.
Eggshell thickness was determined using electronic Kincrome calipers accurate to 0.01 mm. To ensure consistency, shell fragments from the body of the egg were selected, thereby ignoring the slightly thicker shell from the egg poles. Fragments were measured at several points and the average thickness was recorded. Only shells with detailed surface structure were selected to avoid those that may have weathered. The F value, defined as the load required to initiate egg crushing when applied to both poles, was determined using the equation derived by Ar et al. (21). Briefly, egg mass (W, in grams) was calculated with the following equation:
where L is shell thickness in μm. F was then calculated as 50.86W0.915. The two equations were combined to give an F calculation of 0.01469L2.0424, allowing direct determination of yield point force using eggshell thickness only. The safety factor S is determined by simply dividing F by bird mass (B, in grams) and subtracting 1. Negative S values indicate susceptibility to breakage.
Supplementary Material
Acknowledgments
We thank Sara Browne from the Hawke's Bay Museum, Celina Yapp at the Waitomo Caves Discovery Centre, and Ian Smith from Otago University for allowing measurement and sampling of moa eggshells from their collections. We are grateful to D. K. Zelenitsky for allowing reproduction of the scanning electron micrograph in Fig. 5. This study was supported by a Royal Society of New Zealand Marsden Fund grant (“The Molecular Basis of an Extinct Phenotype”; to D.M.L.), Griffith University, the University of Auckland, and Massey University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.C.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0914096107/-/DCSupplemental.
References
- 1.Baker AJ, Huynen LJ, Haddrath O, Millar CD, Lambert DM. Reconstructing the tempo and mode of evolution in an extinct clade of birds with ancient DNA: The giant moas of New Zealand. Proc Natl Acad Sci USA. 2005;102:8257–8262. doi: 10.1073/pnas.0409435102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huynen L, Millar CD, Scofield RP, Lambert DM. Nuclear DNA sequences detect species limits in ancient moa. Nature. 2003;425:175–178. doi: 10.1038/nature01838. [DOI] [PubMed] [Google Scholar]
- 3.Lambert DM, et al. Is a large-scale DNA-based inventory of ancient life possible? J Hered. 2005;96:279–284. doi: 10.1093/jhered/esi035. [DOI] [PubMed] [Google Scholar]
- 4.Worthy TH, Holdaway RN. The Lost World of the Moa. Christchurch, New Zealand: Canterbury Univ Press; 2002. [Google Scholar]
- 5.Bunce M, et al. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature. 2003;425:172–175. doi: 10.1038/nature01871. [DOI] [PubMed] [Google Scholar]
- 6.Bunce M, et al. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proc Natl Acad Sci USA. 2009;106:20646–20651. doi: 10.1073/pnas.0906660106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tennyson A, Martinson P. Extinct Birds of New Zealand. Wellington, New Zealand: Te Papa Press; 2006. [Google Scholar]
- 8.Anderson A. Prodigious Birds; Moas and Moa-Hunting in Prehistoric New Zealand. Cambridge, UK: Cambridge Univ Press; 1989. [Google Scholar]
- 9.Owen R. On the bone of an unknown struthious bird from New Zealand, meeting of November 12, 1839. Proc Zool Soc London. 1840;VII:169–171. [Google Scholar]
- 10.Holdaway RN, Jacomb C. Rapid extinction of the moas (Aves: Dinornithiformes): Model, test, and implications. Science. 2000;287:2250–2254. doi: 10.1126/science.287.5461.2250. [DOI] [PubMed] [Google Scholar]
- 11.Gill BJ. A catalogue of moa eggs (Aves: Dinornithiformes) Records Auckland Museum. 2006;43:55–80. [Google Scholar]
- 12.Gill BJ. Eggshell characteristics of moa eggs (Aves: Dinornithiformes) J R Soc N Z. 2007;37:139–150. [Google Scholar]
- 13.Hector J. Notice of an egg of the great moa (Dinornis gigantea), containing remains of an embryo, found in the province of Otago, New Zealand. Proc Zool Soc London. 1868;3:991–992. [Google Scholar]
- 14.Handford P, Mares MA. The mating systems of ratites and tinamous: An evolutionary perspective. Biol J Linn Soc Lond. 1985;25:77–104. [Google Scholar]
- 15.Davies SJJF. Bird Families of the World. 9. Ratites and Tinamous (Tinamidae, Rheidae, Dromaiidae, Casuariidae, Apterygidae, Struthionidae) Oxford: Oxford Univ Press; 2002. [Google Scholar]
- 16.Oskam CL, et al. Fossil avian eggshell preserves ancient DNA. Proc R Soc B. 2010;277:1991–2000. doi: 10.1098/rspb.2009.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DNeasy Blood and Tissue Kit [product specifications] Available at http://www1.qiagen.com/Products/GenomicDnaStabilizationPurification/DNeasyTissueSystem/DNeasyBloodTissueKit.aspx.
- 18.Gaillard C, Strauss F. Ethanol precipitation of DNA with linear polyacrylamide as carrier. Nucleic Acids Res. 1990;18:378. doi: 10.1093/nar/18.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worthy TH. Rediscovery of the types of Dinornis curtus Owen and Palapteryx geranoides Owen, with a new synonymy (Aves: Dinornithiforms) Records of the Museum of New Zealand Te Papa Tongarewa. 2005;16:33–43. [Google Scholar]
- 20.Duff RS. Pyramid Valley. Christchurch, New Zealand: Association of Friends of the Canterbury Museum; 1949. [Google Scholar]
- 21.Ar A, Rahn H, Paganelli CV. The avian egg: Mass and strength. Condor. 1979;81:331–337. [Google Scholar]
- 22.Schönwetter M. Handbuch der Oologie. Berlin: Akademie-Verlag; 1960-1971. [Google Scholar]
- 23.Dickison MR. Durham, NC: Duke University; 2007. The Allometry of Giant Flightless Birds. PhD thesis. [Google Scholar]
- 24.Davies SJJF. In: Grzimek's Animal Life Encyclopedia. 8 Birds I Tinamous and Ratites to Hoatzins. 2nd Ed. Hutchins M, Murphy JB, Schlager N, editors. Farmington Hills, MI: Gale Group; 2003. pp. 69–101. [Google Scholar]
- 25.Tyler C. Some chemical, physical and structural properties of moa egg shells. J Polynesian Soc. 1957;66:110–130. [Google Scholar]
- 26.Zelenitsky DK, Modesto SP. New information on the eggshell of ratites (Aves) and its phylogenetic implications. Can J Zool. 2003;81:962–970. [Google Scholar]
- 27.Martinson HG. Role of the double-stranded nucleic acid backbone configuration in adsorption interactions. Biochemistry. 1973;12:2737–2746. doi: 10.1021/bi00738a029. [DOI] [PubMed] [Google Scholar]
- 28.Campbell AJ. Nests and Eggs of Australian Birds. Sheffield, UK: Pawson and Brailsford; 1900. [Google Scholar]
- 29.Wood JR. Moa (Aves: Dinornithiformes) nesting material from rockshelters in the semi-arid interior of South Island, New Zealand. J R Soc N Z. 2008;38:115–129. [Google Scholar]
- 30.Hartree WH. A preliminary report on the nesting habits of moas on the East Coast of the North Island. Notornis. 1999;46:457–460. [Google Scholar]
- 31.Binladen J, Gilbert MT, Campos PF, Willerslev E. 5′-tailed sequencing primers improve sequencing quality of PCR products. Biotechniques. 2007;42:174–176. doi: 10.2144/000112316. [DOI] [PubMed] [Google Scholar]
- 32.Cooper A, et al. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 2001;409:704–707. doi: 10.1038/35055536. [DOI] [PubMed] [Google Scholar]
- 33.Taberlet P, et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.