Abstract
DNA replication starts at initiation sites termed replication origins. Metazoan cells contain many more potential origins than are activated (fired) during each S phase. Origin activation is controlled by the ATR checkpoint kinase and its downstream effector kinase Chk1, which suppresses origin firing in response to replication blocks and during normal S phase by inhibiting the cyclin-dependent kinase Cdk2. In addition to increased origin activation, cells deficient in Chk1 activity display reduced rates of replication fork progression. Here we investigate the causal relationship between increased origin firing and reduced replication fork progression. We use the Cdk inhibitor roscovitine or RNAi depletion of Cdc7 to inhibit origin firing in Chk1-inhibited or RNAi-depleted cells. We report that Cdk inhibition and depletion of Cdc7 can alleviate the slow replication fork speeds in Chk1-deficient cells. Our data suggest that increased replication initiation leads to slow replication fork progression and that Chk1 promotes replication fork progression during normal S phase by controlling replication origin activity.
Keywords: Cdc7, Cdk2, roscovitine, S phase checkpoint
DNA replication can only start at defined sites of initiation termed replication origins. Metazoan origins are defined by the loading of the prereplication complex (pre-RC, consisting of ORC, Cdc6, Cdt1, and MCM2-7) onto the DNA in a process called replication licensing (1). Metazoan cells contain many more licensed origins than are fired during each round of replication (2). These origins are organized into initiation zones or clusters that are activated at different times during S phase (3). Eukaryotic origin firing is subject to regulation by the DNA damage response pathways. The S phase checkpoint controlled by the ATR and ATM kinases keeps levels of origin firing relatively low during normal S phase, suggesting that excessive origin firing is detrimental to cells (4). If DNA replication is inhibited by DNA damage or shortage of nucleotides, the checkpoint suppresses origin firing more strongly to halt S phase progression until the replication block is removed (5–8). Somewhat contradictory, it has been shown that if replication fork progression is impaired by nucleotide shortages, more origins can be activated per cluster, and that this compensatory origin firing is important for survival of replication blocks (9–12). These observations have prompted the hypothesis that the checkpoint suppresses origins in inactive clusters more strongly than those in clusters that are already active (2, 12). The balance between checkpoint suppression of origin firing and compensatory origin activation might furthermore be regulated by Polo-like kinase 1 (Plk1), which has been shown to counteract the S phase checkpoint in Xenopus egg extracts (13).
The S phase checkpoint responding to replication blocks is mainly mediated by ATR and its downstream effector kinase Chk1 (6–8, 14). ATR becomes activated by regions of single-stranded DNA at stalled replication forks (15) and activates Chk1 by phosphorylating it on Ser345 and Ser317 (16–18). Chk1 has been proposed to regulate origin firing via two pathways. Firstly it inhibits the cyclin-dependent kinase Cdk2 (in complex with Cyclin A or E) by phosphorylation of its activating phosphatase Cdc25A, thus marking Cdc25A for degradation (19). Secondly Chk1 can phosphorylate and inhibit the initiation kinase Cdc7/Dbf4 (Dbf4-dependent kinase, DDK) (20), although it is currently not established that Cdc7 is down-regulated during replication stress (21). Both Cdk2 and Cdc7 phosphorylate the preRC to facilitate loading of the replicative helicase cofactor Cdc45 and thus origin activation (22). Inhibition of Chk1 leads to accumulation of Cdc25A protein in undamaged cells, consistent with Chk1 regulating Cdc25A and thus Cdk2 activity during normal S phase (23). Both ATR and Chk1 inhibition or depletion increases origin firing in unperturbed cells (4, 24).
In addition to increased origin firing, cells inhibited or depleted of Chk1 display reduced rates of replication fork progression (25) as well as an induction of the DNA damage response and double strand breaks (26). Chk1-deficient cells display stretches of single-stranded DNA at nascent replication forks (25). It is not known how Chk1 regulates replication fork progression, although a requirement for Chk1 to control aberrant homologous recombination (HR) at replication forks has been excluded (25). One interesting possibility is that, just like slow replication fork progression leads to increased origin firing, the opposite relationship might exist and increased origin firing could lead to slow replication fork progression, for example by depleting essential replication factors. Lack of Chk1 activity might thus reduce replication fork speeds by increasing origin firing.
In order to test whether increased origin firing is the underlying cause for reduced replication fork progression in Chk1-deficient cells, we used the Cdk inhibitor roscovitine and siRNA-depletion of Cdc7 to down-regulate origin firing. Here we show that the reduction of origin firing to control levels reverses the slow replication fork speeds in Chk1-inhibited cells. Our data suggest that Chk1 regulates replication fork speeds during normal S phase indirectly by inhibition of excess origin firing.
Results
Roscovitine Treatment Does Not Influence Replication Fork Speed.
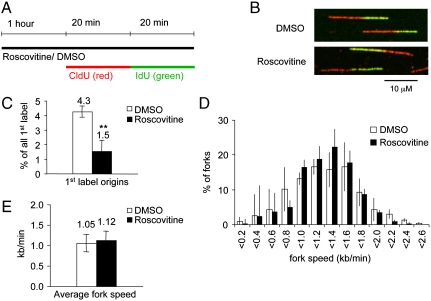
To down-regulate origin firing independently of Chk1 activity, we used the cyclin-dependent kinase (Cdk) inhibitor roscovitine, which efficiently inhibits Cdk1, 2, 5, 7, and 9 (27, 28). U2OS cells were treated with 25 μM roscovitine for 1 h and then in presence of roscovitine pulse-labeled with the thymidine analogues chlorodeoxyuridine (CldU) and iododeoxyuridine (IdU) for 20 min each (Fig. 1A). Cells were lysed, DNA fibers spread out and immunostained using specific antibodies against CldU and IdU (Fig. 1B). This method allows analysis of replication tracts, including measurements of fork speed and origin firing (29) (see Fig. S1A for details). Treatment with roscovitine alone decreased the frequency of origin firing compared to control (Fig. 1C), in agreement with its role as a Cdk inhibitor. A decrease in origin firing will result in delayed S phase progression, unless replication fork speed is increased in order to compensate for the lower amount of replication initiation. However, replication fork speeds were unaffected by roscovitine treatment (Fig. 1 D and E).
Fig. 1.
Effect of the Cdk inhibitor roscovitine on origin firing and replication fork speeds. (A) Labeling protocols for DNA fiber analysis. U2OS cells were pretreated with 25 μM roscovitine (rosc) or an equal volume of DMSO (control) for 1 hour and then pulse labeled with CldU and IdU for 20 minutes each in presence of rosc or DMSO. CldU was detected using a specific primary antibody and a secondary antibody in red. IdU was detected using specific primary antibody and a secondary antibody in green. (B) Representative images of replication tracks from cells treated with DMSO or rosc. (C) Quantification of origin firing in cells treated with DMSO or rosc. First label origins (green-red-green) are shown as percentage of all red (CldU) labeled tracks. (D) Distribution of replication fork speeds in cells treated as in C. (E) Average replication fork speeds in cells treated with DMSO or rosc. Means and standard deviation (S.D.) (bars) of three independent experiments are shown. Values marked with asterisks are significantly different (student’s t-test, ** p < 0.01).
Roscovitine Treatment Increases Replication Fork Speeds in Chk1-Inhibited Cells.
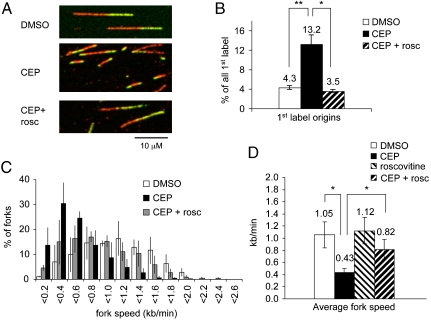
Cells were treated with the specific Chk1 inhibitor CEP-3891 (500 nM) (26) for 1 h, labeled in presence of CEP-3891 as above, and processed for DNA fiber spreads. As expected, CEP-3891 treatment increased the frequency of new origin firing (Fig. 2 A and B). When cells were treated with CEP-3891 and roscovitine together, the frequency of new origin firing was similar to control levels, showing that Cdk inhibition can counteract the increased origin firing induced by Chk1 inhibition (Fig. 2B; see Fig. S1B for more details of replication structures).
Fig. 2.
Cotreatment with roscovitine partially rescues fork slowing Chk1 inhibitor-treated cells. (A) Representative images of replication tracks from cells treated with DMSO, CEP-3891 (CEP), or CEP + roscovitine (rosc). (B) Quantification of origin firing in cells treated with DMSO, CEP, or CEP + rosc. First label origins (green-red-green) are shown as percentage of all red (CldU) labeled tracks. (C) Distribution of replication fork speeds in cells treated as in B. (D) Average replication fork speeds in cells treated with DMSO, CEP, rosc, or CEP + rosc. Means and standard deviation (S.D.) (bars) of three independent experiments are shown. Values marked with asterisks are significantly different (student’s t-test, * p < 0.05, ** p < 0.01).
We then determined replication fork elongation after inhibition of Chk1 and replication origin firing. As expected, CEP-3891 treatment alone also resulted in reduced replication fork speeds (Fig. 2 C and D). Interestingly, if cells were cotreated with roscovitine and CEP-3891, replication fork speeds were higher compared to CEP-3891 treatment alone. Average replication fork speeds in cells treated with CEP-3891 and roscovitine together were intermediate between control- and CEP-3891-treated cells (Fig. 2 C and D). To exclude a contribution of artifacts caused by high frequencies of fork fusion, we decreased pulse labeling times, resulting in shorter labeled replication tracks (see Figs. S2 and S3 and SI Text for details). This yielded results consistent with the experiments using longer pulse labels. Our observations suggest that fork speeds in Chk1-inhibited cells can be partially rescued by inhibiting origin firing, and thus that excessive replication initiation does contribute to the replication fork slowing induced by Chk1 inhibition.
Chk1- and Cdk-Inhibition Have Different Effects on S Phase Progression.
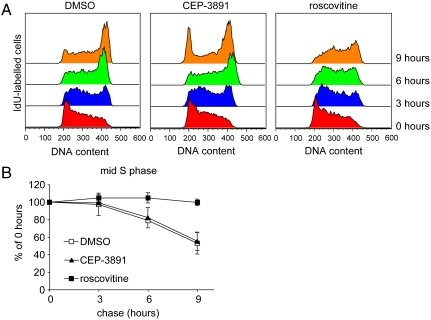
Because it increases origin firing while decreasing fork progression rates, CEP-3891 treatment might not affect overall S phase length if increased initiation was able to compensate for slow fork progression. In contrast, roscovitine treatment decreases origin firing without increasing fork speeds (Fig. 1), which suggested that roscovitine treatment should increase S phase length. To test S phase progression in CEP-3891 or roscovitine-treated cells, we pulse-labeled asynchronously growing U2OS cells with IdU for 20 min and released the cells into IdU-free medium containing DMSO (control), CEP-3891, or roscovitine for 0–9 h. Flow cytometry of nuclei stained for IdU and DNA was used to measure the rate at which IdU-labeled cells progressed through S phase (Fig. 3 and Fig. S4 for details). CEP-3891 treated cells displayed normal S phase progression and an overall accelerated cell-cycle progression, as indicated by the earlier entry into the next cell cycle (Fig. 3A, 9 h). In contrast, roscovitine-treated cells displayed strongly delayed S phase progression (Fig. 3B). In agreement with previous reports (27), roscovitine treatment also induced G2/M arrest (Fig. S4B). These data are in agreement with and support the results from the fiber analyses.
Fig. 3.
Chk1 inhibited cells display normal S phase length while Cdk inhibition slows S phase progression. (A) Flow cytometry profiles of progression through S phase in presence of DMSO, CEP-3891, or roscovitine. Asynchronously growing U2OS cells were pulse labeled with IdU for 20 min and released into medium containing drug or DMSO. Nuclei were immunostained for IdU and stained for DNA using propidium iodide (PI). Profiles show PI distribution of cells stained for IdU (which were in S phase during pulse label) after 0, 3, 6, or 9 hours (see also Fig. S4). (B) IdU-labeled cells in mid-S phase after 3, 6, or 9 hours release from IdU as percentage of fraction at 0 hours. Means and standard deviation (S.D.) (bars) of three independent experiments are shown.
Codepletion of Cdc7 Increases Replication Fork Speeds in Chk1-Depleted Cells.
To use an alternative to roscovitine for inhibiting origin firing, we siRNA-depleted cells of the Cdc7 kinase, which promotes origin firing but is not inhibited by roscovitine (30). Cdc7 depletion was then combined with siRNA to deplete Chk1. U2OS cells were transfected with nontargeting siRNA or siRNA directed against human Chk1 or Cdc7 for 48 h and then subjected to DNA fiber analyses (Fig. 4; see Fig. S5 for more details of replication structures). Interestingly, Cdc7 levels were partly reduced in Chk1-depleted cells and vice versa, suggesting that these two proteins are coregulated (Fig. 4A). We could confirm that Chk1 depletion increased and Cdc7 depletion decreased origin firing (Fig. 4B), although the effects of the siRNA treatments seemed not as pronounced as when using small molecule inhibitors and were not statistically significant. This may be due to difficulties in visualizing changes in origin firing induced by siRNA compared to inhibitors, because inhibitor treatment will increase new origin firing acutely, while siRNA treatment would result in a steady state of elevated origin firing that would be difficult to measure. Next, fiber spreads were analyzed for replication fork speeds. As expected, Chk1 depletion alone reduced replication fork speeds (Fig. 4C), while Cdc7 depletion alone did not affect replication fork speeds (Fig. 4D). Strikingly, codepletion of Cdc7 with Chk1 restored replication fork speeds to nearly control levels (Fig. 4E and F). These observations demonstrate that increased replication initiation is the main cause for slow replication fork rates in Chk1-depleted cells.
Fig. 4.
Cdc7 codepletion rescues fork slowing in Chk1-depleted cells. (A) Protein levels of Cdc7, Chk1, and β-Actin (loading control) in U2OS cells after 48 hours depletion with Cdc7, Chk1, Cdc7, and Chk1 or control siRNA. (B) Quantification of origin firing in Cdc7-, Chk1- or control-depleted cells. First label origins (green-red-green) are shown as percentage of all red (CldU) labeled tracks. (C) Distribution of replication fork speeds in Cdc7- or control-depleted cells. (D) Distribution of replication fork speeds in Chk1- or control-depleted cells. (E) Distribution of replication fork speeds in Cdc7- or Cdc7 and Chk1-depleted cells. (F) Average replication fork speeds in Cdc7-, Chk1-, or control-depleted cells. Means and standard deviation (S.D.) (bars) of three independent experiments are shown. Values marked with asterisks are significantly different (student’s t-test, * p < 0.05).
Discussion
We report that the slow replication fork rates in Chk1-inhibited or depleted cells can be reversed by simultaneous inhibition or depletion of proteins that promote origin firing. Our data therefore suggest that Chk1 effects replication fork speeds during normal S phase by inhibiting excess origin firing. It was previously shown that slowing of replication fork progression by nucleotide shortages can lead to increased origin activation (9–12). To our knowledge, it has not been described so far that increased origin activation can also lead to slowing of replication fork progression. Increased origin firing and reduced replication fork progression are well-described outcomes of ATR signaling defects in mammalian cells (4, 8, 24, 25, 29, 31). Here we show that these two phenotypes are closely linked and that increased origin firing is most likely causing the slowed fork progression. The mechanism of origin control by ATR signaling is known to be largely mediated by Chk1-dependent phosphorylation of Cdc25A to inhibit Cdk2 (19). On the other hand, it is not known whether Chk1 also phosphorylates components of the replication machinery to promote replication fork progression. Our observations suggest that rather than acting directly at the replication fork, Chk1 promotes replication fork progression indirectly by phosphorylating Cdc25A. Our data also suggest that other ways of increasing origin firing, such as overexpression of Cdc7 or depletion of Cdc25A, could lead to a similar reduction in replication fork speeds as in Chk1-deficient cells. Our conclusions agree with previous suggestions that Chk1-mediated control of replication initiation prevents DNA damage during S phase (26). How could increased replication initiation inhibit replication fork progression? One possibility is that the higher density of ongoing replication forks could accelerate depletion of factors that are essential for proper replication fork progression, such as nucleotides. Our previous observations suggested that slow fork speeds in Chk1-deficient cells does not result from nucleotide pool imbalance (25). However, replication forks need other factors for efficient progression, such as histone chaperones (32). Alternatively, excessive origin firing could activate a signaling pathway that down-regulates replication fork speeds. For example, Chk1 inhibition has been shown to activate ATR-mediated checkpoint signaling (26).
There are several caveats to our interpretations: Cdk activity promotes HR, which could be upregulated at destabilized or stalled forks in Chk1-defective cells. Cdk-inhibition could suppress aberrant HR, allowing faster fork progression. However, we previously found that suppression of HR does not rescue fork speeds in Chk1-defective cells, suggesting that aberrant HR is not responsible for this phenotype (25). Cdc7 is not only involved in regulation of origin firing; it is activated by treatment with etoposide or hydroxyurea (21), and it could be possible that Cdc7 actively down-regulates fork progression in response to destabilized forks in Chk1-deficient cells. Furthermore, the rescue of fork speeds by roscovitine treatment or codepletion of Cdc7 appears to be not complete. This suggests that Chk1 may promote fork progression by additional mechanisms, such as direct phosphorylation of the replication machinery. Chk1 stabilizes replication forks stalled by replication inhibitors independently of its role in promoting homologous recombination (6, 14, 33). It seems unlikely that stalled replication forks can be stabilized by control of origin firing, and these data therefore suggest that Chk1 plays other important roles in replication fork stability. However, the near complete rescue of fork speeds using Cdc7 siRNA suggests that such a mechanism would play a relatively minor role during normal DNA replication. It seems likely that fork stalling as caused by replication inhibitors would affect a relatively small subset of forks during normal S phase. This view is supported by a previous report that DNA damage induced by Chk1 inhibition depends on initiation factors, Cdk2 and Cdc45 (26).
Interestingly, Cdc7 protein levels appear to be reduced in Chk1-depleted cells and vice versa (Fig. 4A). The decreased Chk1 levels in Cdc7-depleted cells might be due to the reduced S phase fraction in these cells (34), but changes in cell-cycle distribution can not explain the reduction of Cdc7 in Chk1-depleted cells, because Cdc7 levels are stable throughout the cell cycle (35). Cdc7 might become down-regulated to counteract some of the increased origin firing in Chk1-depleted cells. This observation could also help explain why Chk1 inhibition affects origin firing and fork progression more strongly than Chk1 depletion: Because under the experimental conditions Chk1 inhibitor is present for less than 2 h, it may not induce the same changes in protein levels of Cdc7 (and probably other proteins) as Chk1 depletion during the 48 h of incubation with siRNA. Interestingly, Chk1 protein levels decrease for unknown reasons if cells are treated with roscovitine for prolonged periods (36). However, although the decrease in Chk1 is likely to counteract reduced origin firing in Cdk-inhibited cells, loss of Chk1 in roscovitine-treated cells was nevertheless associated with increased DNA damage (36). The relationship between origin firing, replication fork progression, and DNA damage clearly merits further investigation.
Although it is currently not clear whether Chk1 regulates Cdc7 in vivo (20, 21), Cdc7 activity is known to promote origin firing. Accordingly, Cdc7 depletion had a similar effect to Cdk inhibition in rescuing replication fork speeds in Chk1-deficient cells (Fig. 4). In addition to origin firing, Cdc7 has been implicated in the DNA damage response; it interacts with and phosphorylates Claspin, thus promoting Chk1 phosphorylation (34). As a mediator of Chk1 activation, Cdc7 could be expected to be required for normal replication fork rates like ATR and Claspin. However, our findings show this not be the case. A likely reason for this discrepancy is that Cdc7 depletion also prevents the excess origin firing usually induced by down-regulation of ATR or Chk1, thus reducing the requirement for Chk1 activity to promote fork progression. Although Cdc7 depletion does not change fork progression rates (Fig. 1), and Cdc7 inhibition even increased fork rates (37), it does result in delayed S phase progression, accumulation of DNA damage, and, in tumor cells, cell death, suggesting that reduced origin firing also leads to difficulties with completing replication (38).
In conclusion, we report that the role of Chk1 in suppressing origin firing may also explain its role in promoting replication elongation. Regulation of origin density is thus an important mechanism by which ATR-Chk1 signaling promotes normal DNA replication. Maintenance of origin firing control may be an important mechanism preventing replication stress in mammalian cells.
Methods
Cell Lines and Treatments.
Human U2OS osteosarcoma cells were grown in DMEM supplemented with 10% fetal calf serum in a humidified CO2 atmosphere at 37 °C. The Chk1 inhibitor CEP-3891 was provided by Cephalon, Inc., and used at 500 nM. Roscovitine was purchased from Sigma and used at 25 μM.
RNA Interference.
To knock down human Chk1 and Cdc7 we employed siRNA duplex oligonucleotides (Dharmacon) directed against the Chk1 target sequence (sense): UCGUGAGCGUUUGUUGAAC (39) and the Cdc7 target sequence (sense): GCAGUCAAAGACUGUGGAU (30). Allstars Negative Control siRNA (Qiagen) was used as nontargeting control. Five thousand cells grown in six well plates overnight were transfected with 100 nM siRNA using Dharmafect 1 reagent (Dharmacon) according to manufacturer’s instructions for HeLa cells. Cells were transfected with 100 nM control siRNA, 50 nM targeting + 50 nM control siRNA or 50 nM of each targeting siRNA for codepletion experiments. Cells were lysed in RIPA buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% Nonidet P40, 5% Na-Deoxycholate, 0.1% SDS) containing 1× protease inhibitor cocktail (Roche). Lysates from 4 × 104 cells per lane were resolved by denaturing PAGE and transferred to nitrocellulose. Proteins were detected using rabbit polyclonal anti-Chk1 antibody (Cell Signaling Technologies, 1∶300), mouse monoclonal anti-Cdc7 antibody (Abcam, DCS-341, 1∶5000), and mouse monoclonal anti-β-Actin antibody (Chemicon/Millipore, C4, 1∶4000). Incubations with primary antibodies were performed at 4 °C overnight.
DNA Fiber Experiments.
Exponentially growing cells were pulse labeled with 25 μM CldU followed by 250 μM IdU for 20 min each. Labeled cells were harvested and DNA fiber spreads prepared as previously described (25). For immunodetection of CldU-labeled tracts, acid treated fiber spreads were incubated with rat anti-BrdU monoclonal antibody (AbD Serotec, 1∶1,000) for 1 h at room temperature. Slides were then fixed with 4% paraformaldehyde and incubated with an AlexaFluor 555-conjugated goat antirat IgG (Molecular Probes, 1∶500) for 1.5 h at room temperature. To detect IdU-labeled patches, a mouse monoclonal anti-BrdU antibody (Becton Dickinson, 1∶1,000) was used over night at 4 °C, followed by an AlexaFluor 488-conjugated goat antimouse IgG (Molecular Probes, 1∶500) for 1.5 h at room temperature. Fibers were examined using a Biorad Radiance confocal microscope using a 60× (1.3NA) lens. The lengths of CldU (AF 555, red) and IdU (AF 488, green) labeled patches were measured using the ImageJ software (http://rsb.info.nih.gov/ij/), and μm values were converted into kb using the conversion factor 1 μm = 2.59 kb (40). Termination (red and red-green-red) or origin structures (green and green-red-green) were not measured. Replication structures were quantified using the Cell Counter Plug-in for ImageJ (Kurt De Vos, University of Sheffield, UK). The paired one-tailed student’s t test was used for statistical analyses.
Flow Cytometry.
Asynchronously growing U2OS cells were pulse labeled with 250 μM IdU for 20 min, washed twice with medium, and harvested immediately (0 h) or released into medium containing CEP-3891 (500 nM), roscovitine (25 μM) or an equal volume of DMSO and harvested after 3, 6, or 9 h. After harvesting cells were washed once with PBS and fixed in 70% ethanol over night at 4 °C. Cells were incubated in 2 M HCl, 0.1 mg/ml pepsin at room temperature for 20 mins, washed in PBS, and blocked in PBS containing 0.5% fetal calf serum, 0.5% Tween 20. Nuclei were incubated with mouse monoclonal anti-BrdU antibody (Becton Dickinson, 1∶100) in 2% fetal calf serum for 1.5 h, washed in PBS, and incubated with AlexaFluor 488-conjugated goat antimouse IgG (Molecular Probes, 1∶200) for 1 h. Nuclei were resuspended in 0.5 ml PBS containing 10 μg/ml propidium iodide.
Supplementary Material
Acknowledgments.
We thank Dr. Stephen Trusko for materials and Dr. Christina Bauerschmidt for helpful discussions. We thank the Medical Research Council, Cancer Research UK, the Swedish Cancer Society, the Swedish Children’s Cancer Foundation, the Swedish Research Council, and the Swedish Pain Relief Foundation for supporting this work financially.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1005031107/-/DCSupplemental.
References
- 1.DePamphilis ML, et al. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18(3):231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert DM. Replication origin plasticity, Taylor-made: Inhibition vs recruitment of origins under conditions of replication stress. Chromosoma. 2007;116(4):341–347. doi: 10.1007/s00412-007-0105-9. [DOI] [PubMed] [Google Scholar]
- 3.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6(7):648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 5.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410(6830):842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 6.Feijoo C, et al. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154(5):913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heffernan TP, et al. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol Cell Biol. 2002;22(24):8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao H, Seiler JA, Burhans WC. Regulation of cellular and SV40 virus origins of replication by Chk1-dependent intrinsic and UVC radiation-induced checkpoints. J Biol Chem. 2003;278(6):4295–4304. doi: 10.1074/jbc.M204264200. [DOI] [PubMed] [Google Scholar]
- 9.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114(3):385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 10.Courbet S, et al. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature. 2008;455(7212):557–560. doi: 10.1038/nature07233. [DOI] [PubMed] [Google Scholar]
- 11.Ibarra A, Schwob E, Mendez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA. 2008;105(26):8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21(24):3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trenz K, Errico A, Costanzo V. Plx1 is required for chromosomal DNA replication under stressful conditions. EMBO J. 2008;27(6):876–885. doi: 10.1038/emboj.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zachos G, Rainey MD, Gillespie DA. Chk1-deficient tumour cells are viable but exhibit multiple checkpoint and survival defects. EMBO J. 2003;22(3):713–723. doi: 10.1093/emboj/cdg060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14(21):2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21(13):4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA. 2002;99(23):14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heffernan TP, et al. Cdc7-Dbf4 and the human S checkpoint response to UVC. J Biol Chem. 2007;282(13):9458–9468. doi: 10.1074/jbc.M611292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenca P, et al. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem. 2007;282(1):208–215. doi: 10.1074/jbc.M604457200. [DOI] [PubMed] [Google Scholar]
- 22.Woo RA, Poon RY. Cyclin-dependent kinases and S phase control in mammalian cells. (Translated from eng) Cell Cycle. 2003;2(4):316–324. (in eng) [PubMed] [Google Scholar]
- 23.Sorensen CS, Syljuasen RG, Lukas J, Bartek J. ATR, Claspin and the Rad9-Rad1-Hus1 complex regulate Chk1 and Cdc25A in the absence of DNA damage. Cell Cycle. 2004;3(7):941–945. [PubMed] [Google Scholar]
- 24.Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. EMBO J. 2007;26(11):2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petermann E, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26(8):3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Syljuasen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer L, et al. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur J Biochem. 1997;243(1–2):527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 28.Bach S, et al. Roscovitine targets, protein kinases and pyridoxal kinase. J Biol Chem. 2005;280(35):31208–31219. doi: 10.1074/jbc.M500806200. [DOI] [PubMed] [Google Scholar]
- 29.Wilsker D, Petermann E, Helleday T, Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint and replication control. Proc Natl Acad Sci USA. 2008;105(52):20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montagnoli A, et al. Cdc7 inhibition reveals a p53-dependent replication checkpoint that is defective in cancer cells. Cancer Res. 2004;64(19):7110–7116. doi: 10.1158/0008-5472.CAN-04-1547. [DOI] [PubMed] [Google Scholar]
- 31.Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. (Translated from eng) Mol Biol Cell. 2008;19(6):2373–2378. doi: 10.1091/mbc.E07-10-1035. (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groth A, et al. Regulation of replication fork progression through histone supply and demand. Science. 2007;318(5858):1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 33.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JM, et al. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene. 2008;27(24):3475–3482. doi: 10.1038/sj.onc.1210994. [DOI] [PubMed] [Google Scholar]
- 35.Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94(26):14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maude SL, Enders GH. Cdk inhibition in human cells compromises chk1 function and activates a DNA damage response. Cancer Res. 2005;65(3):780–786. [PubMed] [Google Scholar]
- 37.Montagnoli A, et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol. 2008;4(6):357–365. doi: 10.1038/nchembio.90. [DOI] [PubMed] [Google Scholar]
- 38.Swords R, et al. Cdc7 kinase—A new target for drug development. Eur J Cancer. 2010;46(1):33–40. doi: 10.1016/j.ejca.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 39.Zhu W, Chen Y, Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol. 2004;24(16):7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henry-Mowatt J, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11(4):1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.