Abstract
Prochlorococcus describes a diverse and abundant genus of marine photosynthetic microbes. It is primarily found in oligotrophic waters across the globe and plays a crucial role in energy and nutrient cycling in the ocean ecosystem. The abundance, global distribution, and availability of isolates make Prochlorococcus a model system for understanding marine microbial diversity and biogeochemical cycling. Analysis of 73 metagenomic samples from the Global Ocean Sampling expedition acquired in the Atlantic, Pacific, and Indian Oceans revealed the presence of two uncharacterized Prochlorococcus clades. A phylogenetic analysis using six different genetic markers places the clades close to known lineages adapted to high-light environments. The two uncharacterized clades consistently cooccur and dominate the surface waters of high-temperature, macronutrient-replete, and low-iron regions of the Eastern Equatorial Pacific upwelling and the tropical Indian Ocean. They are genetically distinct from each other and other high-light Prochlorococcus isolates and likely define a previously unrecognized ecotype. Our detailed genomic analysis indicates that these clades comprise organisms that are adapted to iron-depleted environments by reducing their iron quota through the loss of several iron-containing proteins that likely function as electron sinks in the photosynthetic pathway in other Prochlorococcus clades from high-light environments. The presence and inferred physiology of these clades may explain why Prochlorococcus populations from iron-depleted regions do not respond to iron fertilization experiments and further expand our understanding of how phytoplankton adapt to variations in nutrient availability in the ocean.
One of the most abundant photoautotrophs in tropical and subtropical oligotrophic marine environments is Prochlorococcus (1). More than a dozen genetically diverse isolates have been cultivated and sequenced since Prochlorococcus was first discovered (2–4). These isolates can be subdivided into taxonomically and ecologically distinct lineages according to their adaptation to general environmental conditions [e.g., high-light (HL) vs. low-light (LL) (5)] or temperature regimes (6). Across the globe, warm surface waters are dominated by a single clade of Prochlorococcus (eMIT9312) (7, 8). This clade possesses high rates of sequence divergence while maintaining remarkable conservation both in gene content and synteny, despite a global distribution that spans large gradients in the bioavailable concentrations of the macronutrients nitrogen (N) and phosphorus (P).
However, evidence is mounting that adaptation of Prochlorococcus to local nutrient conditions is often mediated by gene cassettes that enable transport and assimilation of the available nutrients (4, 9, 10). Although these cassettes impart a strong selective advantage under the appropriate conditions, they have not been associated with phylogenetically distinct clades (9). This suggests that horizontal gene transfer and strong selective pressures to maintain a minimal genome allows cells to acquire or discard genes as required by local conditions and enables Prochlorococcus to proliferate under a wide range of environmental conditions (3, 11–14).
It is likely that our picture of the genetic diversity of the Prochlorococcus community is incomplete. For one, the number of Prochlorococcus cells counted by flow cytometry often exceeds that detected with PCR using primers based on our current knowledge of genetic variation (15, 16). In addition, most sequencing surveys have focused on Mediterranean (17), Atlantic (18–20), and the Hawaii Ocean time-series program sites (21), leaving the possibility that undescribed clades exist in other regions of the ocean.
To address the possibility that there are uncharacterized Prochlorococcus phylotypes, we queried samples from the Global Ocean Sampling (GOS) expedition. GOS represents the largest metagenomic study to date and includes samples collected from surface waters from the Atlantic, Pacific, and Indian Oceans during a 3-y circumnavigation of the world. In these samples, we discovered two previously undescribed Prochlorococcus clades in high-nutrient, low-chlorophyll (HNLC), iron-depleted waters in the Pacific and Indian Oceans. Reconstruction of consensus genomes and comparative approaches revealed that the gene repertoires of these clades reflect a streamlining of their iron requirements. These results are consistent with the presence of an HL Prochlorococcus ecotype adapted to low iron bioavailability.
Results
Identification of Prochlorococcus Diversity.
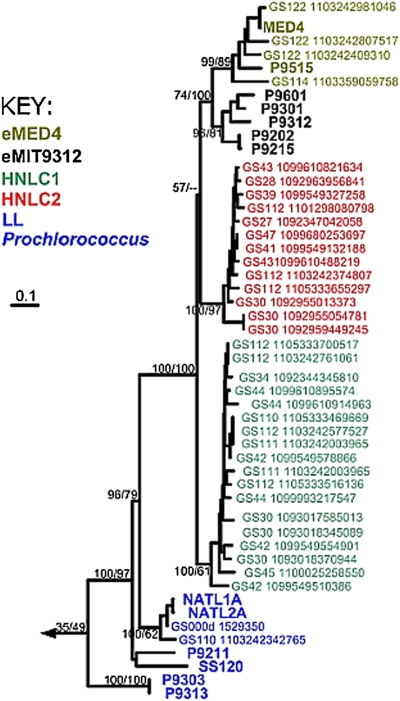
The first evidence for uncharacterized Prochlorococcus lineages was observed as a distinctive pattern of fragment recruitment (Methods) with substantial divergence from HL Prochlorococcus genomes arising from a small number of GOS samples. To further investigate this pattern of Prochlorococcus diversity, we developed phylogenetic markers for Prochlorococcus suitable for analyzing metagenomic libraries. To find markers that are not prone to lateral gene transfer and able to distinguish between closely related phylotypes in metagenomic libraries, we looked for markers that satisfied five criteria: (i) present as a single copy in all Prochlorococcus and marine Synechococcus genomes, (ii) similar phylogeny as the 13 sequenced Prochlorococcus genomes, (iii) high rates of sequence divergence, (iv) variation consisting of substitutions rather than insertions and deletions, and (v) lengths <800 bp so that the entire gene could be identified within a single GOS sequencing read. We identified six genetic markers (rpsT, psb27, purC, nadD, msrA, and PMSR) adhering to these criteria. These genetic markers could resolve most nodes including branch points between strains within the HL-adapted eMIT9312 clade. The only exception was the branching point of LL strains SS120 and MIT9211, which varied among markers. However, other studies have shown that this branch point is difficult to resolve, and the position is still debated (3). Using these genetic markers, we screened the unassembled GOS metagenomic libraries (Table S1). All genetic markers revealed an undescribed Prochlorococcus clade branching between the LL eNATL and the HL ecotypes (Fig. 1 and Fig. S1). This topology was highly supported by both neighbor-joining and maximum likelihood phylogenetic approaches. The branch point is much closer to the HL clades than eNATL. The clade can be further separated with some confidence into two distinct subclades.
Fig. 1.
Phylogenetic analysis of Prochlorococcus rpsT marker gene. This marker identifies two distinct clades of Prochlorococcus (red and green), each equally divergent from the other two recognized HL clades represented by MED4 and MIT9312 (gold and black). The rpsT (30S ribosomal protein S20) gene from sequenced genomes is shown in bold, whereas those from GOS reads show the site and ID of the containing read. Bootstrap values for neighbor-joining over maximum likelihood approaches are provided as percentages on every branch. Additional markers are presented in Fig. S1.
A detailed analysis of the geographic distribution of the newly identified clades shows that they were associated with the Eastern Equatorial Pacific and tropical Indian Ocean (Fig. 2), with relative abundances comparable to eMIT9312 in other GOS samples (Table S1). We then looked at the distribution of different Prochlorococcus clades in the context of various physical and chemical parameters, such as light, temperature, nitrate, and iron, to determine which parameters correlate with the distribution of these newly detected clades (Fig. S2 A–D and Table S2). Iron concentrations were simulated using the ocean biogeochemical element cycling (BEC) model because we were not properly equipped to measure trace metal values (22). Nitrate and phosphate were monthly averages derived from the World Ocean Atlas (23). Because all GOS samples were from surface waters, only the HL ecotypes eMED4, eMIT9312, and the uncharacterized clades were present in noteworthy numbers according to fragment recruitment rates; thus, we restricted the analysis to these lineages. Previous studies have demonstrated that the distribution of eMED4 and eMIT9312 can be distinguished according to temperature (6). Supporting this, we only detected eMED4 at sites with a temperature <21 °C (Fig. S2A). In contrast, both eMIT9312 and the uncharacterized clades are detected in warmer waters and, in particular, the uncharacterized clades are only seen in samples with a temperature above 25 °C. However, the distribution of eMIT9312 and the uncharacterized clades is very different across nitrate concentrations (Fig. S2B). eMIT9312 is found in samples with <1 μM nitrate, whereas the uncharacterized clades are detected in high-nitrate waters. Despite the high N and P concentration, only modest chlorophyll concentrations are observed in these sites; such oceanic regions are historically denoted HNLC. Thus, we denoted the uncharacterized clades HNLC1 and HNLC2.
Fig. 2.
Distribution of Prochlorococcus clades in marine surface water metagenomic samples overlaid on chlorophyll-a measurements. Circles and squares represent sampling sites from Hewson et al. (26) and the GOS (13). Purple, white, and green coloring of the shapes indicates that a site is dominated by the HNLC, eMIT9312, or eMED4 clades, respectively.
Most of the HNLC regions are depleted in dissolved iron relative to other nutrients, specifically N or P, resulting in a unique nutrient limitation scenario (24). Thus, we speculated that the distribution of the HNLC clades is linked to iron availability. On the basis of the simulated iron values, the HNLC clades are mainly found in waters with <0.5 nM dissolved iron, whereas eMIT9312 and eMED4 are found in regions with higher iron concentrations (Fig. S2C). Two samples from the tropical Indian Ocean (GS111 and 112) dominated by HNLC clades presumably had a higher concentration of iron. However, satellite-based measurements of the nonphotochemical quenching-corrected fluorescence quantum yields suggest that the phytoplankton found in this region are indeed iron-stressed (Fig. S2D) (25). Thus, the BEC model may overestimate the iron concentration in the sparsely studied Indian Ocean samples. The HNLC clades are missing from Eastern Pacific samples GS23 and GS25, where the model indicates that iron is low and N is high. However, the model may underestimate the concentration because of coastal influences on iron availability. Using canonical correspondence analysis to summarize the distribution pattern, we observe that the primary habitat of the HNLC clades is low-iron, high-nitrate, and higher-temperature waters, with iron being the predominant factor distinguishing these strains from the HL-adapted eMIT9312 (Fig. 3). However, other unaccounted-for factors may also influence their distribution.
Fig. 3.
Canonical correlation analysis showing abundance of Prochlorococcus ecotypes relative to environmental parameters for temperature (actual), iron (modeled), and nitrate (modeled) abundance.
A recent study produced several metagenomic datasets from the central Pacific Ocean, including both the N-limited North Pacific and the iron-limited Equatorial Pacific Ocean (26). On the basis of the best BLASTX hits to known Prochlorococcus genomes, these samples were reported to contain an unusually high abundance of the cold water–adapted eMED4 ecotype, despite very warm water temperatures (>27 °C). In light of this information, we reexamined these stations and found that the majority of previously designated eMED4 and eMIT9312 sequences instead grouped with the HNLC clades (Fig. 2 and Table S1).
Genome Organization of HNLC Lineages.
For a more detailed analysis of the genome content of HNLC1 and HNLC2 cells, we assembled a consensus genome for each lineage using the Celera Assembler (27). These consensus genomes represent an ensemble view of the genome organization of populations and are not equivalent to a complete or even draft genome assemblies from a clonal isolate. The resulting assembly required minor manual curation to break two chimeric scaffolds and ultimately produced a set of 24 scaffolds corresponding to two HNLC clades. The assemblies had an average depth of coverage of 52 and 21 reads, respectively, for the HNLC1 and HNLC2. Average percentage GC for all scaffolds is ≈30%. The individual scaffolds were separated into HNLC1 and HNLC2 bins according to differences in depth of coverage across sampling sites (Fig. S3). The scaffolds were ordered and oriented so that they best matched the layout found in the closest relatives, eMED4 and eMIT9312, both of which are highly syntenic with each other. This resulted in consensus genomes for HNLC1 and HNLC2 of 1.56 Mb and 1.48 Mb, respectively. The lengths of the assemblies are roughly equivalent in size to other HL Prochlorococcus genomes when one allows for the presence of hypervariable genomic islands that we would not expect to assemble (12, 13). Synteny within each scaffold is highly consistent with the eMIT9312 and eMED4 sequenced genomes (Fig. S4). We also observe that many of the break points are syntenic with the locations of hypervariable and variable islands. Thus, we conclude that the fragmentary nature of the assembly is largely the result of variation present within the HNLC lineages. Overall these results suggest that isolates from these clades, should they become available, will have genomes that are structurally highly syntenic to that of other HL isolates.
Genomic Adaptation.
To elucidate whether and how these HNLC clades might be physiologically distinct, we identified genes that seem to be gained or lost. For this comparative analysis, we checked for the presence of individual genes both in the assemblies and directly in the environmental data (to circumvent potential assembly errors; Dataset S1). Given that the abundance of HNLC clades is strongly associated with predicted low iron availability (Fig. 3), we first examined genes related to iron uptake and stress response. Like most other HL Prochlorococcus strains, the HNLC clades seem to possess the same genes for iron acquisition and response to iron stress (28). Like other HL Prochlorococcus, the HNLC clades seem to lack TonB-like receptors and a siderophore biosynthesis pathway, so there is no evidence that the HNLC lineages possess a high-affinity siderophore-mediated iron uptake system.
Faced with the absence of any new systems for acquiring iron, we instead find at least six iron-containing proteins that seem to be both statistically underrepresented in samples dominated by HNLC1 and HNLC2 and missing in the genome assemblies (Table 1). This is a high number, considering that cyanobacterial genomes of similar size—including those of HL Prochlorococcus—contain between 60 and 65 iron-binding protein domains (29). Thus, the HNLC clade has the lowest number of iron-binding proteins of any cyanobacteria (Fig. S5). Taken together, these results suggest that the HNLC clades have adapted to low-iron environments not by becoming more efficient at acquiring iron but by eliminating a number of their iron-requiring proteins.
Table 1.
Selected genes from Prochlorococcus strain AS9601 that are present or absent from the HNLC consensus genomes and metagenomic samples
 |
*Genes in white are absent from the assemblies, in blue are suspected of being viral, and in purple are core genes present in all cyanobacteria.
†Sites dominated by eMIT9312 before HNLC abundant waters, including GOS sampling sites GS22–23 and GS26.
‡Sites dominated by HNLC clades near South Pacific Gyre, including GOS sampling sites GS40–47.
§Sites dominated by eMIT9312 post Pacific HNLC abundant waters, including GOS sampling sites GS52–53.
¶Sites in Indian Ocean containing a mixture of HNLC and eMIT9312 clades. Consists of GOS sampling sites GS111–112, including 454 titanium data, resulting in higher counts.
The lost iron-containing proteins include plastoquinol terminal oxidase (PTOX), two ferredoxins, and cytochrome Cm (Table 1). Each of these losses would remove a sink for electrons within the photosynthetic pathway. PTOX is hypothesized to catalyze cyclic electron flow around photosystem (PS) II in Synechococcus and Ostreococcus (30, 31), providing an outlet for electron flow when acceptors downstream of PSII are already reduced or unavailable. In both of these organisms, this “electron safety valve” allows for a high ratio of PSII to PSI by alleviating the potential for photodamage with increases in light. PTOX-like activity has also been detected in phytoplankton assemblages from the predominantly P-limited Sargasso Sea and N-limited North Pacific (32). However, it was not detected at the subsurface chlorophyll maximum, consistent with the lack of PTOX in the genomes of the LL Prochlorococcus strains (33). Recently, it has been suggested that the usage of PTOX may be an adaptation to chronic iron limitation (34), although the usage of PTOX in response to iron limitation has not been examined. Although PTOX does contain a paucity of Fe (2 molecules per protein) relative to the cytochrome B6f complex and PSI (18 molecules total), the true determinants for metal requirements, the relative protein concentrations, and protein turnover times are not known. Instead, the biogeography of PTOX in Prochlorococcus genomes suggests that, at least in this genus, PTOX usage may be more related to N, P, or light stress than to adaptation to iron limitation.
Although PTOX is not found in the consensus genomes, it is found in a short 10-kb scaffold from the assembly. The distribution of reads associated with this scaffold is different from the HNLC consensus assemblies, having an association with only a subset of GOS samples in the South Pacific Gyre (Table S1). The percentage GC for this scaffold is 25%, which is significantly lower than the mean of 32.5% for similarly sized subsequences from the consensus HNLC genomes (SD 1.27%). The scaffold contains a number of HL-inducible genes, a phage-associated gene, and a PSI gene (psaC). Potentially, this PTOX-containing scaffold belongs to a Prochlorococcus phage, a possibility supported by the observation of PTOX in other cyanophages (35–38). Alternatively this scaffold may be part of a hypervariable region found within a subset of the HNLC populations (Table S1).
The loss of other iron-containing proteins removes more potential outlets from the photosynthetic electron chain. Cytochrome Cm is thought to be involved in electron transfer from PSI to the cytochrome c oxidase complex during periods of stress (39). Although the exact conduits are unknown, the loss of two ferredoxins likely removes several paths for electrons to move away from or to photosynthetic reaction centers. Finally, the HNLC genomes do not contain genes for the iron-containing enzyme nitrate reductase (10), which is consistent with a physiological strategy of iron requirement minimization. This adaptation removes a potential physiological sink for reducing equivalents and indicates that this organism does not use the abundant compound in the exogenous N pool.
In summary, the HNLC clade has lost proteins implicated in providing alternative pathways for electron transfer and thus the ability to cope with environmental changes, such as shifting light levels. We hypothesize that physiologically the loss of PTOX almost certainly prevents HNLC1 and HNLC2 from maintaining a high ratio of PSII to PSI in the HL environment of the tropical surface ocean. Given the low bioavailability of iron, HNLC1 and HNLC2 may maintain low absolute levels of PSII and PSI at a near even ratio, which would place a substantial limit on the maximal growth rate of these organisms and increase susceptibility to shifts in light intensity. Given that the growth of all other photoautotrophs in these systems is severely limited by iron, such tradeoffs may be warranted yet provide a ready explanation for the lack of the HNLC clades in other ocean regions.
Genomic Diversity in the HNLC Clade.
Although there are some clear cases of genes being absent in the HNLC lineages, there are a number of genes that are present at lower or higher abundances than we would anticipate if the genes were present in every cell. These examples probably indicate further diversity within the accessory proteins of HNLC1 and HNLC2 lineages. One example includes the urease and urea transporter operons that include genes for acquiring and metabolizing urea. Two distinct urease operons were assembled from the metagenomic data and could be tentatively assigned to the HNLC1 and HNLC2 consensus genomes on the basis of their abundances. Both operons occur at significantly lower-than-expected coverage, which averages 24.2 and 8.6 reads per base, respectively. Numerous other genes are less abundant than would be expected. Some of these genes will be part of the accessory genome and thus present in only a subset of each lineage. Others may be present in only one of the two lineages. In total, 45 HNLC1 and 20 HNLC2 genes were found in only one of the consensus genomes, suggesting that they may be present in only one of the lineages (Dataset S1), although this is likely an overestimate. Phage-associated genes may be responsible for some of the genes that are less abundant than expected, or they may lead to higher-than-expected frequencies for genes that are also part of the core genome. We also identified 225 clusters of potentially novel peptides that are abundant only in the waters containing the HNLC clades and that could be linked to the HNLC assemblies (Dataset S2). All of these peptides were uncharacterized, so their functions and potential roles are unknown.
Discussion
Biogeochemical Considerations.
The Equatorial Pacific Upwelling Region sampled here has been shown to have extremely low available iron (≤0.1 nM) and is thought to limit phytoplankton growth (40, 41). Part of the Indian Ocean gyre may also be iron limited (42, 43), although the BEC model suggests otherwise (22). However, the biogeochemistry is less defined here compared with the Pacific and Atlantic Oceans. Satellite measurements of fluorescence quantum yield of phytoplankton has been proposed as a measure of iron stress (44), and using this, Behrenfeld et al. (25) observed extended iron stress in part of the Indian Ocean gyre. Thus, it is conceivable that the iron concentration is low in the Indian Ocean sites, where we detect the HNLC clades of Prochlorococcus. The detection of these clades also supports the observation from satellite data that iron may be low in this region.
Iron addition experiments carried out in the Eastern Equatorial Pacific found that most phytoplankton, including marine Synechococcus, underwent rapid growth upon addition of iron (41, 45–47). The most striking exception was Prochlorococcus, which showed only a minimal response both in growth and fluorescence to iron enrichment. This suggests that Prochlorococcus cells in this region were not iron stressed to the same extent as other phytoplankton lineages. Our data suggest that the HNLC ecotype may have a reduced iron quota at the expense of a variety of pathways for energy generation and stress tolerance, which would explain their lack of growth response to iron addition. However, the tradeoff could be an upper limit to their photosynthetic capacity and cap their growth rate under elevated iron conditions. Thus, we hypothesize that cells from the eMIT9312 clade may have a higher growth rate when iron is more available, allowing them to outcompete HNLC cells in iron replete environments.
Adaptation in Prochlorococcus.
Genomic islands conferring fitness under particular environmental conditions have been previously identified for phosphate and N metabolism in Prochlorococcus (9, 10). In this case, ecotypes adapted to high or low nutrients seem to be associated with microdiverse clusters (14). In contrast, adaptations to high light and low light or to specific water temperature have resulted in phylogenetically distinct clades and thus define the major lineages of Prochlorococcus (5, 6). Our genomic analysis indicates a loss of ≈10% of the iron-containing genes, thus suggesting a reduction of the iron quota in the HNLC clades compared with other HL Prochlorococcus. It is important to note that we do not know the protein expression level of any of these lost proteins, so it is not possible to translate the genomic changes into a specific iron quota reduction. Future research of Prochlorococcus iron quota in conjunction with more precise iron measurements in this and other areas will reveal the effect of these genome variations. The phylogenetic distinctiveness of the HNLC clade suggests that adaptation to low iron availability could involve systemic changes beyond the gain or loss of a few genes. Given that gene content is not dramatically different from other clades, some of these changes may be regulatory in nature. This could include an adjustment of both the absolute expression and the ratio of PSI and -II complexes, which predominantly dictate iron requirements (48).
We are puzzled to observe not one but two coexisting HNLC lineages—each nearly as divergent from each other as they are from eMIT9312 and eMED4. Comparisons of the two genomes have not uncovered any significant gene content differences that would indicate unique physiologies. Perhaps the two lineages carry different adaptations to iron stress that each result in low iron quota. Alternatively, they have slightly different light optima and resulting depth distribution, and mixing would then allow for coexistence. However, more research is needed to identify the underlying factors for this coexistence.
Overall, our data provide evidence for a previously undescribed HL ecotype of Prochlorococcus proliferating in low-iron environments like the Equatorial Pacific and tropical Indian Ocean. These results are likely to influence our understanding of how iron availability affects phytoplankton physiology under light and nutrient stress, ocean biogeochemistry, and diversity now and in a future warmer and acidic ocean.
Methods
Fragment Recruitment.
Fragment recruitment was performed as described previously (13). Recruitment plots to the HNLC assemblies and PTOX scaffold can be accessed at http://gos.jcvi.org/users/gosPhaseII/advancedReferenceViewer.html.
Phylogenetic Analysis.
First, we aligned three different protein families from Prochlorococcus and Synechococcus genomes using ClustalW (49). The peptide ClustalW alignments were used to train HMMER2 hidden Markov models (HMMs) (50). These models were used to search every GOS sequencing read (1e-2 e-value cutoff) in all six reading frames using Logical Depth's accelerated version of HMMER2. Reads with positive hits were filtered to exclude partial hits (<95% of full length) and to remove sequences with better matches to noncyanobacterial genomes according to BLASTN (51). The nucleotide sequences corresponding to these peptide matches were then extracted. The metagenomic nucleotide sequences were combined with the orthologous genes from the sequences Prochlorococcus and Synechococcus genomes. Phylogenetic trees with 100 bootstraps were built with PHYLIP (ver. 3.65) using neighbor-joining and maximum likelihood (52).
Environmental Data.
Temperature was measured during sampling. We retrieved chlorophyll concentration data from MODIS AQUA satellite data (modis.gsfc.nasa.gov). Quenching-corrected fluorescence quantum yields were estimated as described by Behrenfeld et al. (25). We retrieved monthly average values for phosphate and nitrate for each location and depth from the World Ocean Atlas (23). A caveat is that this approach may introduce additional variance in the analysis of correspondence between genome content and phosphate availability, because these values do not represent the exact nutrient concentration at the time of sampling.
Determination of Prochlorococcus Abundance.
The abundance of environmental populations closely related to Prochlorococcus strains AS9601, MED4, MIT9215, and NATL1A, as well as the HNLC1 and HNLC2 genomes, was derived from fragment recruitment data (13). Large segments of each genome were selected because they had normal recruitment patterns and were not associated with variable or hypervariable segments of the genome. Only reads that recruited best to the given reference genome relative to all of the complete and draft genomes available at the National Center for Biotechnology Information as of September 18, 2009 were used. Valid reads also had to recruit to the reference genome at 80% or better identity over their entire length. The relative abundance per sample was derived from the count of base pairs per sample after being normalized for the length of the genomic segment on the reference that was used and normalized for the amount of sequencing that was collected per sample. Values are provided in terms of percentage of reads recruited per megabase pair.
Canonical Correspondence Analysis.
The variance contribution of individual environmental factors on Prochlorococcus clade distribution was determined with partial canonical correspondence analysis (forward selection, α = 0.05 and 9,999 perturbations) using CANOCO (ver. 4.0, Microcomputer Power) (53). Ordination plots were visualized in CanoDraw.
Census of Iron-Binding Proteins.
HMMs from the Superfamily (54) database were used to identify protein structural domains within the fold family (FF) category of the Structural Classification of Proteins Database (SCOP) (55), which categorizes all of the protein structures in the Protein Data Bank (56). The SCOP was manually annotated according to metal-binding and specificity (29). At the FF hierarchy, 90% of the metal-binding FFs bind a single metal (57). In Fig. S5, only the counts of the known Fe-binding FFs are presented. These methods of metal-binding protein identification have been vetted through manual curation (29, 57–59).
Supplementary Material
Acknowledgments
We thank Dr. Keith Moore (University of California, Irvine, CA) for his biogeochemical element cycling model data; Dr. Michael Behrenfeld (Oregon State University, Corvallis, OR) for his iron stress data; Charles H. Howard, captain of the Sorcerer II, and fellow crew members for their time and effort in support of this research; numerous individuals at the J. Craig Venter Institute for their support in processing and sequencing the libraries used in this analysis; and Drs. Nyree West and Marcelino Suzuki for their comments and suggestions on the paper. Funding for this study was supplied by the Department of Energy, Office of Science, and Office of Biological and Environmental Research (DE-FG02-02ER63453), the National Science Foundation, the Gordon and Betty Moore Foundation, and the J. Craig Venter Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: This Whole Genome Shotgun project has been deposited at DNA Data Base in Japan/European Molecular Biology Laboratory/GenBank under accession no. ADHP00000000. The version described in this article is the first version, ADHP00000000.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009513107/-/DCSupplemental.
References
- 1.Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dufresne A, et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc Natl Acad Sci USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kettler GC, et al. Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocap G, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 5.Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 6.Johnson ZI, et al. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 7.Bouman HA, et al. Oceanographic basis of the global surface distribution of Prochlorococcus ecotypes. Science. 2006;312:918–921. doi: 10.1126/science.1122692. [DOI] [PubMed] [Google Scholar]
- 8.Zwirglmaier K, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 9.Martiny AC, Coleman ML, Chisholm SW. Phosphate acquisition genes in Prochlorococcus ecotypes: Evidence for genome-wide adaptation. Proc Natl Acad Sci USA. 2006;103:12552–12557. doi: 10.1073/pnas.0601301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martiny AC, Kathuria S, Berube PM. Widespread metabolic potential for nitrite and nitrate assimilation among Prochlorococcus ecotypes. Proc Natl Acad Sci USA. 2009;106:10787–10792. doi: 10.1073/pnas.0902532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scanlan DJ. Physiological diversity and niche adaptation in marine Synechococcus. Adv Microb Physiol. 2003;47:1–64. doi: 10.1016/s0065-2911(03)47001-x. [DOI] [PubMed] [Google Scholar]
- 12.Coleman ML, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 13.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martiny AC, Tai AP, Veneziano D, Primeau F, Chisholm SW. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- 15.Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 16.Zinser ER, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garczarek L, et al. High vertical and low horizontal diversity of Prochlorococcus ecotypes in the Mediterranean Sea in summer. FEMS Microbiol Ecol. 2007;60:189–206. doi: 10.1111/j.1574-6941.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- 18.Jameson E, Joint I, Mann NH, Mühling M. Detailed analysis of the microdiversity of Prochlorococcus populations along a North-South Atlantic Ocean transect. Environ Microbiol. 2009;12:156–171. doi: 10.1111/j.1462-2920.2009.02057.x. [DOI] [PubMed] [Google Scholar]
- 19.Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. 2009;11:1340–1347. doi: 10.1111/j.1462-2920.2009.01860.x. [DOI] [PubMed] [Google Scholar]
- 20.Zwirglmaier K, et al. Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- 21.DeLong EF, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 22.Moore JK, Doney SC, Glover DM, Fung IY. Iron cycling and nutrient-limitation patterns in surface waters of the World Ocean. Deep Sea Res Pt II. 2002;49:463–507. [Google Scholar]
- 23.Boyer TP, et al. World Ocean Database 2005. Washington, DC: US Government Printing Office; 2006. p. 190. [Google Scholar]
- 24.de Baar HJW, et al. Synthesis of iron fertilization experiments: From the iron age in the age of enlightenment. J Geophys Res—Oceans. 2005 10.1029/2004JC002601. [Google Scholar]
- 25.Behrenfeld MJ, et al. Satellite-detected fluorescence reveals global physiology of ocean phytoplankton. Biogeosciences. 2009;6:779–794. [Google Scholar]
- 26.Hewson I, Paerl RW, Tripp HJ, Zehr JP, Karl DM. Metagenomic potential of microbial assemblages in the surface waters of the central Pacific Ocean tracks variability in oceanic habitat. Limnol Oceanogr. 2009;54:1981–1994. [Google Scholar]
- 27.Miller JR, et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics. 2008;24:2818–2824. doi: 10.1093/bioinformatics/btn548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivers AR, Jakuba RW, Webb EA. Iron stress genes in marine Synechococcus and the development of a flow cytometric iron stress assay. Environ Microbiol. 2009;11:382–396. doi: 10.1111/j.1462-2920.2008.01778.x. [DOI] [PubMed] [Google Scholar]
- 29.Dupont CL, Yang S, Palenik B, Bourne PE. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc Natl Acad Sci USA. 2006;103:17822–17827. doi: 10.1073/pnas.0605798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey S, et al. Alternative photosynthetic electron flow to oxygen in marine Synechococcus. Biochim Biophys Acta. 2008;1777:269–276. doi: 10.1016/j.bbabio.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Cardol P, et al. An original adaptation of photosynthesis in the marine green alga Ostreococcus. Proc Natl Acad Sci USA. 2008;105:7881–7886. doi: 10.1073/pnas.0802762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackey KRM, Paytan A, Grossman AR, Bailey S. A photosynthetic strategy for coping in a high-light, low-nutrient environment. Limnol Oceanogr. 2008;53:900–913. [Google Scholar]
- 33.Dufresne A, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zehr JP, Kudela RM. Ocean science. Photosynthesis in the open ocean. Science. 2009;326:945–946. doi: 10.1126/science.1181277. [DOI] [PubMed] [Google Scholar]
- 35.Malakhov MP, Malakhova OA, Murata N. Balanced regulation of expression of the gene for cytochrome cM and that of genes for plastocyanin and cytochrome c6 in Synechocystis. FEBS Lett. 1999;444:281–284. doi: 10.1016/s0014-5793(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 36.Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: Implications for mechanisms of cyanophage evolution. Environ Microbiol. 2009;11:2370–2387. doi: 10.1111/j.1462-2920.2009.01966.x. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weigele PR, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. 2007;9:1675–1695. doi: 10.1111/j.1462-2920.2007.01285.x. [DOI] [PubMed] [Google Scholar]
- 39.Bernroitner M, et al. Cyanobacterial cytochrome c(M): Probing its role as electron donor for Cu(A) of cytochrome c oxidase. Biochim Biophys Acta. 2009;1787:135–143. doi: 10.1016/j.bbabio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Blain S, Bonnet S, Guieu C. Dissolved iron distribution in the tropical and sub tropical South Eastern Pacific. Biogeosciences. 2008;5:269–280. [Google Scholar]
- 41.Bonnet S, et al. Nutrient limitation of primary productivity in the Southeast Pacific (BIOSOPE cruise) Biogeosciences. 2008;5:215–225. [Google Scholar]
- 42.Wiggert JD, Murtugudde RG. The sensitivity of the southwest monsoon phytoplankton bloom to variations in aeolian iron deposition over the Arabian Sea. J Geophys Res-Oceans. 2007 10.1029/2006JC003514. [Google Scholar]
- 43.Wiggert JD, Murtugudde RG, Christian JR. Annual ecosystem variability in the tropical Indian Ocean: Results of a coupled bio-physical ocean general circulation model. Deep Sea Res Pt II. 2006;53:644–676. [Google Scholar]
- 44.Behrenfeld MJ, et al. Controls on tropical Pacific Ocean productivity revealed through nutrient stress diagnostics. Nature. 2006;442:1025–1028. doi: 10.1038/nature05083. [DOI] [PubMed] [Google Scholar]
- 45.Cavender-Bares KK, Mann EL, Chisholm SW, Ondrusek ME, Bidigare RR. Differential response of equatorial Pacific phytoplankton to iron fertilization. Limnol Oceanogr. 1999;44:237–246. [Google Scholar]
- 46.Coale KH, et al. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial Pacific Ocean. Nature. 1996;383:495–501. doi: 10.1038/383495a0. [DOI] [PubMed] [Google Scholar]
- 47.Martin JH, et al. Testing the iron hypothesis in ecosystems of the equatorial Pacific Ocean. Nature. 1994;371:123–129. [Google Scholar]
- 48.Raven JA, Evans MCW, Korb RE. The role of trace metals in photosynthetic electron transport in O-2-evolving organisms. Photosynth Res. 1999;60:111–149. [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 51.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 52.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 53.ter Braak CJF. Canonical correspondence analysis: A new eigenvector method for multivariate direct gradient analysis. Ecology. 1986;67:1167–1179. [Google Scholar]
- 54.Gough J. Genomic scale sub-family assignment of protein domains. Nucleic Acids Res. 2006;34:3625–3633. doi: 10.1093/nar/gkl484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 56.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dupont CL, Butcher A, Valas RE, Bourne PE, Caetano-Anolles G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc Natl Acad Sci USA. 2010;107:10567–10572. doi: 10.1073/pnas.0912491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palenik B, et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palenik B, et al. Genome sequence of Synechococcus CC9311: Insights into adaptation to a coastal environment. Proc Natl Acad Sci USA. 2006;103:13555–13559. doi: 10.1073/pnas.0602963103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.