Abstract
The molecular chaperone heat shock protein 90 (Hsp90) is an important and abundant protein in eukaryotic cells, essential for the activation of a large set of signal transduction and regulatory proteins. During the functional cycle, the Hsp90 dimer performs large conformational rearrangements. The transient N-terminal dimerization of Hsp90 has been extensively investigated, under the assumption that the C-terminal interface is stably dimerized. Using a fluorescence-based single molecule assay and Hsp90 dimers caged in lipid vesicles, we were able to separately observe and kinetically analyze N- and C-terminal dimerizations. Surprisingly, the C-terminal dimer opens and closes with fast kinetics. The occupancy of the unexpected C-terminal open conformation can be modulated by nucleotides bound to the N-terminal domain and by N-terminal deletion mutations, clearly showing a communication between the two terminal domains. Moreover our findings suggest that the C- and N-terminal dimerizations are anticorrelated. This changes our view on the conformational cycle of Hsp90 and shows the interaction of two dimerization domains.
Keywords: FRET, cooperativity, vesicles, intramelocular communication
In eukaryotes heat-shock protein 90 (Hsp90) is a molecular chaperone required for survival of the cell under physiological conditions and during heat shock and for the activation of a large set of client proteins. It consists of two chains with dimerization interfaces at the C- and N-terminal ends. Its chaperone function is dependent on ATP binding and hydrolysis as well as N-terminal and C-terminal dimerization [(1, 2) and reviewed in refs. 3–6]. The crystal structure of the full-length Hsp90 from yeast bound to adenosine 5′-[β,γ-imido]triphosphate (AMP-PNP) and p23 revealed a compact homodimeric structure where both the N- and C-terminal domains are dimerized (7). Recent studies in solution with small-angle X-ray scattering, hydrogen exchange mass spectrometry (8–10) and single-molecule fluorescence (11) showed a highly dynamic and stochastic picture of Hsp90, where the system is in equilibrium among different N-terminal open and closed (compact) states. Up to now experiments have not been able to separately observe N- and C-terminal dimerization kinetics, and therefore the C-terminal domain was assumed to be closed on the time scale of experiments [motivated by the low KD value of 60 nM for yeast Hsp90 (12)]. Our single-molecule assay overcomes this experimental limitation and allows the investigation of the C-terminal dimerization interface independently in real time. Contrary to the general assumptions, fast opening and closing kinetics of the C-terminal domains can be found. Furthermore, we clearly detect an unexpected C-terminal open and an N-terminal closed state and a communication in between the C and N termini.
Results
Single-Molecule Fluorescence Resonance Energy Transfer (smFRET) Shows C-terminal Dimerization Kinetics.
To directly monitor the dynamics of the C-terminal domain, we created a variant of yeast Hsp90 in which glutamine at position 560 in the C-terminal domain was exchanged against a cysteine (C560). The single cysteine in one monomer was labeled with the donor fluorophore Atto550, and in the second monomer with the acceptor fluorophore Atto647N. The dimers were then caged in lipid vesicles that were immobilized onto a solid substrate similar to the experiments by Cisse et al. (13) and by Rhoades et al. (14) (see Fig. 1 and SI Methods for details). Functionality of the investigated constructs was tested by an ATPase assay (15, 16) as well as by subunit exchange and p23 binding experiments where suitable (see Fig. S1 and Fig. 7).
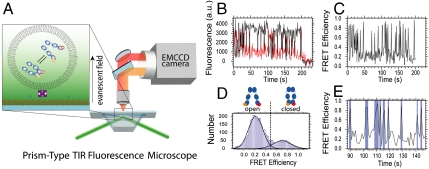
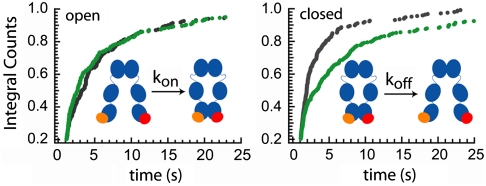
Fig. 1.
Experimental setup. The Hsp90 molecules (size 5–10 nm) were caged in lipid vesicles with a diameter of about 100 nm (not to scale). The vesicles were immobilized via biotinylated lipids onto a solid substrate in a microfluidic chamber and mounted in a prism-type TIR microscope. Single-molecule fluorescence from the donor and acceptor were detected simultaneously by an electron multiplying charge coupled device camera (A). Matching time traces were overlaid (B) and FRET efficiencies determined (C). The cumulated histogram of all FRET efficiencies shows two states that are clearly separated by a threshold (D), which allow us to determine the rate constants from a separation of the time trace into open and closed states (E). See Methods for details. A detailed scheme of the setup is shown in Fig. S2.
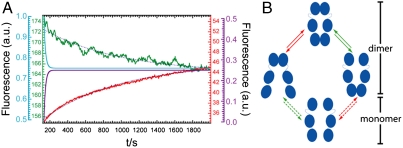
Fig. 7.
Subunit exchange experiment. (A) After mixing of differently labeled Hsp90 homodimers in a spectrofluorimeter subunit exchange leads to an increasing FRET signal; the donor signal (green) decreases whereas the acceptor signal (red) increases in time. Blue dotted lines show global fit for both curves with τ = 850 s. The violet and blue lines show the outcome of the simulation of the subunit exchange assuming no correlation between the N- and C-terminal dimerization, which results in τ = 18 s. (B) Model for the N- and C-terminal opening and closing. The molecule stays mainly in the dimeric states—see main text for details. The reaction scheme for the bulk measurements and the exchange simulation are shown in Fig. S6.
The C-terminal interaction was measured by single-molecule fluorescence resonance energy transfer (smFRET) (17) at 30 °C in a custom-built prism-type total-internal reflection fluorescence microscope as described before (11) (see Fig. 1, Methods, SI Methods, and Fig. S2 for details). We observe C-terminal open-closed dynamics on the time scale of seconds, in contrast to current models of Hsp90 that assume the C terminus to be closed on time scales of 100 s (5, 18). Several bulk experiments (10) and the single-molecule experiments in ref. 11 show that the N-terminal domain is very stably dimerized in the presence of AMP-PNP (Fig. 2A). Surprisingly, even under these conditions the C terminus shows opening and closing dynamics (Fig. 2B)—clearly demonstrating that an N-terminal closed state does not result in a C-terminal closed state. In contrast, addition of the cochaperone Sti1 keeps the C termini mainly closed (Fig. S3).
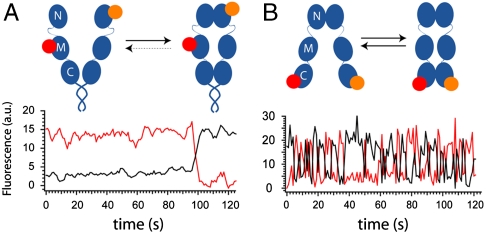
Fig. 2.
Comparison of N- and C-terminal dynamics of Hsp90. Exemplary fluorescence time trace of the N-terminal (A) (11) and C-terminal (B) dimerization kinetics of Hsp90 with 2 mM AMPPNP. (Top) Schematics of the various conformations. To investigate the N-terminal kinetics, a coiled coil motive was introduced at the C terminus to allow for longer observation times; this did not change the dynamics (11). (Bottom) Fluorescence signal of the donor (black trace) and the acceptor (red trace). The N terminus stays closed (high acceptor signal) on the time scale of the experiment in A until the acceptor bleaches after around 100 s.
The FRET efficiencies allow us to estimate the distances between the fluorophores in the C-terminally open and closed state with assumptions on their orientation (κ2 = 2/3). This assumption is reasonable for systems with anisotropies of less than 0.2 (19). Here we have an anisotropy of around 0.23 for the donor and 0.18 for the acceptor dye, and the following values should therefore be taken only as a rough estimate. The calculated distances of the FRET pair are in all cases around 6.4 nm for the open and 4.3 nm for the closed state. The length of the linker between the cystein and the fluorophore is 0.8 nm in both cases. These distances are in very good agreement with the crystal structure where the distance between the two cysteins is around 3.5 nm (in the closed state without linker). For determination of the Förster radius and further discussion, see SI Methods. As the vesicle size is on the order of 100 nm, complete dissociation of the dimer (N- and C-terminal open) would result in a FRET efficiency of zero and can therefore be excluded on the time scale of the experiment (Fig. S3).
Influence of Nucleotides on C-terminal Open and Closed States.
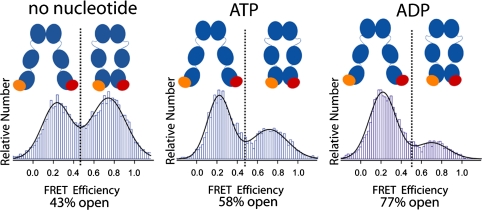
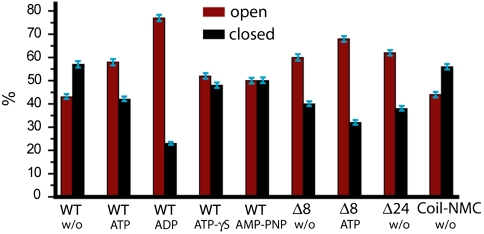
After observing surprisingly fast dynamics in the C-terminal domain, the question arises whether this dimerization is regulated. Because Hsp90 function critically depends on ATP binding and hydrolysis, we repeated the described experiment under different nucleotide conditions. First, we used the natural nucleotides ATP and ADP and compared the results to the situation without nucleotide. We compiled the FRET efficiencies into histograms like the one depicted in Fig. 1D and described in Methods. The area underneath the two peaks in these FRET histograms corresponds to the occupancy of the two states: Low FRET indicates that the C terminus is in an open state, and high FRET indicates that the C terminus is in a closed state. As can be seen in Fig. 3, without nucleotide the C-terminal domains are about 43% open and 57% closed. In the presence of ATP, this distribution is almost reversed, and with ADP the equilibrium is even more shifted to the open state. These observations are contrary to the situation at the N-terminal domain where no strong influence of ATP or ADP on the dimerization equilibrium was found (11, 20).
Fig. 3.
Effect of nucleotide on the C-terminal dimerization. Relative amount of observed FRET efficiencies between donor (orange) and acceptor molecule (red) at the C-terminal domain for different nucleotide conditions. High FRET values correspond to the C-terminal closed state, and low FRET values to the C-terminal open state. The equilibrium distribution of the open and closed states was determined by a Gaussian fit. The effects of ATP analogues are shown in Fig. S4.
What causes these shifts in equilibrium? To answer that question we determined the dwell-time distributions of the dimerization kinetics. The FRET efficiency at the minimum between the two peaks in the FRET efficiency histograms shown in Fig. 3 (dotted line) defines the separation between open and closed states. Now each time trace is separated into parts with open and closed states (Fig. 1E), and the dwell times in these states can be determined (11). The distributions of these dwell times give quantitative kinetic information. Fig. 4 shows such integrated dwell-time distributions in the absence (green) and in the presence of ATP (black). Whereas the on-rate constants of the C-terminal dimerization (determined from the time in the open state) are about the same (Fig. 4 Left), the off-rate constants increase in the presence of ATP by more than a factor of 2 (Fig. 4 Right). Thus, the shift to the more open conformation is mainly caused by the destabilization of the C-terminally dimerized complex upon ATP binding and cannot be explained by a potential N-terminal-induced spatial proximity (which would affect the on-rate constants).
Fig. 4.
Effect of nucleotide on C-terminal dynamics. Integrated dwell-time distributions in the open (Left) and closed (Right) states are shown in the presence of ATP (black) and in the absence of nucleotide (green). In all cases double exponential fits are necessary, suggesting a complex binding mechanism. The rate constants and amplitudes are listed in Fig. S4.
To investigate the effects of nucleotides in more detail, we used the ATP analogues ATP-γS and AMP-PNP (Fig. 5 and Fig. S4). ATP-γS is slowly hydrolized by Hsp90 [about a factor of 7 slower than ATP (20)], and AMP-PNP is not hydrolized at all on the time scale of the experiments. In the presence of ATP-γS, the obtained dwell times for the closed state are similar to those for ATP, which suggests that the hydrolysis of the nucleotide does not lead to C-terminal destabilization. This is further underlined by the at least 1 order of magnitude different time scales for the dimerization and hydrolysis kinetics. AMP-PNP is known to force Hsp90 into a permanently N-terminally closed state (11); however, this is not the case for the C-terminal domains. In the presence of AMP-PNP, Hsp90 still shows C-terminal dynamics with about an equal population of the open and closed states (Fig. 5) similar to the situation without nucleotide, but with slightly different open and closed times (Fig. S4). This observation shows that the C-terminal opening and closing takes place even when the N-terminal domain is fixed in the closed state.
Fig. 5.
Effect of N-terminal mutation on C-terminal dimerization. Relative amount of observed C-terminal FRET efficiencies (%) determined from the single-molecule experiments for the different nucleotide conditions and N-terminal mutations. The equilibrium distributions are shown in Fig. 3 and Figs. S4 and S5. Assuming a confidence interval corresponding to 1 standard deviation, the uncertainty of all values is less than 5% of their value, i.e., usually around 2.5% for the relative amount of observed FRET efficiencies.
Effect of N-Terminal Mutations on C-Terminal Open and Closed States.
To address the structural basis of the observed effects, we created several N-terminal Hsp90 mutants and investigated their effect on C-terminal dimerization. We chose two mutants with deletions of either 8 (Δ8) or 24 (Δ24) amino acids at the very N-terminal end. These mutants have been characterized in the past (21, 22). Δ8 shows a higher ATPase rate than the wild-type protein, whereas Δ24 has no ATPase activity, because those amino acids engaged in the cross stimulation of the N-terminal domains have been removed (22). In the third mutant, a coiled coil motif had been fused to the N-terminal end of Hsp90 to keep the N-terminal ends in close proximity (henceforth called Coil-NMC). This leads to a four times faster ATPase rate compared to the wild-type protein (Fig. S1). The obtained FRET histograms for these mutants are shown in the supplement (Fig. S5). As can be seen in Fig. 5, the equilibrium of the C-terminal dimerization is shifted toward a more open state for the deletion mutants compared to the wild-type Hsp90.
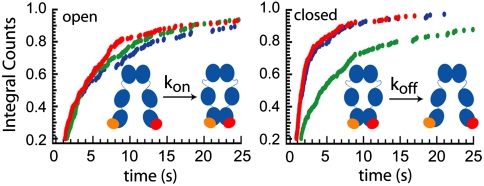
To further understand this shift toward a more C-terminal open state, we determined the kinetics of the Δ8 and Δ24 Hsp90 deletion mutants as well as the Coil-NMC. The Δ8 and Δ24 mutants show a similar occurrence of open and closed states (Fig. 5) and kinetics (Fig. 6, red and blue) in the absence of ATP. This means that already the deletion of the first 8 amino acids destabilizes the C-terminal interaction; the deletion of 16 more amino acids does not have an additional effect in the nucleotide-free state. These two mutants have faster off-rate constants (shorter dwell times of the closed state) than the wild type (Fig. 6, green), whereas the on-rate constants (proportional to one over the dwell times of the open state) are quite similar to the wild type. It is also interesting to note that both the occurrence and the kinetics of these two mutants in the absence of nucleotides are quite similar to the wild-type protein in the presence of ATP (Fig. 5, Fig. S4, and Fig. S5).
Fig. 6.
Effect of N-terminal mutations on C-terminal dynamics. Integrated dwell-time distributions in the open (Left) and closed (Right) states are shown for the Δ8 (red), for the Δ24 (blue), and the wild-type Hsp90 (green) in the absence of ATP. Further kinetic data and fit values are shown in Fig. S5.
N-terminal dimerization should keep the C-terminal domains in closer proximity and therefore cause an increase in kon if only geometric effects play a role. To investigate this, we compared the kon values of the Δ24 mutant (which is thought to show less stable N-terminal dimerization), the wild type without ATP (which can be closed and open), and the Δ8 mutant in the presence of ATP (which is permanently closed at the N-terminal domains) (Fig. S5). The kon values are the same within the experimental uncertainty. This shows that the proximity of the N-terminal domains has little influence on C-terminal kinetics, in contrast to the nucleotide state, which has a strong effect.
We tested if the N-terminal deletion and ATP binding cause the same (structural) effect at the C-terminal domain or if the observed effects are additive. Fig. 5 shows that both the presence of ATP and the Δ8 mutation cause a larger population of the C-terminally dissociated state, suggesting a common mechanism. On the other hand, there is still an additional effect of ATP on the Δ8 mutant, because the population is even more shifted to the C-terminally open state.
Coordination between N- and C-Terminal Dynamics.
Finally, we investigated how the above-described C-terminal kinetics can explain bulk monomer exchange experiments. The bulk monomer exchange rates of yeast Hsp90 were determined in a spectrofluorimeter with the same fluorescently labeled constructs used for the single-molecule experiments and result in around 1 exchange per 1,000 s in the nucleotide-free state (Fig. 7A; for a detailed description of the experiment, see SI Methods and Fig. S6). In addition, we estimated the monomer exchange rates by kinetic simulations under the assumption that N- and C-terminal dimerization are independent—i.e., not coordinated (Fig. S6 shows the underlying reaction pathway). The data for the N-terminal dimerization kinetics are taken from Mickler et al. (11) and for the C-terminal dimerization kinetics from above. For determining an upper (slow) limit for the monomer exchange time from single-molecule experiments, we took the slowest rate constants, which are around 0.1 per second for both C-terminal and N-terminal opening and closing. The equilibrium constants depicted by the green and red arrows in Fig. 7B should be identical for uncoordinated C- and N-terminal dynamics, which makes the simulation straightforward (see Methods). The results for the simulation of such a scenario are depicted in Fig. 7A (blue and violet trace). They are clearly at least an order of magnitude faster than the bulk experimental results (green and red trace). Therefore, the rate constants for C-terminal opening are not independent from the N-terminal dimerization. The molecules stay mainly in the dimeric states; i.e., they perform many C- or N-terminal open-close cycles before dissociation (assuming two dimerization domains). In other words, the observed exchange rate constant cannot be explained by an independent movement of the N- and C-terminal ends, which necessitates cooperativity between N- and C-terminal open and closed states. Even more, the N-terminal dimerization has to have some anticorrelation on the C-terminal dimerization, which then slows down the dissociation of the Hsp90 dimer (see Movie S1). Middle-domain contact as a reason for the slow subunit can be excluded because middle and N-terminal domains show the same kinetics, thus moving as one (Fig. S7).
Discussion
The single-molecule FRET system presented here allowed us to dissect the C-terminal open-close kinetics of Hsp90. Of particular interest is the coordination of this C-terminal kinetics with the N-terminal kinetics and particularly with nucleotide binding at the N-terminal domain.
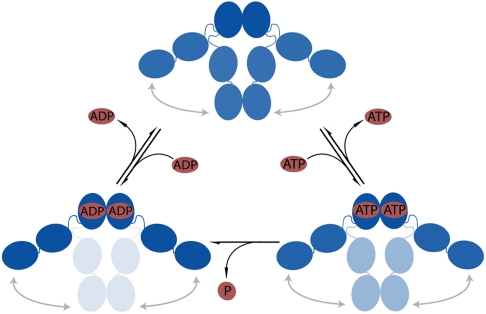
We observe a strong influence of ATP and ADP on the C-terminal dimerization equilibrium and dynamics. The binding of ATP and ADP leads to a destabilization of the C-terminal closed state. Thus, during the ATP hydrolysis cycle, Hsp90 is successively shifted to a more C-terminally open conformation, and we can align these three states in a reaction cycle (Fig. 8) in which binding and hydrolysis of ATP will facilitate the opening of the C-terminal domains.
Fig. 8.
Model for the nucleotide dependent C-terminal association and dissociation. Dark blue represents high occupancy of the state, and medium and light blue represent a lower occupancy of the respective state. When ATP or ADP is bound, the equilibrium between the C-terminal open and closed states is shifted toward the open state. Additional schemes for the N-terminal mutants are shown in Fig. S8.
Because the C-terminal region is far away from the nucleotide binding pocket, the observed effects can be explained only by a long distance communication through the whole protein. This is in line with molecular dynamic simulations (23) where correlated changes between residues in the N- and C-terminal domains after binding of ATP or ADP were found.
Experimentally, Retzlaff et al. recently showed that point mutations at position 597 in the C-terminal domain have a pronounced effect on ATP hydrolysis (24), i.e., a communication in exactly the opposite direction from the C- to the N-terminal domain. Similar observations were made for C-terminal deletion mutants (25). In addition, hydrogen exchange experiments demonstrated ATP-induced conformational changes throughout the N- and middle domains in yeast Hsp90 (26) and the bacterial homologue HtpG (10). Such a coordination between the N- and C-terminal domains is surprising because of the presence of a long flexible linker between the middle and the N-terminal domain. This linker seems to play an important role in the regulation of Hsp90 dynamics and function (27, 28).
The N-terminal deletion mutations give further insight into structural changes that cause the signaling from the N- to the C-terminal domains. The first 8 amino acids are known to intrinsically inhibit the ATPase activity of Hsp90 by an interaction with the ATPase lid of the nucleotide binding pocket (21, 22). Their deletion should therefore abolish this inhibition and the loss of intrinsic inhibition would then be connected to the C-terminal effects observed (Fig. S8). This notion is supported by the fact that ATP has a smaller effect on the Δ8 Hsp90 mutant than on the wild-type C-terminal dimerization equilibrium. In fact, the C-terminal dimerization equilibrium of the wild-type protein in the presence of ATP and the N-terminal deletion mutants Δ8 and Δ24 in the absence of nucleotide are almost the same (Fig. 5). On the other hand, there is still a detectable effect of ATP on the Δ8 mutant (Fig. 5). This is consistent with ATP binding and the deletion of the N-terminal 8 amino acids having a common (but not identical) impact on the C-terminal opening and closing.
As both ATP and Δ8 shift Hsp90 toward the N-terminally closed state, the question arises whether keeping the N-terminal domains in close proximity results in a shift of the C-terminal dimerization equilibrium to the open state. The results obtained for the Coil-NMC construct tell us that this is not the case. Although the N-terminal domains are in close proximity in this construct, there is no effect on the C-terminal dimerization equilibrium compared to the wild-type protein without nucleotide (Fig. 5). Therefore, structural changes induced by ATP binding or deletion of the N-terminal segment are necessary to shift this equilibrium.
There are a couple of reasons why previous methods have not revealed a C-terminal opening and closing: (i) In bulk experiments synchronization is required to follow such a movement; this has recently been applied to the N-terminal movement (20), but so far not to the C-terminal movement. (ii) In bulk experiments the dissociation is difficult to distinguish from opening, and separation of C- and N-terminal opening is very involved (and with the published data not possible); the same holds true for published electron microscopy data.
Surprisingly, the monomer exchange rates of Hsp90 are much slower than expected in the case of independent C- and N-terminal dynamics (Fig. 7). Thus, there must be some coordination between these two processes, which manifests itself in an anticorrelated opening and closing of the N- and C-terminal domains. We think that this is relevant in vivo, because N-terminal opening (which has been shown to be relevant) and C-terminal opening are coupled. The presence of both N-terminal and C-terminal open dimers might allow for a reinterpretation of several ensemble studies, where only N-terminally open and closed conformations were used to fit the experimental data.
Conclusion
The results show that single-molecule FRET experiments on proteins in immobilized lipid vesicles provide unique insights into their dynamics. In particular for protein complexes, the dynamics of single dimerization sites can be separately investigated.
This is demonstrated on the example of the N- and C-terminal dimerization of the homodimer Hsp90. Surprisingly we discovered coordination between the two dimerization interfaces, which allows us to propose a completely revised picture of Hsp90 dynamics.
Methods
Protein Expression and Purification.
The N-terminal deletion mutants were generated as described in Richter et al. (22). The cysteine point mutation at position 560 was created with the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene) in the case of Hsp90 wild type and with the Finnzymes Site-Directed Mutagenesis Kit in the case of Δ8-Hsp90, Δ24-Hsp90, and Coil-NMC. Protein expression and purification were in all cases done according to the protocol described in ref. 29. The Coil-NMC mutant was a kind gift from Dan Bolon and designed like the mutants described in ref. 30.
Labeling Efficiency.
The fluorescence molecules (AttoTec) were attached to the Hsp90 cysteine mutants by maleimide chemistry. The labeling efficiency was determined by measuring the absorption spectra in a Nanodrop ND-1000 UV-Vis spectrometer. Finally, a labeling ratio of one fluorophore per Hsp90 monomer was achieved (± 15%).
Experimental Setup.
All single-molecule fluorescence measurements were performed in a custom-built prism-type total internal reflection fluorescence microscope equipped with two lasers (532 nm Compass 215 M 75 mW, Coherent Inc.; and 635 nm LPM635-25C 25 mW, Newport) allowing alternating laser excitation. The typical lifetime of the Atto647N fluorophore attached to Hsp90 is about 30 s in our system. Therefore, we excited the fluorophores only for 100 ms every second if not otherwise stated. This allowed for observation times of several minutes. The two colors for donor and acceptor fluorescence were separated and simultaneously recorded on an Andor DV887 (Andor Technology) camera (Fig. 1A). The movies were analyzed with a program from the Hugel Lab based on Igor Pro 6.01 (WaveMetrics): First the time traces of single fluorophores were extracted from the movie with a threshold criterion. Then the two colors were overlaid and the corrected FRET efficiency was calculated (Fig. 1C) (31). Blinking of the donor is excluded by the stability of the total fluorescence intensity, whereas the blinking of the acceptor as a source for this dynamics is excluded by direct excitation of the acceptor and alternating excitation (Fig. S9). Further controls to distinguish protein dynamics and dye photophysics (32) and a more detailed description of the setup are given in the SI Methods and Fig. S9.
These FRET efficiencies (every single data point) are then cumulated in a histogram as shown in Fig. 1D. As fluorescence is a stochastic process, the FRET efficiency of a single state is expected to have a Gaussian distribution. Our efficiencies can very nicely be fit with two Gaussian distributions and can therefore be divided into two states separated at a transfer efficiency of around 0.5. The overlap of the two distributions results in a misassignment of less than 1%. Every crossing of this threshold corresponds to a transition between open/close. The dwell times for the open and close events have been plotted in integrated plots, which overcome the necessity of data binning (33).
The kinetics were all fitted with the following double exponential:
the obtained amplitudes Ai and rate constants τi are summarized in Figs. S4 and S5.
Simulations of Monomer Exchange.
The process of subunit exchange has been simulated by numerically iteratively solving the underlying differential equations for the rate constants and concentrations. The concentration of a state i at the time t + Δt is given by
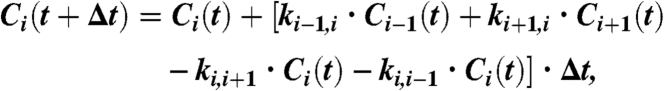 |
where ki,i+1 is the rate constant of the process depopulating state i and populating state i + 1. The scheme depicted in Fig. S6 describing the subunit exchange can therefore be represented by a set of coupled differential equations. We have the seven states: homodimer with closed N terminus or closed C terminus or both, heterodimer with N terminus closed or C terminus closed or both, and finally a monomeric state.
In an exchange experiment two species of homodimers are added and the formation of heterodimers is observed (SI Methods). Accordingly, we start the simulation with the four states without heterodimers: a monomeric, a N-terminal dimerized, a C-terminal dimerized, and a C- and N-terminal dimerized state. The equilibrium concentrations for these states are given by the N- and C-terminal dimerization kinetics. To calculate an upper limit for the exchange kinetics, we took the slowest rate constants measured for the N terminus (11) and the C terminus (Fig. S4) in the absence of nucleotide, namely, 1/10 s-1. Finally, we assume that the probability to form a heterodimer upon dissociation of a dimer is 50%.
Supplementary Material
Acknowledgments.
We thank Dan Bolon, Markus Götz, Don Lamb, and Matthias Rief for helpful discussions and critical reading of the manuscript. We thank the referees for very helpful comments. T.H and J.B. thank the German Science Foundation (DFG), Nanosystems Initiative Munich (NIM), and the Fonds der Chemischen Industrie for financial support. C.R. is supported by the International Doctorate Program NanoBioTechnology (IDK-NBT) program of the Elitenetzwerk Bayern.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000916107/-/DCSupplemental.
References
- 1.Wegele H, Muschler P, Bunck M, Reinstein J, Buchner J. Dissection of the contribution of individual domains to the ATPase mechanism of Hsp90. J Biol Chem. 2003;278:39303–39310. doi: 10.1074/jbc.M305751200. [DOI] [PubMed] [Google Scholar]
- 2.Wayne N, Bolon DN. Dimerization of Hsp90 is required for in vivo function. Design and analysis of monomers and dimers. J Biol Chem. 2007;282:35386–35395. doi: 10.1074/jbc.M703844200. [DOI] [PubMed] [Google Scholar]
- 3.Young JC, Moarefi I, Hartl FU. Hsp90: A specialized but essential protein-folding tool. J Cell Biol. 2001;154:267–273. doi: 10.1083/jcb.200104079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- 5.Richter K, Buchner J. Hsp90: Twist and fold. Cell. 2006;127:251–253. doi: 10.1016/j.cell.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Richter K, Reinstein J, Buchner J. A Grp on the Hsp90 mechanism. Mol Cell. 2007;28:177–179. doi: 10.1016/j.molcel.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Ali MM, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bron P, et al. Apo-Hsp90 coexists in two open conformational states in solution. Biol Cell. 2008;100:413–425. doi: 10.1042/BC20070149. [DOI] [PubMed] [Google Scholar]
- 9.Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: Insights into the conformational dynamics of Hsp. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graf C, Stankiewicz M, Kramer G, Mayer MP. Spatially and kinetically resolved changes in the conformational dynamics of the Hsp90 chaperone machine. EMBO J. 2009;28:602–613. doi: 10.1038/emboj.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mickler M, Hessling M, Ratzke C, Buchner J, Hugel T. The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nat Struct Mol Biol. 2009;16:281–286. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 12.Richter K, Muschler P, Hainzl O, Buchner J. Coordinated ATP hydrolysis by the Hsp90 dimer. J Biol Chem. 2001;276:33689–33696. doi: 10.1074/jbc.M103832200. [DOI] [PubMed] [Google Scholar]
- 13.Cisse I, Okumus B, Joo C, Ha T. Fueling protein DNA interactions inside porous nanocontainers. Proc Natl Acad Sci USA. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhoades E, Cohen M, Schuler B, Haran G. Two-state folding observed in individual protein molecules. J Am Chem Soc. 2004;126:14686–14687. doi: 10.1021/ja046209k. [DOI] [PubMed] [Google Scholar]
- 15.Ali MMU, et al. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panaretou B, Piper P. Isolation of yeast plasma membranes. Methods Mol Biol. 1996;53:117–121. doi: 10.1385/0-89603-319-8:117. [DOI] [PubMed] [Google Scholar]
- 17.Ha T, et al. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–18477. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 19.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 20.Hessling M, Richter K, Buchner J. Dissection of the ATP-induced conformational cycle of the molecular chaperone Hsp90. Nat Struct Mol Biol. 2009;16:287–293. doi: 10.1038/nsmb.1565. [DOI] [PubMed] [Google Scholar]
- 21.Richter K, et al. Intrinsic inhibition of the Hsp90 ATPase activity. J Biol Chem. 2006;281:11301–11311. doi: 10.1074/jbc.M510142200. [DOI] [PubMed] [Google Scholar]
- 22.Richter K, Reinstein J, Buchner J. N-terminal residues regulate the catalytic efficiency of the Hsp90 ATPase cycle. J Biol Chem. 2002;277:44905–44910. doi: 10.1074/jbc.M208457200. [DOI] [PubMed] [Google Scholar]
- 23.Morra G, Verkhivker G, Colombo G. Modeling signal propagation mechanisms and ligand-based conformational dynamics of the Hsp90 molecular chaperone full-length dimer. PLoS Comput Biol. 2009;5:e1000323. doi: 10.1371/journal.pcbi.1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Retzlaff M, et al. Hsp90 is regulated by a switch point in the C-terminal domain. EMBO Rep. 2009;10:1147–1153. doi: 10.1038/embor.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Owen BA, Sullivan WP, Felts SJ, Toft DO. Regulation of heat shock protein 90 ATPase activity by sequences in the carboxyl terminus. J Biol Chem. 2002;277:7086–7091. doi: 10.1074/jbc.M111450200. [DOI] [PubMed] [Google Scholar]
- 26.Phillips JJ, et al. Conformational dynamics of the molecular chaperone Hsp90 in complexes with a co-chaperone and anticancer drugs. J Mol Biol. 2007;372:1189–1203. doi: 10.1016/j.jmb.2007.04.059. [DOI] [PubMed] [Google Scholar]
- 27.Tsutsumi S, et al. Hsp90 charged-linker truncation reverses the functional consequences of weakened hydrophobic contacts in the N domain. Nat Struct Mol Biol. 2009;16:1141–1147. doi: 10.1038/nsmb.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hainzl O, Lapina MC, Buchner J, Richter K. The charged linker region is an important regulator of Hsp90 function. J Biol Chem. 2009;284:22559–22567. doi: 10.1074/jbc.M109.031658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter K, Buchner J. Hsp90: Chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 30.Wayne N, Lai Y, Pullen L, Bolon DN. Modular control of cross-oligomerization: Analysis of superstabilized Hsp90 homodimers in vivo. J Biol Chem. 2010;285:234–241. doi: 10.1074/jbc.M109.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy D, et al. Vernakalant hydrochloride for rapid conversion of atrial fibrillation: A phase 3, randomized, placebo-controlled trial. Circulation. 2008;117:1518–1525. doi: 10.1161/CIRCULATIONAHA.107.723866. [DOI] [PubMed] [Google Scholar]
- 32.Chung HS, Louis JM, Eaton WA. Distinguishing between protein dynamics and dye photophysics in single-molecule FRET experiments. Biophys J. 98:696–706. doi: 10.1016/j.bpj.2009.12.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebhardt JC, Clemen AE, Jaud J, Rief M. Myosin-V is a mechanical ratchet. Proc Natl Acad Sci USA. 2006;103:8680–8685. doi: 10.1073/pnas.0510191103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.