Abstract
EGF induces the translocation of EGF receptor (EGFR) from the cell surface to the nucleus where EGFR activates gene transcription through its binding to an AT-rich sequence (ATRS) of the target gene promoter. However, how EGFR, without a DNA-binding domain, can bind to the gene promoter is unclear. In the present study, we show that RNA helicase A (RHA) is an important mediator for EGFR-induced gene transactivation. EGF stimulates the interaction of EGFR with RHA in the nucleus of cancer cells. The EGFR/RHA complex then associates with the target gene promoter through binding of RHA to the ATRS of the target gene promoter to activate its transcription. Knockdown of RHA expression in cancer cells abrogates the binding of EGFR to the target gene promoter, thereby reducing EGF/EGFR-induced gene expression. In addition, interruption of EGFR–RHA interaction decreases the EGFR-induced promoter activity. Consistently, we observed a positive correlation of the nuclear expression of EGFR, RHA, and cyclin D1 in human breast cancer samples. These results indicate that RHA is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus.
Keywords: cyclin D1, nuclear translocation, inducible nitric oxide synthase, transcription
Cell surface EGF receptor (EGFR) has been shown to be localized in the nucleus (1–4). Nuclear EGFR has been demonstrated to contribute to cancer cell resistance to cetuximab and radiation treatment (5, 6) and to be negatively correlated with overall survival of patients with multiple cancer types (7–11). Moreover, nuclear EGFR interacts with signal transducer and activator of transcription 3 (STAT3), signal transducer and activator of transcription 5A (STAT5A), E2F1, DNA-dependent protein kinase (DNA-PK), and proliferating cell nuclear antigen (PCNA) and plays important roles in cell transformation, proliferation, and DNA repair and replication (12–16). Nuclear EGFR regulates gene expression by binding to an AT-rich sequence (ATRS) of the gene's promoter (13, 16, 17). Additionally, a recent unbiased protein-DNA interactome study indicates that EGFR is a DNA-binding protein (18). However, EGFR does not contain a DNA-binding domain, and evidence supporting direct binding of EGFR to the specific DNA sequence is lacking. Thus, identifying the DNA-binding partner for EGFR is crucial for understanding how EGFR regulates gene transcription in the nucleus.
RNA helicase A (RHA), the human homolog of Drosophila maleless (MLE) that increases the transcription of male X-linked genes (19), is a multifunctional protein and is conserved in Drosophila and mammals (20–22). RHA belongs to the aspartate-glutamate-alanine-aspartate (DEAD) box family of proteins and has the ability to bind to RNA and DNA (23, 24). RHA regulates gene transcription by interacting with transcription factors (22) or by binding directly to the target gene promoter (25). Moreover, Drosophila MLE activates rox2 transcription by binding to an AT-rich region of the gene promoter (26). Interestingly, this AT-rich region contains the previously reported EGFR-binding sequence, an ATRS in the promoter regions of cyclin D1 (17) and inducible NOS (iNOS) (13), raising the very interesting question of whether RHA serves as a DNA-binding partner for nuclear EGFR to activate gene transcription.
Here, we report that RHA is a DNA-binding partner for EGFR in regulating its target gene transcription in the nucleus of cancer cells.
Results
Nuclear Interaction Between EGFR and RHA.
To understand the functionality of nuclear EGFR, nano-liquid chromatography (LC)/MS/MS was used to identify proteins with the potential to interact with EGFR in the nuclei of cancer cells. As shown in Fig. S1A and Table S1, we identified several RNA helicase proteins, and RHA in particular caught our attention because it is a well-known transcriptional activator (22) and its Drosophila homolog MLE has been shown to bind to the ATRS-containing sequence of rox2 gene promoter (26). Thus, we hypothesized that RHA is a DNA-binding partner for EGFR-mediated gene transcription in the nucleus.
To determine whether RHA indeed partners with EGFR, we first confirmed that EGFR and RHA interact in vivo. As shown in Fig. 1 A and B, endogenous association of EGFR with RHA in response to EGF treatment was detected mainly in the nuclei but not in the cytoplasm in multiple cell lines. In addition, EGF-induced EGFR–RHA interaction was time dependent on the EGFR nuclear translocation, and the maximum association of EGFR with RHA was observed at 30-min treatment with EGF (Fig. 1C). Consistent with the biochemical results, confocal microscopy showed that the EGF-induced colocalization of EGFR (green) and RHA (red) was observed in the nuclei of both MDA-MB-468 (Fig. 1D) and HeLa cells (Fig. S2A). To confirm further the nuclear location of EGFR/RHA complexes, sequential photosections of a nucleus were examined, and EGFR/RHA complexes were clearly detected in middle sections (i.e., planes 11–14) in both MDA-MB-468 and HeLa cells (Fig. S2 B and C). Taken together, these results suggest that EGFR and RHA interact mainly in the nucleus and that this interaction is greatly increased upon EGF treatment.
Fig. 1.
Association of EGFR with RHA in the nucleus. (A) Endogenous association of EGFR with RHA in A431 cells. Cells with 80–85% confluence were serum starved overnight before EGF treatment. Equal amounts of cellular fractionated proteins were immunoprecipitated with an anti-EGFR antibody and loaded for Western blotting. Input samples from equal amounts of proteins blotted for EGFR, RHA, lamin B, and α-tubulin are shown as loading and fractionation controls. C, cytoplasmic fraction; N, nuclear fraction; −, without EGF treatment; +, with EGF treatment. (B) Endogenous association of EGFR with RHA in MDA-MB-468 cells. Quantification of the band's density was performed using ImageJ 1.41 (National Institutes of Health). The density of the band in lane 5 was set as 1. The numbers under the band in lane 6 indicate the relative density of that band as compared with the density of the band in lane 5. (C) Time-dependent association of EGFR with RHA in the nucleus. Nuclear proteins from A431 were immunoprecipitated with an anti-EGFR antibody and then were immunoblotted to detect RHA. Input nuclear fraction samples blotted for EGFR, RHA, lamin B, and tubulin are shown as the loading and fractionation controls. (D) (Left) Colocalization of EGFR and RHA in MDA-MB-468 cells. Cells treated with EGF (50 ng/mL for 30 min) or left untreated were stained with indicated antibodies. Colocalization of EGFR and RHA is shown as yellow in the merged image and is indicated by arrows in the Inset. Scale bar, 10 μm. (Right) The bar graph shows the percentage of the 50 counted cells with colocalized EGFR and RHA.
To study whether EGFR can bind directly to RHA, an in vitro pull-down assay was performed using in vitro translated EGFR and purified GST-RHA fragments. As shown in Fig. S1B, EGFR was pulled down by two RHA fragments, RHA623–960 and RHA961–1270, but not by GST alone, RHA1–303, or RHA304–622, indicating a direct interaction between EGFR and the C-terminal domain of RHA in vitro.
Regulation of Gene Expression by EGFR/RHA Complex in Vivo.
To determine whether EGFR regulates gene expression in the nucleus through its association with RHA, we performed luciferase assays using either a cyclin D1 promoter (pCCD1-Luc) or an iNOS promoter (piNOS-Luc) in HeLa cells. As shown in Fig. 2A, EGFR (lanes 3 and 4) and RHA (lanes 5 and 6) each increased the luciferase expression, and the combination of EGFR and RHA (lanes 7 and 8) significantly enhanced the activity of these promoters both at basal level and with EGF treatment, suggesting that EGFR and RHA transactivate a target gene promoter in a cooperative fashion.
Fig. 2.
Coactivation of gene expression by EGFR and RHA. (A) Costimulation of cyclin D1 (pCCD1-Luc, Left) and iNOS (piNOS-Luc, Right) promoter activity by EGFR and RHA. P values calculated from Student's t test are shown above paired bars. (B) Knockdown of RHA expression diminishes EGFR-induced promoter activity. HeLa cells with stable expression of indicated shRNAs were transfected with plasmids. Luciferase assay was performed after 5 h EGF stimulation. The expression levels of EGFR and RHA are shown in the lower panel. The density of the RHA band was quantified using ImageJ with the density of the basal level band (i.e., lane 1 without EGF) from control shRNA set as 100. The numbers under other bands are the relative band densities as compared with the density of lane 1. Ctrl, control; IB, immunoblotting. (C) ATRS-dependent activation of the cyclin D1 promoter by EGFR and RHA. Relative luciferase activities (i.e., percentage of wild-type ATRS promoter activity) are presented as the mean results ± SD (n = 3). (D) Association of EGFR/RHA with the cyclin D1 gene promoter. (Top) Binding of EGFR and RHA to the cyclin D1 gene promoter in A431 cells. The band's density was quantified using ImageJ with the band density in lane 3 set as 1. The numbers under other bands are the relative densities as compared with the density of the band in lane 3. (Middle) Sequential ChIP-PCR analysis of the association of EGFR and RHA with the cyclin D1 promoter. (Bottom) Reduced binding of EGFR to the cyclin D1 promoter after RHA knockdown. (E) Reduced association of EGFR with the iNOS promoter in A431 cells after RNA knockdown. (Upper) Normal ChIP-PCR. (Lower) Quantitative ChIP-PCR.
The role of RHA in nuclear EGFR-induced gene expression was investigated further in HeLa cells by the knockdown of RHA expression with shRNA and siRNA. Knockdown of RHA expression significantly decreased EGFR-induced cyclin D1 promoter activity (Fig. 2B, Left, and Fig. S3 A–C). Moreover, the down-regulated cyclin D1 promoter activity could be rescued by reexpression of RHA in the RHA knocked-down cells (Fig. S3D, lanes 9 and 10 vs. 7 and 8, respectively). Knocking down RHA consistently reduced the level of endogenous cyclin D1 protein (Fig. S3 E and F) and suppressed EGF-stimulated cell growth (Fig. S3G). Similarly, knockdown of RHA expression also reduced the EGFR-induced promoter activities of iNOS (Fig. 2B, Right) and c-fos (Fig. S4A), which are known to be regulated by nuclear EGFR (13), but did not reduce the promoter activity of c-Jun (Fig. S4B), which is known to be regulated by traditional EGFR downstream pathways (27, 28). Taken together, these results support the involvement of RHA in nuclear EGFR-induced gene expression.
To determine whether RHA regulates cyclin D1 gene transcription through its binding to the ATRS of the promoter (similar to the binding of its Drosophila homolog MLE to the ATRS-containing sequence of rox2 promoter) and, if so, whether RHA is the protein through which nuclear EGFR binds to the cyclin D1 gene promoter to regulate its transcription, we performed promoter-reporter assays using cyclin D1 promoter constructs with wild-type or mutated ATRS. Compared with the promoter containing wild-type ATRS, mutation of ATRS in the cyclin D1 promoter decreased the EGFR-stimulated luciferase activity (lane 2 in Fig. 2C or lane 4 vs. lane 3 in Fig. S5A), a finding consistent with the results of a previous study (17), and also blocked the effect of RHA in stimulating the cyclin D1 promoter activity (lane 3 in Fig. 2C or lane 6 vs. lane 5 in Fig. S5A). Moreover, the ATRS mutation reduced the luciferase activity induced by coexpression of EGFR and RHA (lane 4 in Fig. 2C and lane 8 vs. lane 7 in Fig. S5A). The activity of cyclin D1 promoter with ATRS mutation still was stimulated moderately by EGFR (lane 4 vs. lane 2 in Fig. S5A) but not by RHA (lane 6 vs. lane 2 in Fig. S5A), suggesting that activated traditional downstream signaling pathways of EGFR might contribute to the increased luciferase activity. Similarly, a moderate increase of the ATRS-mutated cyclin D1 promoter activity by the EGFR/RHA complex was observed (compare lane 8 with lane 4 in Fig. S5A), again suggesting that the activated traditional signaling pathways of EGFR and/or a pathway of EGFR/RHA independent of ATRS result in the observed luciferase activity. Taken together, the results suggest that the stimulatory effect of the EGFR/RHA complex on cyclin D1 promoter activity depends mainly on the ATRS of the gene promoter.
We next asked whether the EGFR/RHA complex can bind to the region of the cyclin D1 promoter containing ATRS in vivo. As shown in Fig. 2D, Top, ChIP-PCR analysis indicated that both EGFR (lanes 3 and 4) and RHA (lanes 5 and 6) are associated with the cyclin D1 promoter. This association was enhanced by EGF treatment and was not detected by normal IgG (lanes 1 and 2). Sequential ChIP-PCR analysis revealed that EGFR and RHA associate to bind to the cyclin D1 promoter upon EGF stimulation (Fig. 2D, Middle), suggesting that the EGFR/RHA complex binds to the cyclin D1 promoter in vivo. This notion was supported further by an experiment in which knockdown of RHA expression abolished the binding of EGFR to the cyclin D1 promoter, indicating that RHA is required for EGFR binding to the cyclin D1 promoter in vivo (Fig. 2D, Bottom). A consistent result also was obtained from EMSA (Fig. S5B) in which the association of the ATRS probe and the nuclear extract (lane 4) could be blocked by pretreatment of the nuclear extract with antibodies against EGFR (lane 8), RHA (lane 9), or both (lane 10) but not by IgG (lane 7). However, RHA knockdown reduced but could not abrogate the binding of EGFR to iNOS promoter upon EGF treatment (Fig. 2E), probably because, in addition to RHA, EGFR interacts with STAT3, which binds to STAT3-binding site in iNOS promoter. Thus, recruitment of EGFR to the iNOS gene promoter through STAT3 may occur (13). Taken together, these results suggest that EGFR associates with its target gene promoter through RHA in vivo.
Tyrosine Kinase-Dependent Activation of Cyclin D1 Promoter by EGFR/RHA Complex.
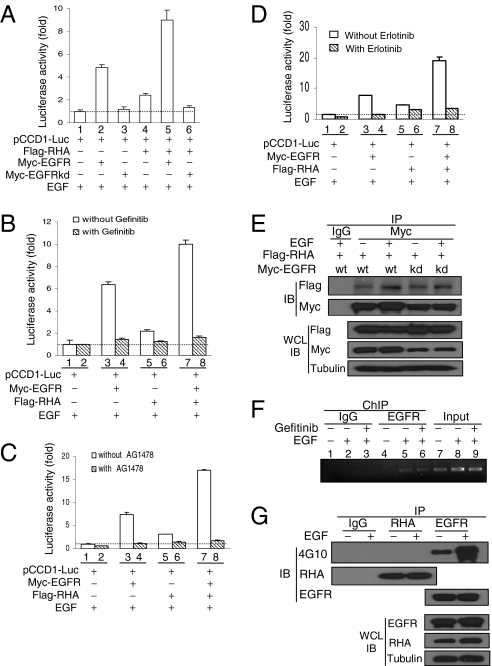
To study whether the stimulatory effect of EGFR/RHA complex on cyclin D1 promoter is dependent on the tyrosine kinase activity of EGFR, an EGFR kinase dead mutant (Myc-EGFRkd) was used for the promoter assay. As shown in Fig. 3A, EGFRkd not only abrogated its stimulatory effect on the cyclin D1 promoter activity (lane 3 vs. lane 2) but also blocked the RHA-induced cyclin D1 promoter activity (lane 6 vs. lane 4). Treatment of the cells with EGFR tyrosine kinase inhibitors (genfitinib, AG1478, or erlotinib) consistently blocked the stimulating effects of both EGFR and RHA on the promoter activity (lanes 4, 6, and 8 vs. lanes 3, 5, and 7 in Fig. 3 B, C, and D, respectively), suggesting that EGFR tyrosine kinase activity is important for EGFR/RHA complex-induced cyclin D1 gene transcription.
Fig. 3.
Requirement of EGFR tyrosine kinase for EGFR/RHA-induced promoter activity. (A) Abrogation of EGFR/RHA-induced cyclin D1 promoter activity by EGFRkd mutant. (B) Abrogation of EGFR/RHA-induced promoter activity by Gefitinib (10 μM) treatment. (C) Abrogation of EGFR/RHA-induced promoter activity by AG1478 (10 μM) treatment. (D) Abrogation of EGFR/RHA-induced promoter activity by Erlotinib (2.5 μM) treatment. (E) EGFR tyrosine kinase-independent interaction between EGFR and RHA. A total cell lysate from HEK293T cells cotransfected with indicated plasmids was immunoprecipitated with an anti-Myc antibody followed by blotting with an anti-Flag antibody. IB, immunoblotting; IP, immunoprecipitation; WCL, whole-cell lysate. (F) Effects of Gefitinib treatment on the binding of EGFR to cyclin D1 promoter. A431 cells treated with EGF without or with Gefitinib were crosslinked, fractionated, and submitted to ChIP-PCR analysis. (G) There was no detectable tyrosine phosphorylation of endogenous RHA upon EGF stimulation. A431 cell lysate was immunoprecipitated with an anti-EGFR or anti-RHA antibody followed by detection with a tyrosine phosphorylation antibody 4G10.
We then asked if the tyrosine kinase activity of EGFR is required for its association with RHA in vivo. The results shown in Fig. 3E indicate that the interaction between EGFR and RHA does not require EGFR tyrosine kinase activity, and this notion was supported by the treatment of MDA-MB-468 cells with EGFR tyrosine kinase inhibitor (Fig. S6A). Moreover, treatment with Gefitinib did not reduce the binding of EGFR to the cyclin D1 promoter in A431 cells (Fig. 3F, lane 6 vs. lane 5). We then investigated whether EGFR can phosphorylate RHA and found no tyrosine phosphorylation of RHA in A431 cells treated with EGF (Fig. 3G). Moreover, using MS, we failed to identify any tyrosine phosphorylation site of RHA, although we did identify two previously reported serine phosphorylation sites, Ser87 and Ser321 (Fig. S7) (29, 30). These results suggest that the major function of RHA in EGFR/RHA-induced cyclin D1 gene transcription is to bring EGFR to the cyclin D1 promoter to form a transcription complex. The requirement of EGFR tyrosine kinase activity for EGFR/RHA-induced cyclin D1 gene transcription and our inability to detect tyrosine phosphorylation of RHA by EGFR suggest that another component or other components in the EGFR/RHA complex may be phosphorylated by EGFR and could play a role in EGFR transcriptional activation in the nucleus.
On the other hand, we investigated whether EGFR/RHA-induced cyclin D1 gene expression is dependent on the ATPase/helicase activity of RHA. The RHA mutant with loss of ATPase/helicase activity (RHAK417R) (31) had effects on cyclin D1 promoter activity similar to those of wild-type RHA (Fig. S8A), suggesting that the ATPase/helicase activity of RHA is not required for RHA to activate cyclin D1 gene transcription. Consistently, loss of ATPase/helicase activity did not change the interaction of RHA with EGFR (Fig. S8B).
Reduction of EGFR/RHA Complex-Induced Promoter Activity by Interrupting EGFR--RHA Association.
To investigate further the effect of the EGFR–RHA interaction on the EGFR-induced gene transcription, we generated several RHA and EGFR mutation constructs to disturb the interaction of EGFR and RHA and to examine the outcome of the constructs on the EGFR-induced cyclin D1 promoter activity. We first constructed several RHA mutants with deletion of RHA at its N-terminal RNA-binding domain (RHA360–1270 and RHA676–1270), central helicase domain (RHAΔ378–585, RHAΔ378–767, and RHAΔ378–946) and C-terminal Arg-Gly-Gly (RGG) box (RHA1–771, RHA1–809, RHA1–920, and RHA1–1076) (Fig. 4A). We found that deletion of the central helicase domain of RHA abrogated its interaction with EGFR (Fig. 4B), but all the RHA mutants with helicase domain deletion were still translocated into the nucleus (Fig. S9 A and B), ruling out the possibility that the RHA mutants’ lack of interaction with EGFR results from a lack of nuclear translocation. We then investigated the effects of the RHA mutants (RHAΔ378–585, RHAΔ378–767, and RHAΔ378–946) on EGFR-induced cyclin D1 promoter activity in HeLa cells. To reduce the background from the endogenous RHA, the recipient HeLa cells were stably transfected with RHA shRNA (targeting 3′-UTR) to knock down the endogenous RHA expression (Fig. 2B). These helicase-domain deletion mutants of RHA, which cannot interact with EGFR, greatly reduced the EGFR-induced activity of the cyclin D1 promoter (Fig. 4C, lanes 16 vs. 14, 18 vs. 14, and 20 vs. 14). Because ATPase/helicase activity of RHA per se did not affect EGFR-induced promoter activity (Fig. S8A), the reduced promoter activity by the RHA mutants probably resulted from the interruption of the RHA–EGFR interaction. In addition, we constructed several EGFR mutants and found that deletion of the EGFR intracellular domain (EGFR1–644; Fig. S9C, lane 3) or mutation of the EGFR nuclear localization signal (NLS) (EGFRmNLS, Fig. 4D, lane 3), previously shown defective to enter the nucleus (32, 33), abrogated its interaction with RHA in vivo. Accordingly, interrupting EGFR–RHA interaction by the EGFR mutants EGFR1–644 and EGFRmNLS significantly reduced EGFR/RHA-induced cyclin D1 promoter activity (lanes 13 and 14 vs. 11 and 12, respectively, in Fig. S9D and lanes 11 and 12 vs. 9 and 10, respectively, in Fig. 4E). These data, taken together, support the notion that the interaction between EGFR and RHA is required for the gene transactivation induced by the EGFR/RHA complex. It is worth noting that EGFR645–1186, which showed strong ability to interact with RHA in vivo (lane 4 in Fig. S9C), did not have the ability to activate cyclin D1 gene promoter either by itself (lanes 7 and 8 vs. 3 and 4, respectively, in Fig. S9D) or in combination with RHA (lanes 15 and 16 vs. 11 and 12, respectively, in Fig. S9D), suggesting that the interaction of EGFR and RHA is not sufficient and the full length of EGFR is required for the EGFR/RHA complex-activated cyclin D1 gene expression.
Fig. 4.
Reducing EGFR-induced cyclin D1 promoter activity by interruption of EGFR–RHA Interaction. (A) Schematic of RHA deletion constructs. +, binding to EGFR; −, no binding to EGFR. RGG, RHA RGG domain. (B) Disturbance of EGFR–RHA interaction by deleting the helicase domain of RHA. HEK293T cells were cotransfected with indicated plasmids followed by cell lysis, immunoprecipitation, and Western blot. Expression levels of whole-cell lysates blotted for RHA (Flag), EGFR, and tubulin are shown. IB, immunoblotting; IP, immunoprecipitation; WCL, whole-cell lysate. (C) Reduction of EGFR-induced cyclin D1 promoter activity by deleting the helicase domain of RHA. Luciferase assay was performed in HeLa cells with stable knockdown of endogenous RHA. (D) Reduction of EGFR–RHA interaction by EGFRmNLS. HEK293T cells were transfected with indicated plasmids and lysed for immunoprecipitation-Western blot. (E) Abrogation of EGFR/RHA-induced cyclin D1 promoter activity by EGFRmNLS.
Positive Correlation of the Nuclear Expression of EGFR, RHA, and Cyclin D1 in Breast Cancer Cells.
To determine the pathologic relevance of the relationship among nuclear EGFR, RHA, and cyclin D1, we analyzed the expression of these proteins in 51 human breast tumor samples. As shown in Fig. 5, the expression level of nuclear EGFR is correlated strongly with the expression level of nuclear RHA (Fig. 5A). Moreover, the expression of both nuclear EGFR and RHA is correlated positively with that of cyclin D1 (Fig. 5 A–C), further supporting our hypothesis that the nuclear EGFR/RHA complex regulates cyclin D1 gene expression in cancer cells.
Fig. 5.
Correlation of the expression of nuclear EGFR, RHA, and cyclin D1 in human breast tumor samples. (A) Relationships among the expression of nuclear EGFR, RHA, and cyclin D1 in human breast tumor samples. High, high level of nuclear staining (>31% for RHA and >21% for cyclin D1); Low&−, low level of nuclear staining (0–30% for RHA and 0–20% for cyclin D1); M+/−, nuclear staining negative but membrane staining either positive or negative for EGFR; N+, nuclear staining positive for EGFR. (B) Relationship between the nuclear expression levels of RHA and cyclin D1 in these tumor samples. (C) Representative human breast tumor samples showing positive correlation among the expression levels of nuclear EGFR (nEGFR), RHA (nRHA), and cyclin D1 (nCCD1). (Magnification: 400×.)
Discussion
In this study, we explored the potential mechanism for EGFR-transactivated gene expression in the nucleus. We found that EGF stimulates the nuclear translocation of EGFR and its interaction with RHA. We then investigated whether the EGFR/RHA complex is associated with the cyclin D1 gene promoter in vivo. Knockdown of RHA expression abrogates EGFR binding to the cyclin D1 gene promoter and therefore reduces cyclin D1 gene expression. The importance of RHA in EGFR-mediated cyclin D1 gene expression is supported further by the study in which interruption of EGFR–RHA interaction significantly reduced EGFR-induced activity of the cyclin D1 promoter. Interestingly, the stimulatory effect of the EGFR/RHA complex on cyclin D1 transcription is dependent on the kinase activity of EGFR but is independent of the helicase activity of RHA. The model described above is not specific to cyclin D1, because RHA also mediates EGFR-induced iNOS promoter activity, suggesting that RHA is a DNA-binding partner for nuclear EGFR in regulating its target gene expression.
Notwithstanding our finding of RHA binding to the ATRS, RHA has been shown to bind specifically to a GC-rich sequence in p16INK4a promoter to regulate its transcription (25). The different sequence-binding specificities of RHA may result from differences in its posttranslational modification or in the context of the transcriptosome containing RHA. Indeed, acetylation of RHA can regulate its binding to the p16INK4a promoter (25). Our data indicate that the tyrosine kinase activity of EGFR is not required for its interaction with RHA but is required for EGFR/RHA-induced cyclin D1 gene transcription. Because we did not observe the tyrosine phosphorylation of RHA in an immunoprecipitation-Western blot or identify any tyrosyl phosphorylation sites by MS, our results suggest that tyrosyl phosphorylation of RHA, if it occurs at all, is very minor in vivo. The requirement of EGFR tyrosine kinase activity for EGFR/RHA-induced target gene expression suggests that other unidentified component(s) in the EGFR/RHA complex may be phosphorylated by EGFR and play a role in EGFR/RHA-induced gene transcription. In line with this notion, the data shown in Fig. S9 C and D indicate that interaction between EGFR and RHA is required but not sufficient to activate the promoter activity, supporting the possibility that other component(s) may be involved in the EGFR/RHA complex. Further studies are needed to identify these components.
It is worth noting that the association between EGFR and RHA was found to be independent of EGFR tyrosine kinase in MDA-MB-468 cells (Fig. S6A), in which EGFR is overexpressed (1.9 × 106/cell) (34). However, the interaction of EGFR and RHA was inhibited by EGFR tyrosine kinase inhibitor in HeLa cells (Fig. S6D, Top), in which the level of EGFR expression is normal (1.4 × 105/cell) (35). The impaired association of EGFR with RHA in HeLa cells upon treatment with tyrosine kinase inhibitor is caused by the absence of EGF/EGFR internalization and EGFR nuclear translocation in the presence of EGFR tyrosine kinase inhibitor (Fig. S6 C and D). In MDA-MB-468 cells, by contrast, treatment with tyrosine kinase inhibitor showed little inhibitory effect on EGFR nuclear translocation (Fig. S6B). Therefore, it is likely that the EGFR–RHA association may be dependent upon the cell type and the level of EGFR expression in the cells. In systems in which EGFR is overexpressed, such as MDA-MB-468 and A431 cells, the internalization of EGFR could occur through a noncoated-pit mechanism (36, 37) and has been demonstrated to be independent of tyrosine kinase (38, 39). Accordingly, we found that EGFR–RNA association is independent of kinase in these two cell types. However, in HeLa cells, which express a normal range of EGFR, the internalization is known to occur mainly through a coated-pit mechanism (37, 40, 41) that can be blocked by the EGFR tyrosine kinase inhibitor, and therefore the EGFR–RHA association becomes kinase dependent.
Experimental Procedures
The detail procedures are described in SI Experimental Procedures. Briefly, we used cellular fractionation, IP/Western blot, and confocal microscopy to determine the association and co-localization of EGFR and RHA in cancer cells, ChIP to investigate the binding of EGFR/RHA at target gene promoter, transient transfection and luciferase reporter assay to test the promoter activity, IHC to study the relationship among nuclear expression of EGFR, RHA and cyclin D1 in 51 human breast tissue samples.
Supplementary Material
Acknowledgments
We thank Dr. Stephanie A. Miller, Katy Hale, and Donald Norwood for editing this manuscript. We thank Dr. Mong-Hong Lee (M.D. Anderson Cancer Center, Houston) for providing pcJun.Luc plasmid. This study was supported in part by Grants R01 109311, P01 099031, and CCSG CA16672 from the National Institutes of Health, grants from the National Breast Cancer Foundation, Inc., the Patel Memorial Breast Cancer Research Fund, and the University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (to M.-C. Hung), Grants DOH99-TD-C-111-005 from Department of Health, Taiwan (to M.-C. Hung and L.-Y.L.), NSC 97-3111-B-039 (to M.-C. Hung, L.-Y.L., and Y.-L.Y.), and DOH98-TD-I-111-TM002, and NHRI-EX98-9603BC from National Health Research Institutes, Taiwan, (to L.-Y.L.), and NSC 97-3111-B-039 (to M.-C. Hung, L.-Y.L., and Y.-L.Y.). The study also was supported by the Lupe C. Garcia Fellowship in Cancer Research (to L.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000743107/-/DCSupplemental.
References
- 1.Linggi B, Carpenter G. ErbB receptors: New insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 2.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: Linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94:184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: An emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 4.Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15(21):6484–6489. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das AK, et al. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 2007;67:5267–5274. doi: 10.1158/0008-5472.CAN-07-0242. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lo HW, et al. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 8.Psyrri A, et al. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino M, et al. Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 10.Edwards J, et al. The role of HER1-HER4 and EGFRvIII in hormone-refractory prostate cancer. Clin Cancer Res. 2006;12:123–130. doi: 10.1158/1078-0432.CCR-05-1445. [DOI] [PubMed] [Google Scholar]
- 11.Xia W, et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang SC, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- 13.Lo HW, et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Hanada N, et al. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- 15.Chen DJ, Nirodi CS. The epidermal growth factor receptor: A role in repair of radiation-induced DNA damage. Clin Cancer Res. 2007;13:6555–6560. doi: 10.1158/1078-0432.CCR-07-1610. [DOI] [PubMed] [Google Scholar]
- 16.Hung LY, et al. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin SY, et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 18.Hu S, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS. The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell. 1991;66:935–947. doi: 10.1016/0092-8674(91)90439-6. [DOI] [PubMed] [Google Scholar]
- 20.Lee CG, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 21.Lee CG, Eki T, Okumura K, da Costa Soares V, Hurwitz J. Molecular analysis of the cDNA and genomic DNA encoding mouse RNA helicase A. Genomics. 1998;47:365–371. doi: 10.1006/geno.1997.5139. [DOI] [PubMed] [Google Scholar]
- 22.Fuller-Pace FV. DExD/H box RNA helicases: Multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Grosse F. Nuclear DNA helicase II unwinds both DNA and RNA. Biochemistry. 1994;33:3906–3912. doi: 10.1021/bi00179a016. [DOI] [PubMed] [Google Scholar]
- 24.Hung ML, Chao P, Chang KY. dsRBM1 and a proline-rich domain of RNA helicase A can form a composite binder to recognize a specific dsDNA. Nucleic Acids Res. 2003;31:5741–5753. doi: 10.1093/nar/gkg759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myöhänen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 26.Lee CG, Reichman TW, Baik T, Mathews MB. MLE functions as a transcriptional regulator of the roX2 gene. J Biol Chem. 2004;279:47740–47745. doi: 10.1074/jbc.M408207200. [DOI] [PubMed] [Google Scholar]
- 27.Clarke N, Arenzana N, Hai T, Minden A, Prywes R. Epidermal growth factor induction of the c-jun promoter by a Rac pathway. Mol Cell Biol. 1998;18:1065–1073. doi: 10.1128/mcb.18.2.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kayahara M, Wang X, Tournier C. Selective regulation of c-jun gene expression by mitogen-activated protein kinases via the 12-O-tetradecanoylphorbol-13-acetate- responsive element and myocyte enhancer factor 2 binding sites. Mol Cell Biol. 2005;25:3784–3792. doi: 10.1128/MCB.25.9.3784-3792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 30.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tetsuka T, et al. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 32.Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- 33.Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. American Journal of Translational Research. 2009;1:249–258. [PMC free article] [PubMed] [Google Scholar]
- 34.Filmus J, Pollak MN, Cailleau R, Buick RN. MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun. 1985;128:898–905. doi: 10.1016/0006-291x(85)90131-7. [DOI] [PubMed] [Google Scholar]
- 35.Oksvold MP, Skarpen E, Lindeman B, Roos N, Huitfeldt HS. Immunocytochemical localization of Shc and activated EGF receptor in early endosomes after EGF stimulation of HeLa cells. J Histochem Cytochem. 2000;48:21–33. doi: 10.1177/002215540004800103. [DOI] [PubMed] [Google Scholar]
- 36.Carpenter G. Nuclear localization and possible functions of receptor tyrosine kinases. Curr Opin Cell Biol. 2003;15:143–148. doi: 10.1016/s0955-0674(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 37.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 38.Bailey KE, et al. Epidermal growth factor receptor inhibition modulates the nuclear localization and cytotoxicity of the Auger electron emitting radiopharmaceutical 111In-DTPA human epidermal growth factor. J Nucl Med. 2007;48:1562–1570. doi: 10.2967/jnumed.107.044073. [DOI] [PubMed] [Google Scholar]
- 39.Sundberg AL, et al. Combined effect of gefitinib (‘Iressa’, ZD1839) and targeted radiotherapy with 211At-EGF. Eur J Nucl Med Mol Imaging. 2003;30:1348–1356. doi: 10.1007/s00259-003-1308-9. [DOI] [PubMed] [Google Scholar]
- 40.Carpenter G. The EGF receptor: A nexus for trafficking and signaling. Bioessays. 2000;22:697–707. doi: 10.1002/1521-1878(200008)22:8<697::AID-BIES3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.