Abstract
Transgenic manipulation of subsets of brain cells is increasingly used for studying behaviors and their underlying neural circuits. In Drosophila, the GAL4–upstream activating sequence (UAS) binary system is powerful for gene manipulation, but GAL4 expression is often too broad for fine mapping of neural circuits. Here, we describe the development of unique molecular genetic tools to restrict GAL4 expression patterns. Building on the GAL4-UAS system, our method adds two components: a collection of enhancer-trap recombinase, Flippase (ET-FLP), transgenic lines that provide inheritable, reproducible, and tissue-specific FLP and an FRT-dependent GAL80 “flip-in” construct that converts FLP expression into tissue-specific repression of GAL4 by GAL80. By including a UAS-encoded fluorescent protein, circuit morphology can be simultaneously marked while the circuit function is assessed using another UAS transgene. In a proof-of-principle analysis, we applied this ET-FLP-induced intersectional GAL80/GAL4 repression (FINGR) method to map the neural circuitry underlying fly wing inflation. The FINGR system is versatile and powerful in combination with the vast collection of GAL4 lines for neural circuit mapping as well as for clonal analysis based on the infusion of the yeast-derived FRT/FLP system of mitotic recombination into Drosophila. The strategies and tactics underlying our FINGR system are also applicable to other genetically amenable organisms in which transgenes including the GAL4, UAS, GAL80, and FLP factors can be applied.
Keywords: behavior, intersection, Flippase mosaicism, GAL4, GAL80
Understanding the neural substrates underlying behavior remains a major goal in neuroscience. One effective approach is altering behavioral outputs in animals through genetic manipulation of neurons or neural circuits. This approach is exemplified by numerous studies carried out in mice, zebrafish, Caenorhabditis elegans, and Drosophila (1). In Drosophila, extensive studies of learning, memory, circadian rhythm, courtship, wing inflation, and climbing have provided valuable insights into the neural substrates of these behaviors (2–8). These fly activities are easily observed, are amenable to genetic perturbations, and can be quantitatively analyzed to correlate manipulations of circuits with changes in behavior.
Manipulating neural structures underlying these behaviors has principally depended on imaginative designs and applications of molecular-genetic entities. The yeast-derived GAL4–upstream activating sequence (UAS) binary transgene expression system laid the foundation for many of these manipulations (reviewed in ref. 9). In this system, GAL4 can be expressed in a tissue-specific manner via either enhancer-traps or gene-specific promoters. Wherever GAL4 is made active, a protein-coding sequence under the control of UAS is also expressed (10, 11) (Fig. 1A). Thousands of tissue-specific GAL4 enhancer-trap lines have been created.
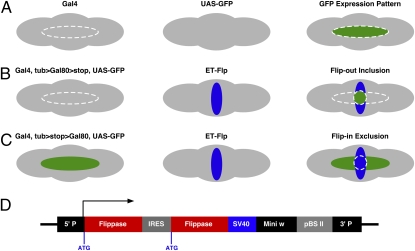
Fig. 1.
Strategies for Flippase-induced GAL4 and GAL80 intersectional methods. (A–C) The expression patterns in the fly brain are illustrated for three genotypes from Left to Right. Right represents the F1 progeny from the cross between Left and Center. (A) A hypothetical expression pattern of a GAL4 is illustrated in the fly brain outlined in the dashed oval (Left). Without GAL4, UAS-GFP is not expressed (Center). In the progeny GFP expression is defined by the expression pattern of the GAL4 (Right, green). (B) In the presence of a tubulin promoter-driven, FRT-flanked GAL80 construct (tubP>GAL80>stop), GAL4 activity is repressed by GAL80 such that GFP is not expressed (Left). An ET-FLP line displays a specific expression pattern (Center, blue). Once the GAL80 is flipped out by the ET-FLP, GAL4 activity and hence GFP expression are restored in the GAL4 and ET-FLP intersected region (Right, circled green). (C) In the reciprocal to the previous configuration, tubulin promoter-driven GAL80 is not expressed due to the inserted FRT-flanked “stop” codon (tubP>stop>GAL80). In the absence of GAL80, GAL4 drives the expression of GFP (Left, green). Once ET-FLP-mediated recombination takes place, GAL80 is flipped in and expressed in Flp-containing cells. Consequently, GAL4 activity is repressed and no GFP is expressed where GAL4 and ET-FLP intersect (Right). (D) An internal ribosome entry site (IRES) is used to enhance expression levels of ET-FLP (hence called ET-FLPx2).
Despite the enormous diversity of available tissue-specific lines, GAL4 expression patterns are rarely restrictive enough to map key elements of neural circuitry. Hence, most brain-behavior studies have used intersectional strategies to focus on subsets of the circuitry of interest (reviewed in ref. 12). Subtractive restriction uses tissue-specific promoter-driven GAL80 to repress GAL4 expression patterns in subcircuits (4, 13). Combinatorial restriction uses a bipartite “split-GAL4” approach to limit the number of GAL4-active cells (7). Random restriction uses patches of expression of the recombinase Flippase (FLP) to catalyze removal of sequences between a tandem pair of FLP recognition target sequences (FRT, which are symbolized “>” in descriptions of upcoming genotypes) (14, 15) (reviewed in ref. 16). A recent tactic used the intersection of the FRT flanked GAL80 (tubP>GAL80>) (17) (Fig. 1B) and heat-shock FLP (hs-FLP) to restrict GAL4 expression and map a motoneuron in proboscis extension response to sweet solutions (17). This recent method complements the mosaic analysis with a repressible cell marker (MARCM) method, which also uses hs-FLP to generate clones in a mitotically restricted manner (18). However, most existing intersectional methods fall short in that promoter-driven GAL80 may fluctuate over time and that FLP mosaics are usually generated in random patterns using hs-FLP.
Here, we describe the development of a transgenic system for dissecting a GAL4-defined neural circuit into subcomponents designed to avoid aforementioned shortcomings. It employs an expression pattern of FLP to restrict overlapping GAL4 activity (Fig. 1C). For this, we developed two resources: (i) a set of 1,000 enhancer-trap (ET)-FLPx2 [constructed as FLP–an internal ribosome entry site (IRES)-FLP] lines, with enhancer-trapping tissue-specific expression of FLP, and (ii) a flip-in GAL80 construct (tubP>stop>GAL80), to convert FLP expression into GAL80-mediated repression of GAL4. Restriction and functional analysis within the GAL4 expression pattern can be created by crossing the ET-FLPx2 collection to a transgenic combination containing at least three transgenes: a flip-in GAL80, a GAL4, and a UAS transgene.
Application of our FLPx2-induced intersectional GAL80/GAL4 repression (FINGR) system enhances neural circuit mapping by pointing to relevant subcircuits underlying a behavior. By contrast to approaches where a heat-shock promoter activates FLP expression (reviewed in ref. 16), here an ET-FLPx2 transgenic line leads to a unique expression pattern of GAL80. Importantly, that pattern can be regenerated in subsequent histological or behavioral tests. As a proof of principle, we applied the FINGR system to mapping the neural circuitry that contains Drosophila’s analog of crustacean cardioactive peptide (CCAP), a circuit containing a superset of cells required for wing inflation (7, 19–23). We also tested our ET-FLPx2 lines with the tubP>GAL80> construct and showed that the flip-out strategy complemented our flip-in system by generating the opposite subset of circuitry. Our findings demonstrate the versatility and a general applicability of this unique intersectional strategy to the dissection of circuits underlying fly behavior.
Results
ET-FLPx2 Directs Heritable Flippase Expression That Is Spatially Unique for a Given Transgenic Line.
A key rate-limiting reagent for FLP-based clonal analysis is the availability of Drosophila transgenic strains in which the yeast-derived FLP is present in a variety of tissues. To overcome this inadequacy and to boost FLP expression levels, we created an enhancer-trap Flippase (ET-FLPx2) construct containing IRES followed by a tandem copy of an FLP-encoding sequence (FLP-IRES-FLP) (24) (Fig. 1D). After generating the initial transgene and then mobilizing it into autosomes (SI Materials and Methods), we mated a series of derived lines to y w, actinP>cd2>gal4; UAS-gfp (25). The latter pair of transgenes acts as a Flippase reporter because GFP is expressed only when Flippase catalyzes the expression of GAL4. In our initial analysis, we focused only on the expression patterns of a subset of ET-FLPx2 lines in the central nervous system (CNS) and particularly those with a selected phenotype (wing inflation, see below).
As one would expect for any collection of enhancer-trapped strains, each ET-FLPx2 line produced a distinct spatial pattern of FLP-induced marker expression. Lines 173A, 276B, 398A, 567A, 820A, 757A, and 1143A display robust (Fig. 2) and repeatable expression patterns (Figs. S1 and S2). At gross anatomical levels, FLPx2 expression in most lines examined was bilaterally symmetric within the nervous system and it generally ranged from widespread to highly restrictive. However, those lines with sparse expression (467A, 57, 420A, and 505A) tended to vary among individuals.
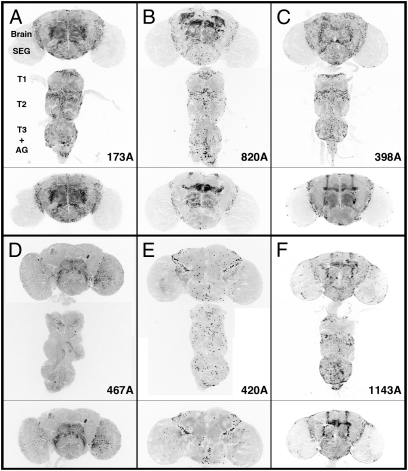
Fig. 2.
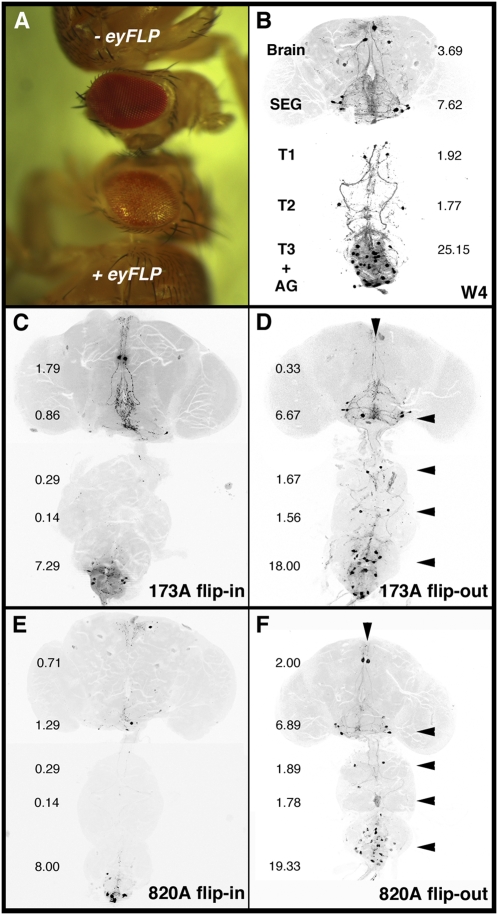
Expression patterns of select ET-FLPx2 lines in the central nervous system of adults. These images depict the CNS (brain and ventral nerve cord, VNC) dissected from those 1- to 5 d-old adult flies that were offspring produced from mating y w, actinP>CD2>GAL4; UAS-GFP parents to a given ET-FLPx2 line. By recombining out the >CD2> cassette, FLP activates actinP>GAL4 expression, which in turn drives expression of UAS-GFP. Hence, the GFP patterns report FLP-expressing cells. A–F show examples of progeny from six different ET-FLPx2 lines. Each Lower CNS image shows the most prominent internal brain regions from the same fly. Note the unique expression patterns for a given ET-FLPx2 line. Six replicates of these and additional ET-FLPx2 expression patterns are shown in Figs. S1 and S2. SEG, subesophageal ganglion. T, thoracic ganglia. AG, abdominal ganglion. The images of the brain and the ventral nerve cord were taken from the same animal, but some in different orientations and then combined.
A close examination of the CNS reveals that lines 173A, 820A, 398A, and 1143A drive FLP patterns in cells within the mushroom bodies. However, each FLP line shows a unique pattern in other brain regions (such as the central complex) and in the ventral nerve cord (VNC). Line 467A shows expression limited to cells in the optic and olfactory lobes of the adult brain and VNC. Further examples of the expression patterns for a small subset of the newly generated lines are shown in replicates in Figs. S1 and S2. These figures indicate a reasonably good reproducibility for patterns of inheritance for most ET-FLPx2 lines. Hence, these transgenic lines can provide a source of inheritable FLP for mosaic analysis.
A GAL80 Repressor System That Functions Both Pre- and Postmitotically to Restrict GAL4 Expression.
To apply the FINGR component of our system, we developed an FLP target transgene consisting of tubP>stop>GAL80, in which an FRT-flanked stop cassette was inserted between a strong constitutive (α-tubulin) promoter and a gene encoding GAL80. In cells expressing FLP, the stop cassette is excised, leading to tubP-promoted production of GAL80. The excision is permanent, resulting in a strong GAL80 repression of any UAS-gene product within a subset of the existing GAL4 pattern (Figs. 1C and 3A).
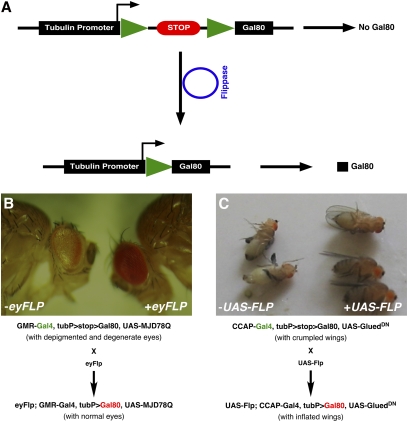
Fig. 3.
Construction and testing of a tubulin promoter-driven FRT-flanked flip-in GAL80 construct. (A) In flies containing the tubP>stop>GAL80 construct, GAL80 is not expressed. GAL80 expression is turned on by removing the stop cassette through flippase-mediated recombination. Green triangles represent FRT sites. (B) The tubP>stop>GAL80 flip-in system functions premitotically to restrict GAL4 expression. In GMR-GAL4, UAS-MJD78Q; tubP>stop>GAL80 flies, photoreceptor cells in adult flies are degenerate and depigmented (left fly). Following the introduction of eyFLP, GAL80 is expressed in photoreceptor cells, resulting in full repression of GAL4 and MJD78Q activity and flies with normal eyes (right fly). Forty-four were observed of 44 expected genotype in F1 flies. (C) The flip-in GAL80 repressor system functions postmitotically to restrict GAL4 expression. In CCAP-GAL4, tubP>stop>GAL80, UAS-GluedDN flies, wings fail to inflate after adults emerge from metamorphosis (Left). In the presence of UAS-FLP, CCAP-GAL4–generated FLP leads to strong GAL80 expression in CCAP cells. These repress CCAP-GAL4 and result in full inflation of the adult wings (Right).
To test the efficacy of the tubP>stop>GAL80 construct, we first examined the ability of eyFLP in inducing GAL80 expression and a consequent repression of eye-specific GMR-GAL4. In this assay, we used eye degeneration in GMR-GAL4, UAS-MJDQ78 flies as a reporter (26, 27). TubP>stop>GAL80 showed no or minimal leaky GAL80 expression because its presence did not affect the level of eye depigmentation in the GMR-GAL4, UAS-MJD78Q flies (Fig. 3B, left). The addition of the premitotic source of eyFLP (28), however, resulted in a full suppression of degeneration in the triple transgenic fly (GMR-GAL4, UAS-MJDQ78; tubP>stop>GAL80) (Fig. 3B, right). The efficiency for this suppression was 100% (44 observed of 44 expected genotype in F1’s). Therefore, FLP-induced recombination within the tubP>stop>GAL80 construct was sufficient to generate robust expression of GAL80 and effectively repress GMR-GAL4.
In a second test, we assessed the ability of tubP>stop>GAL80 to generate mosaics postmitotically. For this, we combined a CCAP-GAL4 transgene (19) with a UAS-GluedDN transgene and examined the effects of these factors on wing expansion. GluedDN causes axonal transport defects of the dynein complex and blocks retrograde signals and secondarily reduces synaptic function (29–31). We found the above combination disrupted wing inflation and resulted in crumpled wings. Further, we observed this crumpled wing phenotype with or without tubP>stop>GAL80 (Fig. 3C, left). Moreover, wing inflation was fully “rescued” when we added a UAS-FLP construct to the triply transgenic combination (which included tubP>stop>GAL80) (Fig. 3C, right). As production of CCAP is a late-stage feature of neuronal differentiation (19), GAL4-driven production of FLP is likely postmitotic. Hence, we conclude that the effect of this recombinase on tubP>stop>GAL80-mediated expression of GAL80 occurred after neuronal cell division ceased.
Application of the FINGR System in Mapping Wing Inflation Circuitry.
Taking advantage of the fly stock with crumpled wings (CCAP-GAL4, UAS-GluedDN, tubP>stop>GAL80), we added UAS-mCD8GFP to help visualize the circuit defined by ET-FLPx2–restricted CCAP-GAL4 expression patterns. The quadruple transgenic combination, hence called W4, was viable and had crumpled wings (Fig. S3). The W4 stock was easily maintained, and individuals within it manifested no gross behavioral or morphological abnormalities aside from noninflated wings. GFP readily marked the circuit defined by CCAP-GAL4 in W4 flies (Fig. 4A).
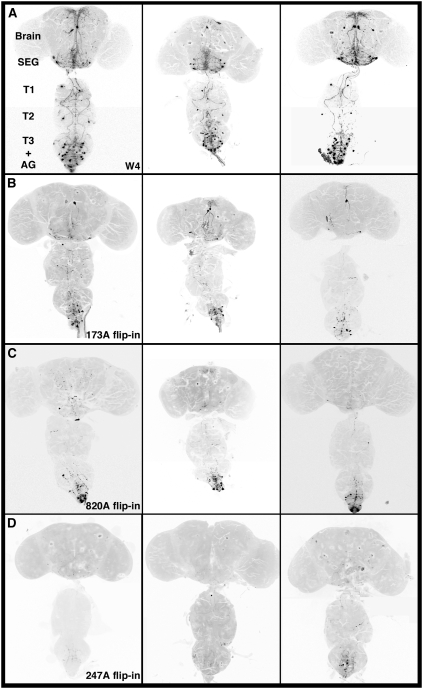
Fig. 4.
Mapping the neural circuits involved in fly wing inflation with tubP>stop>GAL80. These images represent the adult CNS. The fly in A carried the W4 genotype and had crumpled wings (Fig. S3). The flies in B–D are a series of progeny from W4 crossed to each specific ET-FLPx2 line; they all had fully inflated wings. (A) mCD8GFP expression pattern in the W4 parental line containing UAS-mCD8GFP; CCAP-GAL4, tubP>stop>GAL80, and UAS-GluedDN. The expression pattern of the W4 parental line containing UAS-mCD8GFP; CCAP-GAL4, tubP>stop>GAL80 and UAS-GluedDN is shown. Additional replicates to the three shown here can be found in Fig. S4. On average, each W4 fly CNS has 40.15 ± 1.06 (n = 13) GFP-positive CCAP cells. (B–D) Cells retaining GFP and GluedDN expression after GAL80 flip-in for three ET-FLPx2 lines are shown. The FLP-containing enhancer trap in line 173A is broadly expressed in the CNS (Fig. 2A and Fig. S1A). As a result of FLP actions, GAL4 activity in most CCAP neurons was repressed; hence they no longer drove the expression of mCD8GFP and GluedDN (B). These GFP-negative cells are sufficient to mediate wing inflation. There are 10.36 ± 1.29 (n = 14) GFP-positive CCAP neurons visible following 173A flipping GAL80 in. Line 820A also expressed broadly in the CNS (Fig. 2B) and hence showed most overlap with CCAP-GAL4 (C). Only 8.0 ± 0.62 (n = 7) CCAP cells within the posterior VNC (abdominal ganglion) retained mCD8GFP and GluedDN expression. Line 247A is capable of flipping in GAL80 in nearly all CCAP neurons to repress CCAP-GAL4 activity. Although this line is not informative for the wing inflation circuit, it exemplifies the overall versatility of the ET-FLPx2 transgenic types and the FINGR method. The images of the brain and the ventral nerve cord were taken from the same animal, but some in different orientations and then combined.
By combining flies with the W4 transgenes with a series of ET-FLPx2 enhancer traps we screened for trapped lines with relevant overlapping patterns of FLP. Such transgene combinations rescued wing inflation and removed the marking of the relevant neural subcircuit within the brain and the thorax. In particular, these enhancer-trapped FLPs generated sufficient GAL80 repression in a subset of the circuitry to ameliorate the effect of earlier expression of CCAP-GAL4–driven UAS-GluedDN. In the case of a particular ET-FLPx2 line, flies with inflated wings displayed mCD8GFP-marked neuronal subsets that did not include those critical for the behavior. Cells lacking GFP expressed FLP and successfully removed the stop cassette in tubP>stop>GAL80. Hence, the remaining GFP-marked cells in the CCAP pattern are dispensable for wing inflation.
We screened through part of the collection of the ET-FLPx2 lines to identify those that were able to restore wing inflation in the W4 background. From 500 lines we identified 25 lines that inflated the wings. The efficiency or penetrance of these ET-FLPx2 lines in wing inflation ranged from 44 to 100% (Table S1). Despite differences in penetrance, it should be stressed that flies containing this mosaic pattern can be reconstituted as the ET-FLPx2 is inheritable and F1 flies can be rescreened. Selected ET-FLPx2 enhancer traps from this primary screen are shown in Fig. S3.
We next focused on a subset of ET-FLPx2 enhancer traps to examine overlap patterns of GFP-positive CCAP neurons. Fig. 4 shows examples of the reproducibility in three lines with consistent wing inflation (Table S1, Fig. S4). Line 247A caused GAL80 repression of CCAP-GAL4 in nearly all of the CCAP cells, and thus it is least informative for mapping the wing expansion circuit. Nonetheless, it demonstrates the effectiveness as well as the repeatability of the FINGR system in circuit mapping. Two other lines, however, were more selective in repressing a subset of neurons within the CCAP circuit. Notably, on average 10.36 ± 1.29 (n = 14) and 10.43 ± 0.87 (n = 7) cells remain GFP positive in 173A and 820A F1, respectively (Table S2). This result represents ≈74% reduction in GFP-positive cells compared with W4 flies (40.15 ± 1.06, n = 13). In this GAL80 flip-in paradigm, within the population of cells no longer positive for GFP and GluedDN are those sufficient to mediate wing inflation.
We also considered the overlap pattern with the newly constructed tubP>GAL80> line (17), which operates on the opposite principle from our tubP>stop>GAL80 design. In this construction, GAL80 suppresses CCAP-GAL4–expressed GluedDN until ET-FLPx2 relieves repression in those cells expressing Flippase (henceforth this GAL80 construction is referred to as “flip-out”; Fig. 1B). This construction is predicted to generate the opposite pattern from tubP>stop>GAL80 in crosses to an enhancer-trap FLPx2. Results confirming this prediction are shown in Fig. 5. Flies containing GMR-GAL4, UAS-MJD78Q, and tubP>GAL80> had normal eyes, but became depigmented when eyFLP was introduced (Fig. 5A; 51 observed of 51 expected genotype in F1’s). In flies with CCAP-GAL4, UAS-GluedDN, UAS-mCD8GFP, and tubP>GAL80>, the wings were normal. When crossing this flip-out design to lines 173A, 49% (22/45) of the F1 flies failed to inflate their wings following eclosion. For line 820A 64% (39/61) of the F1’s did not inflate their wings.
Fig. 5.
TubP>GAL80> constructs generate an inverse pattern of suppression compared with the tubP>stop>GAL80 construct. (A) Flies with GMR-GAL4, UAS-MJD78Q; tubP>GAL80> constructs have normal compound eyes (top fly). By catalyzing the removal of GAL80 repression in the eyes using eyFLP, these flies now have degenerating eyes and show depigmentation (bottom fly, 51 observed of 51 expected in F1’s). Note that this is the reciprocal of the one found in Fig. 3B. (B–F) Cells in the adult fly CNS expressing mCD8GFP and GluedDN within the CCAP neurons in W4 (B, which has crumpled wings), W4/173A (C, which has fully inflated wings), and W4/820A (E, which has fully inflated wings) flies. These images are similar to those shown in Fig. 4; they are presented here to contrast the patterns revealed in flies whose GAL80 has been flipped out (D and F). These flip-out flies failed to inflate wings. The flip-in images show that cells within the CCAP circuit negative for GFP and GluedDN are sufficient to inflate the wings. Flipping GAL80 out in the CCAP circuit reveals cells expressing GFP and GluedDN necessary for wing inflation. The average number of cells positive for GFP in the brain, SEG, T1 and T2, and T3 + AG is shown next to the CNS area. Arrowheads point to the CNS areas that are inverse to areas in the flip-in flies. The images of the brain and the ventral nerve cord were taken from the same animal, but some in different orientations and then combined.
Examination of the CNS of the F1 flies failing to inflate their wings revealed that after flipping GAL80 out, the number of cells that express GFP was reduced to 28.22 ± 1.45 (n = 9) and 31.89 ± 1.32 (n = 9) by lines 173A and 820A, respectively (Fig. 5 D and F, Fig. S5, and Table S2). It should be noted that although these two lines show similar numbers of GFP-positive cells, their patterns differ. This difference is difficult to discern in the third thoracic ganglion and abdominal ganglion, but it is apparent in the brain, the subesophageal ganglion, and the first and second thoracic ganglia (compare Fig. 5D and 5F).
Furthermore, comparison of patterns emerging from both tubP>stop>GAL80 and tubP>GAL80> paradigms allows us to distinguish those cells necessary and sufficient for wing inflation. For example, 173A flip-in inflated wings and their CNS consistently had cells remaining GFP positive in the midline of the brain and in the terminal abdominal ganglion whereas the subesophageal ganglion and thoracic ganglia lacked expression (Fig. 5C and Table S2). By contrast, in 173A flip-out, flies had crumpled wings and their CNS did not express in the midline of the brain but did express in the subesophageal ganglion and in the thoracic ganglia (Fig. 5D). This pattern both for flip-in and flip-out is similar for line 820A. GFP-positive cells remained near the posterior tip of the abdominal ganglion and in one pair of cells of the subesophageal ganglion (Fig. 5E). Cells in these regions expressed GFP in 820A flip-out flies. These flip-out patterns are highly reproducible among individuals (Fig. S5), demonstrating the necessity of these cells for wing inflation. Importantly, images from tubP>GAL80> flip-out are the inverse of those produced by the tubP>stop>GAL80 flip-in method (Fig. 5C vs. 5D and 5E vs. 5F). The preliminary patterns of cells mapped through our FINGR method support earlier findings with cervical ligation (32) and split-GAL4 (21–23) that cells in the subesophageal ganglion are required for wing inflation and tanning. Hence, our newly produced toolkit of enhancer traps succeeded in the production of FLP recombinase, and thus of GAL80, in a variety of patterns that were useful to cut across the “background” pattern of GAL4 production.
Discussion
In this study we developed, tested, and used a unique set of molecular genetic tools to restrict the number of active neurons within two circuits in the fly CNS. These results demonstrate that the FINGR system should be fully compatible with the rich collection of various GAL4 lines already available in the fly community. It offers additional versatility, allowing GAL80 repression of GAL4 to be mediated by tissue-specific FLP. Hence, the FINGR system adds a powerful tool to the arsenal at the disposal of researchers for mapping neural circuits and for general clonal analysis in Drosophila. The principles and methods developed here should also be applicable to other genetic organisms in which GAL4/UAS, GAL80, and FRT/FLP are used.
One key innovation is the generation of tissue-specific ET-FLPx2 transgenic flies. As an improvement from a previous single-copy FLP (ET-FLP), doubling of the FLP dosage with the help of IRES results in a higher percentage of ET-FLPx2 lines that show unique and symmetrical expression patterns when examined from animal to animal. Although not ideal for all, these ET-Flpx2 lines can generate an inheritable phenotype with high penetrance. Consequently, the efficiency of ET-FLPx2 in inflating fly wings is significantly higher compared with the ET-FLP lines. Five percent (25/500) of the ET-FLPx2 lines screened turned out to be positive for wing inflation with a penetrance ranging from 44 to 100%. In contrast, only 0.1% (1/1,000) first-generation ET-FLP lines resulted in inflating the wings. Low expression and its resulting stochastic ability to mediate recombination will not be fully avoided by using the ET-FLPx2 construct as endogenous promoters of various strengths determine FLP levels. This is indeed the case for some of our lines. Nonetheless, we believe that these enhanced ET-FLPx2 lines will be valuable and effective for neural circuit mapping.
As a general reagent, our ET-FLPx2 lines can also be useful to Drosophila researchers in areas other than neurobiology. Indeed, preliminary mapping of the ET-FLPx2 lines reveals that FLP is expressed in a variety of nonneuronal tissues, including muscles, intestines, autophagic cells, trachea, and glia. Hence, the ET-FLPx2 lines can be used in replacement of hs-FLP to generate cell-specific clones in conjunction with FRT-marked chromosomes already in use. As a resource of FLP, ET-FLPx2 should also be compatible with the powerful MARCM method (18). With further expansion of the ET-FLPx2 collection and characterization it is possible to facilitate various clonal analyses routinely conducted to address the biology of Drosophila.
Along with the recent work of Gordon and Scott (17) and Shang et al. (33), we have aided in establishing a full set of genetic GAL80-converting tools, tubP>stop>GAL80 (flip-in) and its Venn opposite tubP>GAL80> (flip-out), for flipping GAL80 in or out in FLP-expressing cells, respectively. Both of these approaches can be readily used with the existing GAL4 and UAS lines. These intersectional approaches could also be combined with promoter-driven GAL80 lines for further circuit restriction or with temporally conditional GAL80 and GAL4 (33–36). In addition, both constructs use a strong constitutive tubulin promoter (tubP) and thus avoid the problem of potential variations in GAL80 levels resulting from promoter fluctuation at different developmental stages. Once FLP-mediated recombination takes place, GAL80 is constitutively turned on or off. Finally, tubP>stop>GAL80 and tubP>GAL80> are complementary to each other, allowing one to define the role of two Venn opposites within a known circuitry for mediating a specific behavior, and beyond what we showed for eye development and wing inflation (Fig. 5). In a GAL4-defined circuit, FLP could inactivate (or activate) two nonoverlapping subsets of the circuit with the use of these oppositely designed GAL80 switches to reveal the role of each subcircuit in a behavior.
However, there are differences between the tubP>stop>GAL80 and tubP>GAL80> systems. The tubP>GAL80> construct may be more sluggish to respond as GAL80 expressed before FLP mediates recombination may show perdurance—lasting after the GAL80 cassette has been excised from tubP>GAL80>. We speculate that this difference may account for the lower percentage of flies that failed to have crumpled wings when 173A and 820A were used to flip GAL80 out in UAS-GluedDN, CCAP-GAL4, UAS-mCD8GFP, and tubP>GAL80> flies than in the opposite design. Hence, tubP>stop>GAL80 may be more responsive to reporting the activity of FLP.
The tubP>stop>GAL80 system, however, has a potential drawback in that the UAS gene is constitutively expressed until FRT recombination takes place. This drawback could present a problem for some GAL4/UAS combinations. UAS transgenes such as UAS-Kir2.1 or UAS-TNT that cause lethality or severe developmental defects with a GAL4 may not be suitable for mapping some circuits with tubP>stop>GAL80. Such a weakness could be overcome by using a temperature-sensitive UAS effector such as UAS-Shits.
Applications of the FINGR system include a number of “necessary” and “sufficient” experiments. In cell-specific rescue of a mutant phenotype, one may test whether the flip-in cells are necessary for the rescue with a UAS wild-type gene. Simultaneously, this test determines whether the remaining cells in the circuit are sufficient to rescue the mutant phenotype. In experiments involving the silencing of neuronal excitability with UAS-Kir2.1 or blocking synaptic function with UAS-Shits, it is possible to test whether flip-in cells with tubP>stop>GAL80 (Fig. 1C) are sufficient for restoring a behavior. A further confirmation of the necessity of these same cells can be conducted with the use of tubP>GAL80>flip-out tests (Fig. 1B). In contrast, with effectors that enhance neuronal excitability, tubP>stop>GAL80 can be used to demonstrate whether flip-in cells are necessary to alter a specific behavior. Then, tubP>GAL80> would examine the sufficiency of the same cells in the same behavior. Finally, different FLP lines can be combined and used with these GAL80-flipping systems to further restrict the number of cells in a GAL4 circuit.
Materials and Methods
Strains of D. melanogaster and Their Basic Applications.
Maintenance of stocks and genetic crosses were carried out on cornmeal, molasses, yeast, and agar medium at ambient temperatures. Flies to be tested for wing inflation were maintained at 25–27 °C and 70% relative humidity.
Existing stocks of the following types were used: ccap-gal4 (19), UAS-GluedDN (29), UAS-mcd8gfp (18), yw, actinP>cd2>gal4; UAS-gfp (25), GMR-gal4 (26), and UAS-mjd78Q (27). The following stocks supplied by Bloomington Drosophila Stock Center were also used: UAS-flp (37) and w, Dr/ In(3LR)TMS,P(Δ2–3)99B (38).
Generation of Transgene Constructs.
To construct enhancer-trap Flippase containing FLP-IRES-FLP (ET-FLPx2), an FLP-encoding DNA was generated by PCR from eyFLP-containing flies (28) and cloned into the Topo Blunt vector (Invitrogen), containing a Bsp120I site at the 5′ end and a NotI site at the 3′ end. The fragment was used to replace the NotI-flanked GAL4 in pGawB (11); cloning into pGawB destroyed the 5′ NotI site but retained the 3′ site. Into this NotI site, a derivative of the initial FLP fragment, containing a NotI-flanked IRES-flippase-SV40 terminator fragment, was inserted. This NotI fragment was generated by introducing the IRES sequence from the Ubx region from within a preexisting construct called GFP-IRES-GFP (39) and the SV40 terminator from tubGAL80SV40 (18) as bookends.
The tubP>stop>GAL80 construct was made by cloning a NotI fragment containing FRTstopFRT from UAS>stop>wg into the NotI site of the tubGAL80SV40 vector (18).
Assay for ET-FLPx2-Mediated Wing Expansion.
A strain carrying UAS-mcd8gfp (which encodes a membrane-bound form of GFP) on the X and the second chromosome carrying three inserted transgenics, tubP>stop>gal80, ccap-gal4, and UAS-GluedDN, was generated and hereafter called W4. W4 flies had crumpled wings and were maintained as a stock. W4 females were mated in series to males taken from the ET-FLPx2 strain stocks. F1 flies were scored for fully expanded wings. The central nervous system of these F1 flies was analyzed histologically (see below) or by observing green fluorescence. In the complementary approach of testing the effectiveness of FLP with tubP>gal80>, we crossed homozygous flies carrying UAS-GluedDN; tubP>gal80> to the following second, third combination maintained over a compound balancer T(2,3) In(2LR)SM6b-In(3LR)TM6B: 173A (or 820A);CCAP-GAL4, UAS-mCD8GFP flies. Offspring were scored for those that lacked curly and tubby and failed to inflate their wings.
Supplementary Material
Acknowledgments
We thank Doug Allan (University of British Columbia, Vancouver), Rich Binari (Harvard Medical School, Boston), Marc Halfon (University of Buffalo, Buffalo, NY), Norbert Perrimon (Harvard Medical School), Francesca Pignoni (Upstate Medical University, Syracuse, NY), David Sinclair (Harvard Medical School), Kristin Scott (University of California, Berkeley, CA), Gary Struhl (Columbia University, New York), and Ben White (National Institute of Mental Health, Bethesda) for flies and reagents; Greg Books, Kyomi Cho, Lawrence Chu, Ysabel Milton, Molly Sichio, and Ian Tran (Brandeis University) for initial contributions; Luke Carter, Dennis Chang, Randy Hewes, Mac Hooks, John Tauber, Phillip Vanlandingham, and Ben White for discussions or comments on the manuscript; and Luke Carter for helping with brain imaging. We especially thank Jeffrey Hall for his guidance and insightful suggestions made on the manuscript. An internal fund (to B.Z.) from the University of Oklahoma and a grant from the National Science Foundation (IOS-1025556, to B.Z. and R.A.B.) supported R.A.B. and this research. National Institutes of Health Grant R01NS060878 (to B.Z.) partially supported R.A.B.; and R.A.B. acknowledges a National Institutes of Health National Research Service Award (T32 NS07292) training grant awarded to Brandeis University and a separate National Institutes of Health Grant R01 GM21473 awarded to Jeffrey Hall at Brandeis, which provided support for the early stages of this study.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004669107/-/DCSupplemental.
References
- 1.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 3.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 4.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 5.Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- 6.Manoli DS, et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 7.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez VG, et al. Age-related changes in climbing behavior and neural circuit physiology in Drosophila. Dev Neurobiol. 2007;67:778–791. doi: 10.1002/dneu.20388. [DOI] [PubMed] [Google Scholar]
- 9.Duffy JB. GAL4 system in Drosophila: A fly geneticist's Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 10.Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.White BH, Peabody NC. Neurotrapping: Cellular screens to identify the neural substrates of behavior in Drosophila. Front Mol Neurosci. 2009;2 doi: 10.3389/neuro.02.020.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villella A, Ferri SL, Krystal JD, Hall JC. Functional analysis of fruitless gene expression by transgenic manipulations of Drosophila courtship. Proc Natl Acad Sci USA. 2005;102:16550–16557. doi: 10.1073/pnas.0507056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- 15.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 16.Blair SS. mosaic techniques for studying Drosophila development. Development. 2003;130:5065–5072. doi: 10.1242/dev.00774. Genetic. [DOI] [PubMed] [Google Scholar]
- 17.Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–384. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 19.Park JH, Schroeder AJ, Helfrich-Förster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- 20.Honegger HW, Dewey EM, Ewer J. Bursicon, the tanning hormone of insects: Recent advances following the discovery of its molecular identity. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2008;194:989–1005. doi: 10.1007/s00359-008-0386-3. [DOI] [PubMed] [Google Scholar]
- 21.Luan H, et al. Functional dissection of a neuronal network required for cuticle tanning and wing expansion in Drosophila. J Neurosci. 2006;26:573–584. doi: 10.1523/JNEUROSCI.3916-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peabody NC, et al. Characterization of the decision network for wing expansion in Drosophila using targeted expression of the TRPM8 channel. J Neurosci. 2009;29:3343–3353. doi: 10.1523/JNEUROSCI.4241-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peabody NC, et al. Bursicon functions within the Drosophila CNS to modulate wing expansion behavior, hormone secretion, and cell death. J Neurosci. 2008;28:14379–14391. doi: 10.1523/JNEUROSCI.2842-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart K, Bienz M. A test for cell autonomy, based on di-cistronic messenger translation. Development. 1996;122:747–751. doi: 10.1242/dev.122.3.747. [DOI] [PubMed] [Google Scholar]
- 25.Pignoni F, Zipursky SL. Induction of Drosophila eye development by decapentaplegic. Development. 1997;124:271–278. doi: 10.1242/dev.124.2.271. [DOI] [PubMed] [Google Scholar]
- 26.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 27.Warrick JM, et al. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 28.Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- 29.Allen MJ, et al. Targeted expression of truncated glued disrupts giant fiber synapse formation in Drosophila. J Neurosci. 1999;19:9374–9384. doi: 10.1523/JNEUROSCI.19-21-09374.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eaton BA, Fetter RD, Davis GW. Dynactin is necessary for synapse stabilization. Neuron. 2002;34:729–741. doi: 10.1016/s0896-6273(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 31.Eade KT, Allan DW. Neuronal phenotype in the mature nervous system is maintained by persistent retrograde bone morphogenetic protein signaling. J Neurosci. 2009;29:3852–3864. doi: 10.1523/JNEUROSCI.0213-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura KI, Truman JW. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J Neurosci. 1990;10:403–411. doi: 10.1523/JNEUROSCI.10-02-00403.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci USA. 2008;105:19587–19594. doi: 10.1073/pnas.0809577105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 36.McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: A synthesis of time and space. Trends Genet. 2004;20:384–391. doi: 10.1016/j.tig.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Duffy JB, Harrison DA, Perrimon N. Identifying loci required for follicular patterning using directed mosaics. Development. 1998;125:2263–2271. doi: 10.1242/dev.125.12.2263. [DOI] [PubMed] [Google Scholar]
- 38.Robertson HM, et al. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halfon MS, et al. New fluorescent protein reporters for use with the Drosophila Gal4 expression system and for vital detection of balancer chromosomes. Genesis. 2002;34:135–138. doi: 10.1002/gene.10136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.