Abstract
Segregation of the future germ line defines a crucial cell fate decision during animal development. In Xenopus, germ cells are specified by inheritance of vegetally localized maternal determinants, including a group of specific mRNAs. Here, we show that the vegetal localization elements (LE) of Xenopus Dead end (XDE) and of several other germ-line-specific, vegetally localized transcripts mediate germ cell-specific stabilization and somatic clearance of microinjected reporter mRNA in Xenopus embryos. The part of XDE-LE critical for somatic RNA clearance exhibits homology to zebrafish nanos1 and appears to be targeted by Xenopus miR-18 for somatic mRNA clearance. Xenopus Elr-type proteins of the vegetal localization complex can alleviate somatic RNA clearance of microinjected XDE-LE and endogenous XDE mRNA. ElrB1 synergizes with Xenopus Dead end protein in the stabilization of XDE-LE mRNA. Taken together, our findings unveil a functional link of vegetal mRNA localization and the protection of germ-line mRNAs from somatic clearance.
Keywords: germ cell, microRNA, RNA localization, vegetal, development
The Xenopus germ line is established during oogenesis by formation of a specialized region within the vegetal cytoplasm referred to as the germ plasm (1, 2). Maternal determinants deposited in the germ plasm include specific mRNAs; vegetal RNA localization is achieved via two major pathways named according to their temporal activity during oogenesis as the early and the late transport pathway, respectively. Vegetal transport is driven by cis-acting elements termed localization elements (LEs), mostly residing within the 3′UTR and recognized by specific trans-acting proteins (3, 4). Several mRNAs localized by the early pathway, e.g., Xdazl and Xpat, remain associated with the germ line throughout development and exert important functions during germ cell development (2, 5–7). Xenopus Dead end (XDE) was identified as a late localizing mRNA encoding an RNA binding protein critical for the maintenance of primordial germ cells (PGCs) (8). Despite its late RNA sorting characteristics in the oocyte, XDE mRNA becomes selectively restricted to the germ plasm and thus specifically expressed in the PGCs during embryogenesis. In zebrafish and mice, Dead end (Dnd1) proteins are as well required for germ cell survival (9, 10).
The microRNA-430 family had previously been implicated in the clearance of multiple maternal mRNAs during the phase of maternal-to-zygotic transition of gene expression in early zebrafish development (11). In germ cells, however, certain germ-line-specific mRNAs, such as nanos-1, appear to be protected from miR-430-mediated RNA clearance by an unknown mechanism (12). It was proposed that the Dnd1 protein might mediate the protection of germ-line mRNAs from miR-mediated RNA clearance (13).
On the basis of primary sequence conservation, it has been proposed that Xenopus miR-427 is the ortholog of zebrafish miR-430 (14, 15). Though it was shown that Xenopus miR-427 is indeed involved in the deadenylation of selected maternal mRNAs in early Xenopus embryogenesis, thereby down-regulating their expression (14), it was also revealed that Xenopus miR-427 promotes mesendoderm formation via specifically targeting elements of the nodal signaling pathway (15). It therefore appears that a given miRNA species may have gained diverse activities in different species. In further support of the same notion, it was shown that somatic clearance of maternal mRNAs in Drosophila at the onset of zygotic gene expression is mediated by yet a different species of microRNAs (16).
Here, we report that the XDE mRNA is subject to miR-18–mediated RNA clearance in the soma and specifically protected from degradation in the germ line of Xenopus embryos. The critical regulatory sequence element for this phenomenon is identical to its vegetal LE located within the 3′UTR. Likewise, germ-line restriction activity is exhibited by LEs of several other vegetally localized, germ cell-associated transcripts. In addition, we provide evidence implying that Elr/HuR proteins and thus components of the vegetal RNA transport machinery function in the suppression of miRNA-mediated mRNA clearance. Supporting the idea of a germ cell-specific protective activity of Elr proteins, we find ElrB1 to synergize with Xenopus Dead end protein in mRNA stabilization. In essence, our findings suggest that Elr-type proteins, which have previously been found to be involved in vegetal mRNA transport during oogenesis, can also mediate a different process—namely, protection from miRNA-mediated RNA clearance by binding to one and the same RNA target sequence.
Results
Vegetal Localization Element of Xenopus Dead end Mediates Germ Cell-Specific Protection from Reporter RNA Clearance.
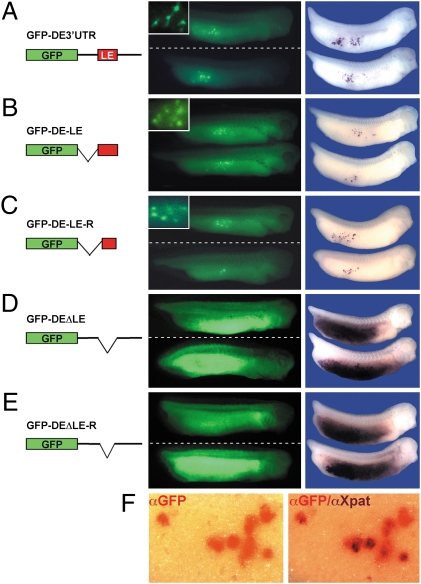
To address the temporal regulation of XDE expression during development, we designed a reporter construct composed of the GFP ORF fused to the XDE 3′UTR. Two-cell-stage Xenopus embryos were injected with synthetic GFP-XDE 3′UTR mRNA into the vegetal side of both blastomeres and monitored for GFP expression at tailbud stage, at which PGCs become detectable at the surface of the embryo. Resembling the endogenous expression pattern of XDE, reporter mRNA-injected embryos exhibited specific GFP expression in the presumptive PGCs (Fig. 1A). To determine if this effect represented a consequence of local translational activation in the germ cells and/or repression elsewhere, we analyzed the distribution of the injected mRNA by GFP whole-mount in situ hybridization. Remarkably, GFP reporter RNA was selectively detected in putative PGCs but absent from somatic cells (Fig. 1A), indicating efficient somatic RNA clearance and/or RNA stabilization in the germ line.
Fig. 1.
The vegetal localization element of XDE mediates germ-line GFP expression by somatic mRNA clearance and PGC-specific RNA accumulation. (A–E) Two-cell-stage embryos were injected either vegetally with reporter mRNAs, cultivated to stage 30–34 and monitored for GFP protein expression (Left), or fixed and analyzed for GFP reporter RNA distribution (Right). PGC-specific GFP protein expression (magnified views in insets) was observed upon injection of 1.2–1.8 ng GFP-XDE 3′UTR (A, 69%, n = 52, N = 2), 1.2 ng GFP-XDE-LE (B, 73%, n = 74, N = 2), and 1.1 ng GFP-XDE-LE-R (C, 71%, n = 28, N = 1). In contrast, ubiquitous GFP protein expression was exhibited by 100% of embryos injected with 1.5 ng GFP-XDEΔLE (D, n = 28, N = 1) or 93% of embryos injected with 1.6 ng GFP-XDEΔLE-R (E, n = 14, N = 1). GFP RNA was specifically detected in PGCs in 72% (A, n = 44, N = 1) for XDE 3′UTR-injected embryos, in 78% (B, n = 82, N = 2) for XDE-LE-injected embryos and 82% (C, n = 74, N = 1) for XDE-LE-R-injected embryos. (D and E) High endodermal levels of GFP RNA were detected in embryos injected with XDEΔLE (D, 97%, n = 113, N = 3) and XDEΔLE-R (E, 100%, n = 69, N = 3). (F) Aspect of a single embryo injected with GFP-XDE-LE and double-stained for GFP (red) and the PGC marker Xpat (purple), photographed after the first (Left) and the second (Right) staining reaction. Although the time for the Xpat staining reaction was reduced, the majority of mGFP5-positive cells also exhibit Xpat signals. n, no. of injected embryos; N, no. of experiments.
To identify the part of the XDE 3′UTR mediating germ-line restriction, we generated a series of truncated XDE 3′UTR fragments, which were fused to the GFP coding sequence (Fig. 1). Interestingly, we found that the 251-nucleotide mRNA segment responsible for germ cell-specific GFP expression and somatic reporter RNA clearance corresponded to the region of the XDE 3′UTR we had previously determined as sufficient to drive vegetal RNA localization in Xenopus oocytes (Fig. 1B) (8). In addition, a 173-nucleotide fragment [LE redefined (LE-R)] central to the previously defined LE also proved to be sufficient for vegetal RNA localization and to specifically restrict GFP reporter mRNA distribution and GFP expression to putative PGCs (Fig. 1C). Upon double whole-mount in situ hybridization, GFP-mRNA positive cells were found to colocalize with the established PGC marker Xpat (6), confirming their identity as PGCs (Fig. 1F).
To determine, if the region of the XDE 3′UTR with vegetal localization activity would also be required for germ cell-specific reporter RNA distribution, we internally deleted the XDE localization element and its redefined form, LE-R, from the full-length XDE 3′UTR GFP reporter. RNAs injected into the vegetal hemisphere of fertilized eggs will be enriched in the endoderm at later stages of development, because the endodermal germ layer forms from the vegetal hemisphere; as to be expected, microinjection of ΔLE as well as ΔLE-R reporter RNAs results in ubiquitous endodermal localization (Fig. 1 D and E). Thus, it is the soma-specific destabilizing activity of the LE that is responsible for the PGC-specific localization of the corresponding microinjected RNA reporter constructs in vivo. Supporting the idea that the absence of reporter RNA in somatic cells involves its clearance, as opposed to its mere spatial redistribution, injection of GFP XDE-LE reporter RNA into the animal pole of Xenopus embryos resulted in the loss of traceable GFP transcripts in the whole embryo (Fig. S1). Thus, protection of the GFP XDE-LE reporter RNA from mRNA clearance might rely on vegetally localized, germ plasm-specific factors.
In summary, these data show that apart from its function in vegetal RNA transport, the XDE-LE is sufficient and required for mRNA clearance in somatic cells and its stabilization in PGCs.
Localization Elements of Other Germ Line-Specific, Vegetally Localizing Transcripts Equally Mediate Somatic Reporter mRNA Destabilization.
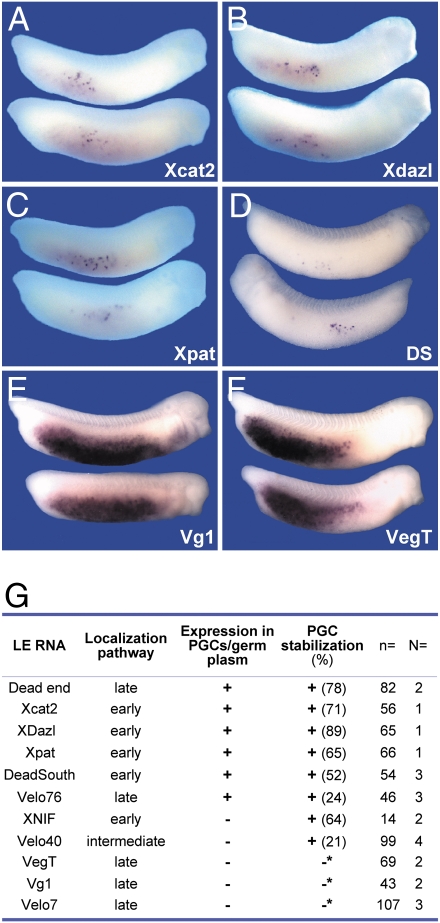
The unexpected interdigitation of vegetal localization and somatic destabilization activities exhibited by the XDE-LE suggests a mechanistic link between both processes. We thus asked if the capacity to mediate somatic destabilization would be a more general property inferred by vegetal localization elements. For this purpose, a set of vegetal localization elements (Fig. 2G) was analyzed by embryonic injection. Not all LEs analyzed appeared capable of targeting lacZ or GFP reporter RNAs to the germ line, notably those derived from the late-localizing, well-studied transcripts Vg1 and VegT (17, 18), which were detected ubiquitously in the endoderm (Fig. 2 E and F). Nevertheless, other LEs exhibited similar characteristics to those observed for the XDE-LE, such as the LEs of Xdazl (5), Xcat2 (19), Xpat (20), and an isoform of XDeadSouth (Fig. 2 A–D); the corresponding endogenous transcripts exhibit germ-line-specific expression. Because both early and late localizing transcripts are represented among those LEs with somatic destabilization activity (Fig. 2G), this activity is not coupled to one particular localization pathway.
Fig. 2.
Somatic RNA clearance is mediated by LEs of other vegetally localized mRNAs expressed in the germ line. (A–F) GFP-tagged LE reporter mRNAs of Xcat2 (A, 800 pg), XDazl (B, 800 pg), Xpat (C, 800 pg), VegT (1.2 ng), Vg1 (1.2 ng), Dead end (XDE-LE, 1.2 ng), or lacZ-tagged LE mRNAs (2 ng each) of the DeadSouth isoform (D) Velo76, XNIF, Velo 40, and Velo7 were injected vegetally into both blastomeres of two-cell-stage embryos. Embryos (stage 30–32) were analyzed for reporter RNA distribution by in situ hybridization using antisense GFP or lacZ probes. (G) LEs of the majority of vegetally localized mRNAs expressed in the germ line were found to mediate germ-line restriction of reporter RNAs irrespective of their transport pathway. Values for PGC stabilization indicate the percentage of embryos exhibiting PGC-specific reporter RNA staining (Fig. S5). Asterisk indicates that for Vg1 (E), VegT (F), and Velo7, PGC staining was detectable in <10% of injected embryos and accompanied by high somatic levels or reporter RNA (100%, 98.6%, and 46.7% of injected embryos, respectively).
Conserved RNA Element Is Critical for the Somatic Destabilization Activity of the XDE-LE.
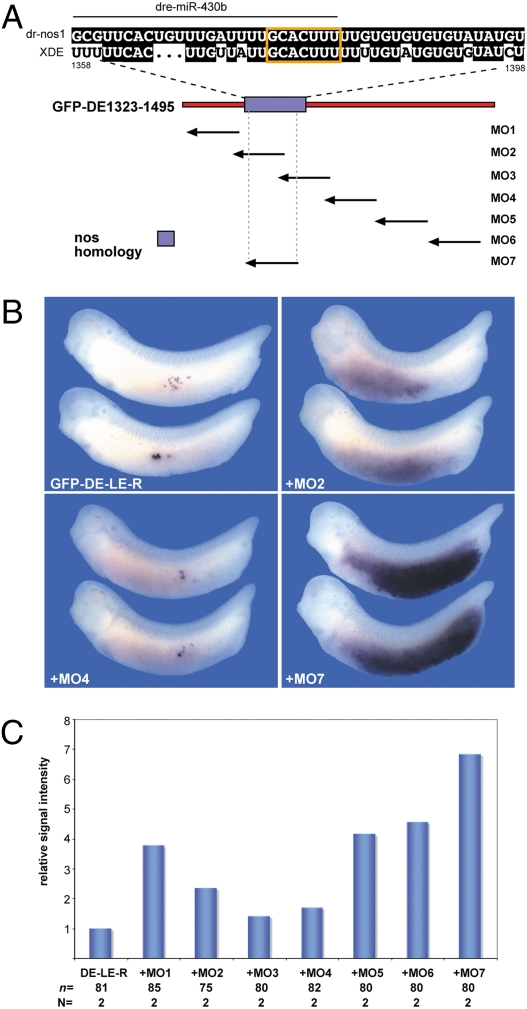
In zebrafish, somatic destabilization of zf-nos1 involves miR-430–mediated deadenylation (12). Interestingly, the zf-nos1 3′UTR and XDE-LE share significant sequence identity in a ∼36-nt stretch of the XDE-LE and the portion of the zebrafish nos1 3′UTR defined as critical for miR-430–mediated deadenylation (12), including a perfect match with the miR-430 target motif (Fig. 3A).
Fig. 3.
Antisense morpholino oligonucleotide-mediated protection of the nanos1 homology element strongly inhibits somatic clearance of XDE-LE. (A) Partial alignment of XDE-LE-R with the zebrafish nanos1 3′UTR reveals a homologous sequence element (XDE nucleotide positions indicated) covering the dre-miR-430b target site (orange box). Identical nucleotides of XDE-LE-R are highlighted in black. The XDE-LE-R is represented by a red rectangle; the position of the nos1 homology domain is marked by a blue box. (B and C) Vegetal coinjection of antisense MOs (1.5 pmol each) with GFP-XDE-LE-R reporter mRNA (1.1 ng) led to inhibition of somatic reporter RNA clearance to variable extent; the strongest effect was achieved by MO7 targeting the nos1 homology region (Table S1). (B) Lateral view of representative embryos stained with a GFP antisense probe. (C) Diagram representing average pixel quantification results of in situ hybridization signals from embryos analyzed on both sides. The statistical significance of the obtained series of values was verified by one-tailed Student's t-test (type 3, P < 0.01; P values: LE-R/MO1: 2.695E-50, LE-R/MO2: 1.299E-14, LE-R/MO3: 0.004, LE-R/MO4: 7.53E-06, LE-R/MO5: 1.133E-47, LE-R/MO6: 3.216E-45, LE-R/MO7: 3.949E-57). Values were adjusted to the score of GFP-XDE-LE alone, which was set to 1.
To assess the functional significance of this element and other parts of the XDE-LE-R for somatic destabilization, we designed a series of antisense morpholino oligonucleotides almost entirely covering XDE-LE-R (Fig. 3A). In addition, one MO (MO7) was generated as complement to 25 central nucleotides of the zf-nos1 homology region. We reasoned that MO binding to the reporter mRNA target could prevent its accessibility for miRNAs and consequently its somatic clearance. Though MO3 and MO4 exhibited no clear effect, the other antisense MOs were capable of interfering at least to some extent with somatic clearance of the XDE-LE-R reporter mRNA. As visualized by hybridization with a GFP riboprobe, followed by quantification of signal intensities (Fig. 3 B and C), the inhibitory activities differed, ranging from 2- to 4-fold (MO1, 2, 5, 6) to ∼7-fold (MO7; Fig. 3C). Therefore, the XDE-RNA element homologous to zf-nos1 seems to be of critical importance for the somatic clearance of the microinjected reporter RNA. Taken together, these observations suggest that the differential regulation of germ-line-associated RNAs observed for zebrafish nos1 and Xenopus DE could rely on a conserved mechanism involving microRNA-mediated RNA degradation via a conserved cis-acting 3′UTR RNA element.
MicroRNA-18 Is Involved in Somatic RNA Clearance of the XDE-LE-R Reporter.
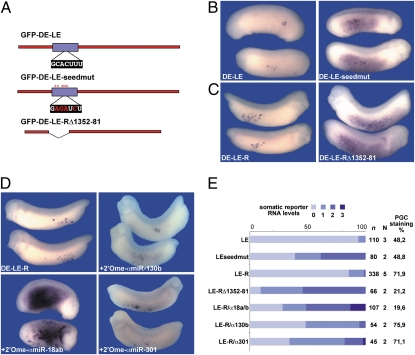
To more directly address the question if miRNAs are involved in somatic mRNA clearance, the XDE-LE-R was scanned for predicted miRNA target sites with the program miRanda (21) using all known, mature Xenopus tropicalis miRNA sequences (22). Various different miRNAs were predicted to bind to the zf-nos1 homology region (Fig. S2) via the target motif GCACUU(U), previously also characterized as miR-430 target sequence in the zebrafish (23). Notably, Xenopus miR-427, proposed to constitute the Xenopus ortholog of zebrafish miR-430 (14), was not among the top predicted miRNAs, even though perfectly matching within the seed sequence. Upon injection of a XDE-LE reporter containing a mutated target motif, elevated levels of reporter mRNA were detected in the endoderm (Fig. 4 A, B, and E). However, PGC restriction of the reporter RNA was not fully inhibited, suggesting that additional elements potentially targeted by miRNAs might contribute to somatic clearance. In another approach to define the importance of the nos1 homology region for germ-line restriction of the reporter RNA, we internally deleted this element from XDE-LE-R. Further underscoring its requirement for somatic destabilization, reporter RNA lacking the part of the nos1 homology region containing the predicted miRNA target motif was detected in the soma at markedly increased levels, whereas PGC restriction was almost fully abolished (Fig. 4 A, C, and E).
Fig. 4.
Somatic clearance of XDE-LE-R reporter RNA is achieved by a microRNA-mediated mechanism. (A) Schematic overview of mutated or deleted reporter constructs. A blue rectangle marks the nos1 homology region; altered positions are marked by asterisks; mutations within the miR target motif are represented in red letters. (B) Mutation of the miR-430 target motif resulted in decreased somatic clearance of the XDE-LE reporter. Upon vegetal injection (2/2 cells) of 1.2 ng XDE-LE mRNA or XDE-LE-seedmut mRNA, somatic levels of reporter mRNAs were ranked visually into four classes: 0 (not detectable, as exemplified in D, Upper Left), 1 (weak; example in D, Lower Right), 2 (medium; example in C, Right), and 3 (very high; example in D, Lower Left). (C) Embryos injected with 1.1 ng XDE-LE-R mRNA or XDE-LE-RΔ-1352–81 mRNA were scored similarly. Partial deletion of the nos1 homology domain also resulted in decreased somatic clearance. (D) Vegetal coinjection of antisense 2′OMeOs amiR-130b (α130b) or amiR-301 (α301) (6.4 pmol each) with 1.1 ng of XDE-LE-R reporter RNA resulted only in slight elevation of somatic reporter RNA levels, whereas coinjection of a mixture of the antisense 2′OMeOs amiR-18a and amiR-18b (α18a/b) (3.2 pmol each) led to strong somatic stabilization of coinjected reporter RNA (Table S1). (E) Comparative diagram of somatic reporter RNA levels (%) scored in the injected embryos; in all cases, embryos compared were stained in parallel for the same time.
To analyze the involvement of specific miRNAs in somatic RNA clearance, antisense inhibition of miRNAs was performed using antisense 2′O-methyl oligoribonucleotides (2′OMeOs). Because several of the miRNAs predicted to bind within the nos1 homology element are closely related in sequence, some of the 2′OMeOs raised were degenerated at several positions, thus targeting more than one miR family member at a time. Coinjection of 2′OMeOs designed to target miR-130b or miR-301 only mildly affected somatic clearance of XDE-LE-R reporter RNA (Fig. 4 D and E). However, coinjection of 2′OMeOs raised against miR-18a and miR-18b clearly alleviated somatic clearance upon coinjection with the XDE-LE-R reporter (Fig. 4 D and E). Thus, a miR-18 family member appears to be involved in the somatic clearance of XDE-LE-R reporter RNA in Xenopus.
Elr Proteins Can Mediate Protection of XDE-LE-R from Somatic RNA Clearance.
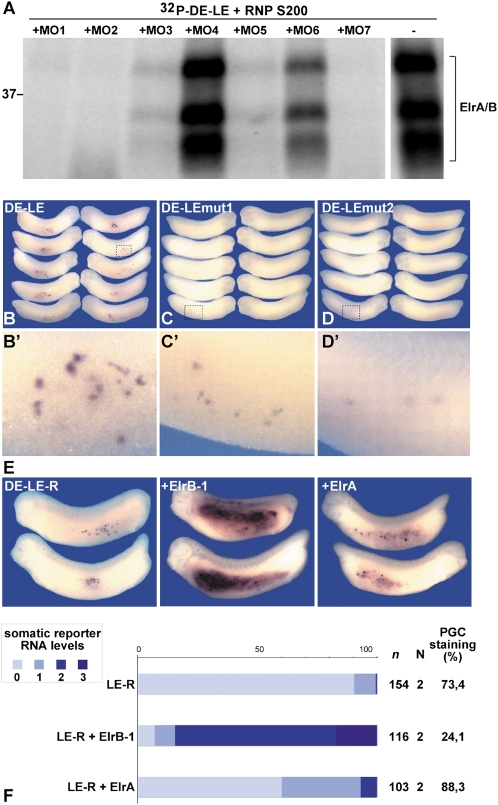
Though miR-18 seemed to mediate the somatic clearance of XDE-LE-R reporter RNA, the mechanism of germ-line-specific protection from RNA clearance still remained unclear. Recently, we could identify Xenopus Elr-type proteins of the Elr/HuR family as components of the vegetal localization complex forming with the XDE-LE (24). The Elr/HuR family of RNA binding proteins has been implicated in various aspects of RNA regulation—e.g., the stabilization of target mRNAs (25) or translational regulation (26). The region within the XDE-LE that is critical for Elr protein binding is located 5′ to and only partially overlapping with the nos1 homology region (Fig. S3). To establish the relationship between Elr protein binding and germ cell-specific suppression of RNA clearance, we took advantage of the same set of MOs described previously (Fig. 3A). UV cross-linking of XDE-LE was performed in the presence or absence of these MOs using a partially purified oocyte RNP fraction enriched in proteins of the vegetal localization complex (24). Similar to our previous observations with coimmunoprecipitation of Elr proteins translated in vitro and fluorescently labeled RNA (24), MOs interfered with XDE-LE cross-linking of Elr proteins to a variable extent, with the strongest inhibitory activities observed for MO1, MO2, and MO7; weak but significant ones for MO3 and MO5; and very weak ones or none for MO6 or MO4, respectively (Fig. 5A). Taken together, the MO effects on Elr binding as measured by UV cross-linking partially overlap with those on somatic RNA clearance (see also Fig. 3C), indicating that Elr proteins might be involved in germ cell-specific RNA protection. However, it seems noteworthy that several MOs, such as MO2 and MO3, clearly affect Elr protein binding, while having no or only moderate effects on RNA clearance.
Fig. 5.
Elr protein binding protects XDE-LE reporter RNA from somatic clearance. (A) UV cross-linking assay using radiolabeled XDE-LE-MO hybrids. 32P-labeled XDE-LE RNA was prehybridized with the antisense MOs indicated and incubated with fractionated oocyte extract before UV irradiation. Cross-linked proteins were separated by SDS/PAGE; the positions of molecular weight markers are indicated. (B–D′) Embryos were injected vegetally (2/2) with 800 pg of wild-type XDE-LE (B and B′, n = 78; N = 1) or mutant reporter mRNAs XDE-LE mut1 (C and C′, n = 71; N = 1) and XDE mut2 (D and D′, n = 88; N = 1). (B′, C′, and D′) Magnified aspects of single embryos marked in B, C, and D. Injection of mutant XDE-LE reporter mRNAs deficient for Elr binding resulted in decreased intensities (B′, C′, and D′) and frequencies of PGC staining (11.3% for XDE-LE mut1, 12.5% for XDE mut2) compared with the wild-type XDE-LE (83.3% PGC staining). (E) Both blastomeres of two-cell-stage embryos were injected vegetally with 1.1 ng of XDE-LE-R reporter mRNA alone or together with 200 pg of MT-ElrB1 mRNA or 400 pg of MT-ElrA mRNA and scored for somatic reporter RNA levels as described (Fig. 4). Coexpression of ElrB1 led to strong somatic stabilization of XDE-LE-R, whereas only slightly elevated levels of somatic reporter levels were detected upon coinjection of ElrA. (F) Diagram of somatic reporter RNA levels (%) scored in ElrB1/ElrA-coninjected embryos. n, no. of injected embryos; N, no. of experiments.
To more directly assess the relevance of Elr protein binding for the stabilization of XDE-LE reporter mRNA in PGCs, we used two mutant forms of XDE-LE containing different combinations of point mutations within A/U-rich elements, known to be important for Elr binding. In UV cross-link competition experiments and electrophoretic mobility shift assays, these mutations had been previously shown to interfere moderately or strongly with ElrA/B binding, respectively [XDE-LE mut1 and XDE-LE mut2 (ref. 24 and Fig. S3)]. Upon injection of these mutant mRNAs, PGC stabilization of the reporter mRNA was almost completely lost (Fig. 5 C, C′, D, D′). Thus, ElrA/B binding ability correlates with PGC-specific protection from reporter mRNA clearance.
To address the question of whether Elr proteins would be sufficient to counteract somatic destabilization, tagged forms of Xenopus ElrB1 and ElrA were overexpressed in Xenopus embryos in the presence of the XDE-LE-R reporter RNA. Ectopic expression of ElrB1 and ElrA resulted in strong and mild endodermal reporter RNA stabilization, respectively (Fig. 5 E and F). Thus, overexpression of Elr proteins, in particular of ElrB1, is sufficient to protect XDE-LE reporter RNA from somatic clearance.
To analyze whether the mechanisms we found to be involved in somatic clearance of XDE-LE reporter RNA would also apply to endogenous XDE mRNA, whole-mount in situ hybridization analysis on MO7-injected embryos was performed. Elevated somatic levels of XDE mRNA were indeed observed in the endoderm upon masking of the nos1 homology region by injection of MO7 (Fig. S4A). We further determined the levels of XDE mRNA at different stages of embryogenesis by quantitative RT-PCR; similar to the time course we had determined for XDE-LE reporter mRNA clearance on the basis of whole-mount in situ hybridization experiments (Fig. S1), the quantity of XDE transcripts strongly decreased after gastrula stages (Fig. S4B). Interestingly, XDE mRNA clearance could be inhibited by the different procedures that we had found to similarly inhibit clearance of the microinjected reporter RNA constructs—by injection of either MO7, anti-miR-18a/b antisense 2′OMeOs, or synthetic ElrB1 mRNA (Fig. S4C). The intensities of the inhibitory effects observed at stage 30 appear to correlate with those for microinjected XDE-LE reporter mRNA constructs (Figs. 3B, 4D, and 5E), with the strongest increase in stability obtained for MO7, and significant but weaker effects for anti-miR-18a/b and ElrB1. Thus, endogenous XDE mRNA clearance appears to be regulated by mechanisms similar to those that we had described for somatic clearance of microinjected XDE-LE reporter mRNA constructs.
In summary, these findings provide strong indications for the involvement of Elr protein binding in germ-line protection of XDE-LE reporter mRNA from miRNA-mediated mRNA clearance.
ElrB1 Synergizes with XDE Protein in XDE-LE Reporter mRNA Stabilization.
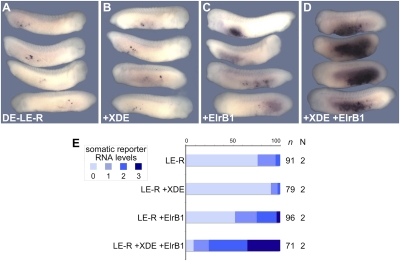
Because Elr proteins are also expressed outside of the germ plasm during embryogenesis (27), the mechanistic basis of their potential germ-line-specific activity is unclear. In the zebrafish system, it was suggested that the Dnd protein itself is involved in germ-line-specific protection of germ-line messenger RNAs from miR-430-mediated RNA clearance (13). To analyze whether Xenopus Dead end could exert a similar effect in the somatic stabilization of XDE-LE, we coexpressed Xenopus Dead end protein together with XDE-LE reporter mRNA. Upon coexpression of moderate amounts of Xenopus Dead end protein (200 pg injected XDE-ORF mRNA), no somatic stabilization of XDE-LE reporter mRNA was observed (Fig. 6B). When we decreased the dosage of MT-ElrB1 injected together with the XDE-LE reporter, we also observed a gradual reduction of the stabilizing activity (Fig. 6 C and E). However, if low doses of both ElrB1 and XDE were combined, XDE-LE stabilization was enhanced in a synergistic manner (Fig. 6 D and E). Thus, the cooperative activities of ElrB1 and XDE proteins might account for germ cell-specific protection of germ-line mRNAs from miR-mediated RNA clearance.
Fig. 6.
ElrB1 and Xenopus Dead end proteins act synergistically in the somatic stabilization of XDE-LE reporter mRNA. (A–E) Whereas vegetal coinjection of FL-XDE mRNA (100 pg, B) or low levels of MT-ElrB1 mRNA (50 pg, C) with GFP-XDE-LE-R reporter mRNA (1.1 ng) led to no traceable or moderate somatic protection of XDE-LE-R, respectively, coexpression of both proteins (same amounts of injected mRNAs) strongly enhanced the stabilizing effect (D and E). (E) Comparison of somatic reporter RNA levels (%) scored in the injected embryos.
Discussion
In this study, we identified a regulatory RNA sequence within the 3′UTR of the late-localizing, PGC-specific mRNA XDE, which exhibits dual activities: it is not only instrumental in directing vegetal localization of XDE mRNA, but it is also critical for somatic RNA clearance, as well as for germ-line-specific protection of the same RNA. LEs of several other germ-line-associated, vegetally localized mRNAs share these same activities. MiRNAs of the miR-18 family participate in somatic RNA clearance, whereas Elr protein binding to the LEs in cooperation with XDE protein mediate RNA stabilization.
Remarkably, the RNA element most critical for germ-line restriction of XDE contains a sequence similar to the one implicated in somatic deadenylation of zebrafish nanos-1 (12), suggesting at least some degree of conservation. However, in contrast to the zebrafish, where miR-430 family members have been implicated in the somatic deadenylation of nanos-1, our findings suggest the involvement of a member of the miR-18 family in Xenopus, acting via the common target motif GCACUU(U). Target sites of xla-miR-427, the predicted miR-430 homolog (14, 15), were not detected within the XDE-LE in this analysis. Interestingly, and in accordance with the findings described here, a function in RNA stability and translational control was previously reported for a 3′UTR region of Xenopus DeadSouth, also representing a vegetally localizing, germ-line-specific mRNA (28). However, apart from the XDeadSouth-LE also exhibiting similarities within the nos-1 homology domain, LEs of the other germ cell-associated mRNAs analyzed here do not reveal any sequence conservation allowing for the derivation of a simple consensus motif. The mere presence of predicted xtr-miR-18a/b target sites does also not define a distinguishing property, because they occur in most LEs analyzed, including those not targeted for somatic clearance, such as Vg1 and VegT. Thus, the determinants for germ-line restriction are likely to involve additional molecular features such as RNA secondary structure and/or the differential recruitment of trans-acting factors.
The onset of xla-miR-18 expression detected at gastrula stage (29) is well compatible with the time course we determined for XDE-LE reporter RNA clearance (Fig. S1) and the clearance of endogenous XDE mRNA (Fig. S4). The relatively weak effect of either mutating the conserved miR-18 target motif or blocking endogenous miR-18 by antisense oligonucleotides could result from redundancy of miR-18 target sites or, alternatively, additional miRNAs binding outside of the nos1 homology region might contribute to the process.
At low levels, Xenopus Dead end mRNA can be detected outside of the germ plasm up to the gastrula stage, but it becomes progressively restricted to PGCs during development (8, 30). Clearance of XDE mRNA from somatic cells is therefore likely to occur and might also account for the observed overall reduction of endogenous XDE mRNA levels taking place during embryogenesis, thus preventing XDE protein expression in the soma. Recently, it has been suggested that in the zebrafish, Dnd1 protein itself is responsible for inhibiting miR-mediated RNA clearance in PGCs by preventing access of miR-430 to its target site (13). In addition, zebrafish Dazl was found to relieve miR-430-mediated translational repression of tdrd7 mRNA by inducing its polyadenylation (31). Findings reported here provide strong evidence that Elr proteins, which we also identified as components of the vegetal RNA localization complex (24), are critical for PGC-specific XDE-LE stabilization. Mutant forms of the XDE-LE reporter RNA deficient in Elr binding are not protected in PGCs. In addition, increased Elr protein levels in the embryo are sufficient for the protection of XDE-LE reporter RNA from miR-mediated clearance. This effect was much more pronounced for ElrB1 than for ElrA; this might rely on the capability of ElrB1 to oligomerize, which is not the case for ElrA (30). However, because Elr expression is also detected in somatic cells (27), the specificity of the protective effect observed in Xenopus embryos is likely to involve additional activities. Our finding that Elr and Dead end proteins can synergize in the stabilization of XDE-LE suggests that both proteins might effectively mediate the protection mechanism at physiological levels. To fully understand the process of germ-line-specific RNP assembly, the profile of translational regulation of XDE needs to be elucidated further. Supporting a general role for Elr proteins in counteracting miRNA-mediated activities, it has been shown previously that the human ELAV-like protein HuR is essential for stress-induced release of target mRNAs from miRNA-mediated translational repression (32). In the future, it will be critical to define the interaction conditions of Elr and XDE proteins with XDE-LE and other vegetally localized mRNAs of the germ line in more detail.
Materials and Methods
Embryo Manipulations.
Embryos from Xenopus laevis adult albino females were obtained by standard procedures. Capped mRNAs synthesized with the mMESSAGE mMACHINE Kit (Ambion) were injected vegetally or animally in a volume of 4 nL into both blastomeres of two-cell-stage embryos. Injected embryos were cultivated at 12.5 °C overnight and at 14 °C for 3 d and fixed at stage 30–34. Upon fixation, embryos were processed for in situ hybridization as described previously (33) using digoxigenin-labeled probes for mGFP5 RNA or lacZ. Alternatively, embryos were anaesthetized in a 0.01% solution of tricaine (MS-222) and microscopically analyzed for GFP expression (SZX12; Olympus). Double whole-mount in situ hybridization was performed with a mixture of a fluorescein-labeled mGFP5 ORF probe and a digoxigenin-labeled Xpat probe (6) detected with Fast Red and NBT/BCIP substrates (Roche), respectively.
Plasmid constructions are described in SI Text, and cloning oligonucleotides are listed in Table S2.
Supplementary Material
Acknowledgments
We thank Ines Eckhardt and Gudrun Kracht (University of Göttingen) for excellent technical assistance; Susanne Koch (University of Göttingen) for mapping XDE-LE-R and the LEs of Velo7 and Velo40; Maike Claussen (University of Göttingen) for mapping the DeadSouth-iso-LE; Erez Raz (Institute of Cell Biology) for pSP64mGFP5-3′UTRnos1; Hugh Woodland (University of Warwick, Coventry, UK) for Xpat; and Andreas Nolte (University of Göttingen) for sequencing and histology. This work was supported by Deutsche Forschungsgemeinschaft Grant Pi159/9-1 (to T.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004401107/-/DCSupplemental.
References
- 1.Kloc M, et al. RNA localization and germ cell determination in Xenopus. Int Rev Cytol. 2001;203:63–91. doi: 10.1016/s0074-7696(01)03004-2. [DOI] [PubMed] [Google Scholar]
- 2.Houston DW, King ML. Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 3.Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121:287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- 4.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 5.Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development. 1998;125:171–180. doi: 10.1242/dev.125.2.171. [DOI] [PubMed] [Google Scholar]
- 6.Hudson C, Woodland HR. Xpat, a gene expressed specifically in germ plasm and primordial germ cells of Xenopus laevis. Mech Dev. 1998;73:159–168. doi: 10.1016/s0925-4773(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 7.Machado RJ, et al. Xenopus Xpat protein is a major component of germ plasm and may function in its organisation and positioning. Dev Biol. 2005;287:289–300. doi: 10.1016/j.ydbio.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol. 2006;291:1–11. doi: 10.1016/j.ydbio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Weidinger G, et al. dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr Biol. 2003;13:1429–1434. doi: 10.1016/s0960-9822(03)00537-2. [DOI] [PubMed] [Google Scholar]
- 10.Youngren KK, et al. The Ter mutation in the dead end gene causes germ cell loss and testicular germ cell tumours. Nature. 2005;435:360–364. doi: 10.1038/nature03595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 12.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 14.Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 17.Rebagliati MR, Weeks DL, Harvey RP, Melton DA. Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell. 1985;42:769–777. doi: 10.1016/0092-8674(85)90273-9. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, King ML. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 1996;122:4119–4129. doi: 10.1242/dev.122.12.4119. [DOI] [PubMed] [Google Scholar]
- 19.Kloc M, Bilinski S, Pui-Yee Chan A, Etkin LD. The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′UTR. Dev Biol. 2000;217:221–229. doi: 10.1006/dbio.1999.9554. [DOI] [PubMed] [Google Scholar]
- 20.Betley JN, Frith MC, Graber JH, Choo S, Deshler JO. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol. 2002;12:1756–1761. doi: 10.1016/s0960-9822(02)01220-4. [DOI] [PubMed] [Google Scholar]
- 21.Enright AJ, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S. miRBase: The microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 23.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 24.Arthur PK, et al. Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem. 2009;284:19982–19992. doi: 10.1074/jbc.M109.009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaman I, et al. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J Biol Chem. 2002;277:41539–41546. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron M, Furrer MP, Wegnez M, Theodore L. Xenopus elav-like genes are differentially expressed during neurogenesis. Mech Dev. 1999;84:139–142. doi: 10.1016/s0925-4773(99)00056-8. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka K, et al. Visualization of the Xenopus primordial germ cells using a green fluorescent protein controlled by cis elements of the 3′ untranslated region of the DEADSouth gene. Mech Dev. 2006;123:746–760. doi: 10.1016/j.mod.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Watanabe T, et al. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 30.Devaux A, Colegrove-Otero LJ, Standart N. Xenopus ElrB, but not ElrA, binds RNA as an oligomer: Possible role of the linker. FEBS Lett. 2006;580:4947–4952. doi: 10.1016/j.febslet.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS ONE. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Hollemann T, Panitz F, Pieler T. In situ hybridization techniques with Xenopus embryos. In: Richter JD, editor. A Comparative Methods Approach to the Study of Oocytes and Embryos. New York: Oxford Univ Press; 1999. pp. 279–290. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.