Abstract
Atg8 is a ubiquitin-like protein that controls the expansion of the phagophore during autophagosome formation. It is recruited to the phagophore during the expansion stage and released upon the completion of the autophagosome. One possible model explaining the function of Atg8 is that it acts as an adaptor of a coat complex. Here, we tested the coat-adaptor model by estimating the area density of Atg8 molecules on the phagophore. We developed a computational process to simulate the random sectioning of vesicles heterogeneous in size. This method can be applied to estimate the original sizes of intracellular vesicles from sizes of their random sections obtained through transmission electron microscopy. Using this method, we found that the estimated area density of Atg8 is comparable with that of proteins that form the COPII coat.
Keywords: Autophagy, lysosome, stress, vacuole, yeast
INTRODUCTION
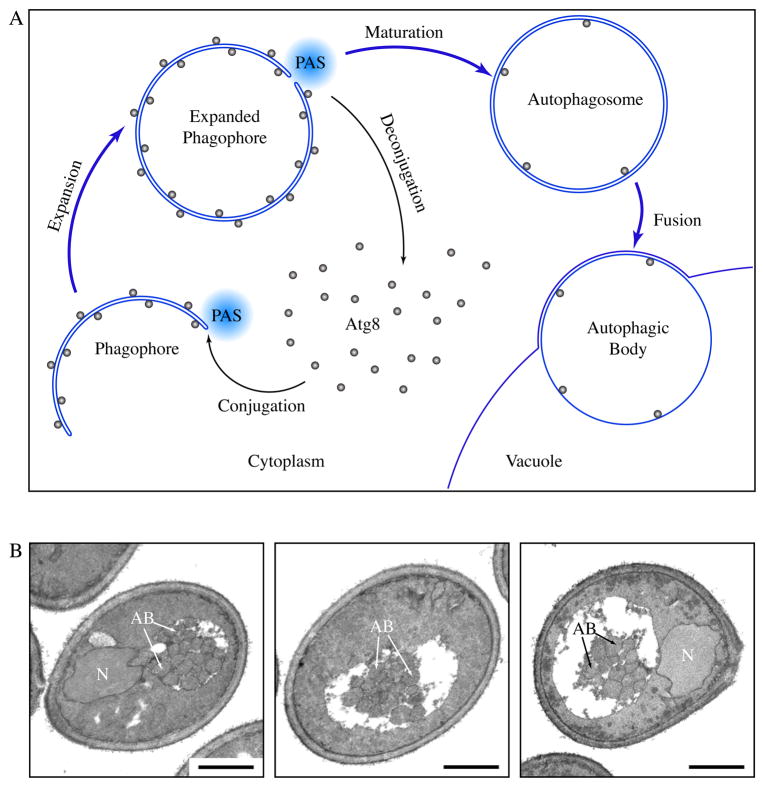
Macroautophagy, hereafter referred to as autophagy, is a pivotal intracellular degradation process.1 During autophagy, cytoplasmic materials are sequestered into an expanding membrane sac, the phagophore, which later matures into a double-membrane vesicle, the autophagosome (Fig. 1A). Each autophagosome eventually fuses with a lysosome, leading to the degradation of the inner membrane and the cargos. The formation of autophagosomes is catalyzed by a set of core machinery proteins at the phagophore assembly site (PAS). The core machinery is supplemented by certain auxiliary proteins, the composition of which varies depending on physiological stimuli.
Figure 1.
Formation of autophagosomes and autophagic bodies. (A) Scheme of autophagy in yeast. Autophagosomes are formed through the expansion and deformation of the phagophore at the PAS. During this process, Atg8 is conjugated and recruited to the PAS. It resides on both sides of the phagophore and controls its expansion. At the end of phagophore expansion, most Atg8 molecules are released back to the cytosol through deconjugation. The fully expanded phagophore then matures into an autophagosome. When autophagosomes fuse with the yeast vacuole, the inner vesicles are released into the vacuole lumen, and are now termed autophagic bodies. In pep4Δ cells, autophagic bodies are not degraded. (B) Representative sections of starving pep4Δ yeast cells. atg8Δ pep4Δ cells expressing GFP-Atg8 and grown to mid-log phase were incubated in nitrogen starvation medium for 4 hours and processed for electron microscopy. Autophagic bodies (AB) accumulated as a cluster of single-membrane vesicles in the yeast vacuole. N, nucleus, Scale bar, 1 μm.
Although autophagosomes and vesicles of the secretory pathway share similar mechanisms in their fusion with corresponding destination compartments,2, 3 it is not clear whether the formation processes of autophagosomes and secretory vesicles also follow the same principles. It is generally accepted that vesicles in the secretory pathway are generated from existing membrane structures by budding.4 This involves the deformation of the membrane by coat complexes, such as the COPII coat for endoplasmic reticulum (ER) to Golgi complex transport, the COPI coat in retrograde intra-Golgi complex and Golgi complex to ER transport, and the clathrin coat in post-Golgi transport.5 Although the participating components vary, these coats share a two-layer scheme of organization, with a membrane-proximal layer of adaptor proteins and a membrane-distal layer of cage proteins. The assembly and disassembly of the coats are regulated by small GTPases. In autophagosome formation, however, the phagophore is considered to be formed de novo instead of from existing organelles.6 A Sar1-like GTPase, which can be targeted to prevent coast disassembly, is absent from the currently known molecular machinery of autophagosome formation. It is therefore unknown whether the determination of the curvature of the phagophore involves a coat cage.
Recently, we discovered that a member of the core autophagosome formation machinery, Atg8, specifically controls the sizes of autophagosomes.7 Atg8 is a ubiquitin-like protein.8, 9 Although it has been suggested to be a tethering factor based on in vitro results,10 properties of Atg8 revealed by in vivo studies possess strong resemblance to those of coat adaptor proteins. During autophagosome formation, Atg8 is conjugated to phosphatidylethanolamine (PE) and initially resides on the surface of the phagophore.11,12 Later, the majority of Atg8 needs to be released by deconjugation prior to the completion of autophagosome formation,7 which mirrors the de-coating stage (Fig. 1A). Similar to known coat adaptors, Atg8 also interacts with cargo receptors.13, 14 Because the phagophore is a membrane sac with two layers of membrane in its planar regions, a transmembrane cargo receptor similar to those in the secretory pathway will not be able to reach coat adaptors on the convex side. This may explain why, unlike other coat adaptors, Atg8 localizes to both sides of the phagophore, with the population on the concave side presumably responsible for interacting with cargo receptor proteins. Functioning as a coat adaptor, Atg8 may restrict the size of the coat cage by limiting the amounts of cage proteins recruited. If this is the case, even though Atg8 does not form a cage by itself, the area density of Atg8 molecules on the membrane should be comparable to that of cage elements. Therefore we tested the coat adaptor model by estimating the area density of Atg8 on the phagophore.
RESULTS AND DISCUSSION
The area density of Atg8 was calculated using two input values: (1) the number of Atg8 molecules recruited to the PAS and (2) the size of the fully expanded phagophore. Our laboratory recently measured the number of Atg8 molecules recruited to the PAS by fluorescence microscopy using GFP-tagged Atg8 expressed in atg8Δ cells under the control of the endogenous ATG8 promoter. Under nitrogen starvation conditions, the average peak number of GFP-Atg8 molecule in each round of Atg8 recruitment and release is found to be approximately 272±9 (mean ± S.E.M., n=100) (Table 1).15
Table 1.
Summary of results in the estimation of the area density of Atg8.
| Estimation of GFP-Atg8 Density | ||
|---|---|---|
| Number of GFP-Atg8 molecules at the PAS | Mean | S.E.M. (n=100) |
| 272 | 9 | |
| Radii of autophagic body sections (EM) | Mean | S.E.M. (n>200) |
| 127 nm | 2 nm | |
| Radii of autophagic bodies (R analysis) | Mean | C.I. |
| 148 nm | 5 nm | |
| Average surface area of one side of expanded phagophore | 2.8×105 nm2 | |
| Area density of GFP-Atg8 | 1 per 2×103 nm2 | |
| Length of edge | 40–50 nm | |
S.E.M., standard error of the mean. C.I., 95% confidence interval.
We then used electron microscopy (EM) to estimate the size of the fully expanded phagophore. The fully expanded phagophore is a transient structure that is difficult to detect by EM because it quickly matures into an autophagosome. The completed autophagosome then fuses with the yeast vacuole (a lysosome analogue), releasing the inner single-membrane vesicle into the vacuole lumen, where it is normally degraded. When the PEP4 gene encoding the vacuolar aspartyl protease Pep4 is knocked out, the inner single-membrane vesicle is stabilized in the vacuole. This vesicle, termed an autophagic body, can be visualized easily in transmission electron microscopy (Fig. 1). There is little inter-membrane space between the two sides of the phagophore, and between the two membranes of an autophagosome.6 The size of the fully expanded phagophore should therefore correspond to the size of the resulting autophagosome, which in turn closely matches the size of the single-membrane autophagic body. Therefore, we quantified the sizes of autophagic bodies to obtain an indirect estimation of phagophore sizes. We found that the average radius of autophagic body cross sections in pep4Δ cells from the aforementioned GFP-Atg8-expressing strain was 127±2 nm (mean ± S.E.M., n>200) (Table 1).
To calculate the actual sizes of these vesicles, however, the biases introduced by the sectioning process need to be corrected. When sectioning a spherical object, the radii of most sections are smaller than that of the sphere, contributing a negative bias. In a population of spheres heterogeneous in size, larger ones have higher probabilities of getting sectioned, contributing a positive bias. In addition to these two major biases, two minor biases are introduced by the following factors: (1) the aggregation of autophagic bodies will obscure each other on the overlaid areas (Fig. 1B); (2) cross sections below a certain size threshold are difficult to recognize because they do not have sufficient details to be distinguished from the background noise.
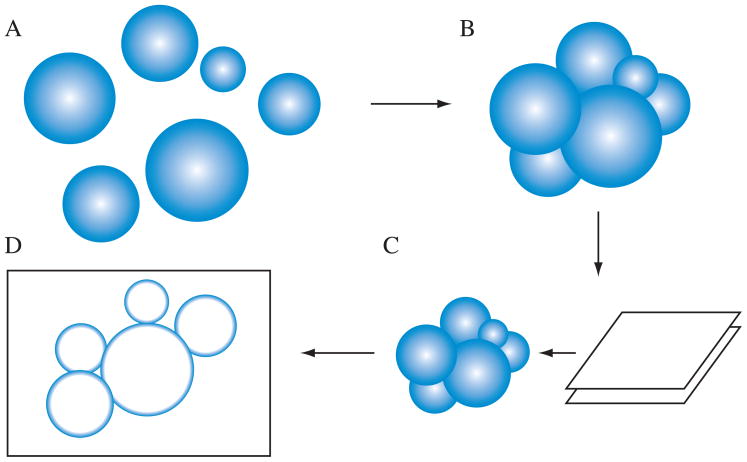
Although similar problems have been studied in the past, we were unable to find a suitable analytical solution. Instead, we developed a computational method using the R statistical software environment ( http://www.r-project.org/) according to the following assumptions (Fig. 2 and Supplementary Data): (a) autophagic bodies are nearly rigid spheres; (b) radii of autophagic bodies are distributed log-normally with unknown parameters, μ and σ, which correspond to the mean and standard deviation of the distribution at a log scale; (c) accumulated autophagic bodies are positioned so that their surfaces are in contact with each other (Fig. 1B); (d) the thickness of each sample section is 70 nm; (e) cross sections with radii less than 50 nm (the recognition threshold in our own experience) are ignored. The simulation process first generates a set of vesicles in silico based on the distribution with a given set of μ and σ values. Next, these vesicles are rearranged to be in contact with each other using an algorithm described by Milenkovic.16 The pile of vesicles is then sectioned, in silico, to produce a 70-nm thick slice, and areas of vesicle cross sections are reported. By varying the parameters of the original distribution, the correlation between the observed cross section distribution and the original distribution can be established by linear regression using the empirical moments.
Figure 2.
Scheme for the computational simulation of sectioning autophagic bodies. (A) First, the program generates a population of vesicles based on the specified parameters. (B) Next, the vesicles are positioned so that their surfaces contact each other. (C) The vesicle cluster is sectioned to produce a 70 nm thick slice. (D) The areas of vesicle sections in the slice is quantified and reported.
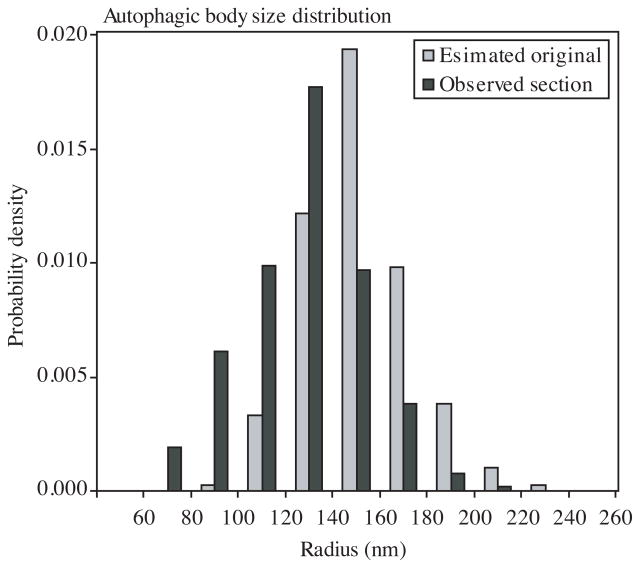
Using this method, we estimated the original autophagic body radii distribution using the data from the electron microscopy analyses (Fig. 3). The estimated average radius was 148±5 nm (mean ± 95% confidence interval) (Table 1). We recently showed that GFP-Atg8 at the PAS corresponds to an active population of the protein that is involved in autophagosome formation.7 We are making an assumption that in each round of autophagosome biogenesis essentially the entire population of Atg8 at the PAS is utilized. As a result of deconjugation (Fig. 1A), most of the Atg8 molecules are recovered and, with some addition for those lost within the autophagic body, each new round of autophagosome formation begins with the same starting number of Atg8 proteins. We are also assuming that the level of GFP-Atg8 at the PAS corresponds to that on the phagophore, and that it is distributed evenly on both the convex and concave sides of the phagophore. Under these conditions, on average 136 Atg8 molecules cover approximately 2.8×105 nm2 of surface area for one side of the membrane, which translates into one Atg8 molecule per 2×103 nm2 of surface area (Table 1). If each Atg8 molecule resides on a vertex of a polyhedral cage, the corresponding length of the edge will be approximately 40–50 nm, and the exact value will depend on the composition of the polyhedron. In comparison, the length of the COPII coat edge is approximately 30 nm.17
Figure 3.
The size distributions of observed sections and estimated original vesicles. Observed section, sizes of actually observed sections of autophagic bodies from EM. Estimated original, the sizes of original autophagic bodies estimated from simulation results.
In summary, we developed a computational simulation process to estimate the sizes of the original vesicles from EM data and calculated the area density of Atg8 on the phagophore. The area density result suggests that a coat adapter role for Atg8 remains a possibility. The key missing piece in the model, however, is the identity of components that constitute the edge of the coat. Previously, the complex formed by Atg12, Atg5 and Atg16 was proposed to be a coat candidate.18 However, the number of Atg16 molecules recruited to the PAS is one order of magnitude lower than that of Atg8,15 ruling out the possibility that this complex can fully cover the phagophore by itself. The function of Atg12–Atg5-Atg16 may instead be limited to recruiting Atg8 and facilitating Atg8 conjugation.19–23
Assessing the sizes of original vesicles from imaged vesicles is an important component of electron microscopy experiments. The methods presented here for autophagic bodies can be applied in general to other intracellular vesicles with similar properties. Prior to adopting this method, potential users need to carefully evaluate whether the assumptions in our model fit the intracellular vesicles under study. Among the five assumptions, the last three (packing, section thickness, recognition threshold) are direct reflections of our actual observations and experimental set-up. The choice of the first two (spheres, log-normal distribution), on the other hand, is a compromise between computational complexity and approximation of reality. Compared with an ellipsoid-based model of the autophagic bodies, which may better resemble what we see in EM samples (Fig. 1B), a sphere-based model reduces the parameters needed to describe their shapes from three radii to just one. Modeling as spheres instead of ellipsoids also eliminates the need to consider rotation of objects in a three-dimensional space, which would otherwise complicate both the sectioning process and their spatial arrangement in a pile. Similarly, making the log-normal distribution assumption allows us to use a simple linear regression-based method instead of solving differential equations, which becomes complicated when using real statistical data. Log-normal distribution is a “normal-like” distribution without a negative tail, frequently used in describing sizes of particles. The vesicle sections generated from our simulations fit well with actual data, indicating that the log-normal distribution is a reasonable approximation of the actual size distribution of autophagic bodies.
Supplementary Material
Footnotes
R source code for the simulation of sectioning of vesicles.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 2.Wang C-W, Stromhaug PE, Shima J, Klionsky DJ. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem. 2002;277:47917–27. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C-W, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–85. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 5.Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–8. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Kovács AL, Pálfia Z, Réz G, Vellai T, Kovács J. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy. 2007;3:655–62. doi: 10.4161/auto.4590. [DOI] [PubMed] [Google Scholar]
- 7.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–8. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–92. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 9.Paz Y, Elazar Z, Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem. 2000;275:25445–50. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- 10.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 11.Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999;147:435–46. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 15.Geng J, Baba M, Nair U, Klionsky DJ. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008;182:129–40. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milenkovic VJ. Position-Based Physics: Simulating the Motion of Many Highly Interacting Spheres and Polyhedra. International Conference on Computer Graphics and Interactive Techniques: Proceedings of the 23rd annual conference on Computer graphics and interactive techniques; 1996. pp. 129–36. [Google Scholar]
- 17.Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci U S A. 2001;98:10704–9. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the ~350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–25. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Huang W-P, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 23.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.