Abstract
At least 5 serotypes of exogenous simian retrovirus type D (SRV/D) have been found in nonhuman primates, but only SRV-1, 2 and 3 have been completely sequenced. SRV-4 was recovered once from cynomolgus macaques in California in 1984, but its genome sequences are unknown. Here we report the second identification of SRV-4 and its complete genome from infected cynomolgus macaques with Indochinese and Indonesian/Indochinese mixed ancestry. Phylogenetic analysis demonstrated that SRV-4 was distantly related to SRV-1, 2, 3, 5, 6 and 7. SRV/D-T, a new SRV/D recovered in 2005 from cynomolgus monkeys at Tsukuba Primate Center in Japan, clustered with the SRV-4 isolates from California and Texas and was shown to be another occurrence of SRV-4 infection. The repeated occurrence of SRV-4 in cynomolgus monkeys in different areas of the world and across 25 years suggests that this species is the natural host of SRV-4.
Keywords: Simian retrovirus 4, Simian retrovirus type D, Cynomolgus macaque, Complete genome sequence, Phylogeny
Introduction
The simian retrovirus type D (SRV/D) is classified in the Betaretrovirus genus and both endogenous and exogenous variants have been identified in nonhuman primates (NHP). The viral genome is a dimer of linear, positive sense, single stranded RNA. Signs of infection vary from no clinical signs to fever, diarrhea, weight loss, lymphadenopathy, retroperitoneal fibromatosis, subcutaneous fibrosarcoma and frequent opportunistic infections secondary to a fatal immune deficiency (Giddens et al., 1985; Henrickson et al., 1984; Lerche and Osborn, 2003). The potential of SRV/D to cause an acquired immune deficiency syndrome in macaques makes SRV/D infection a significant disease with regard to NHP research (Lerche and Osborn, 2003; Morton et al, 2008). Since SRV/D infection interferes with host immune responses in research animals, it can severely compromise research results. SRV/D can be transmitted among monkeys by blood, saliva, urine, feces, or by mother to infant transmission (Fujimoto et al., 2010; Hara et al., 2007; Lerche et al., 1986; Tsai et al., 1990). Cross-species transmission from monkeys to humans is also possible, although no disease has been reported. Two humans who were exposed to SRV/D remained healthy and had no observed disease associated with SRV/D exposure during the two and three year follow-up periods for which data was available (Lerche et al., 2001).
Viral sequences of endogenous and exogenous SRV/D integrate in the host genome and can be either expressed or not expressed due to mutations and deletions. Several simian endogenous betaretroviruses have been found, such as langur type D retrovirus PO-1-Lu (Todaro et al., 1978), squirrel monkey retrovirus (SMRV) (Heberling et al., 1977) and simian endogenous retrovirus (SERV) in baboons (van der Kuyl et al., 1997). Exogenous SRV/Ds have been found in diverse macaque species (genus Macaca) (Lerche and Osborn, 2003). Five serotypes of exogenous SRV/D, SRV-1 to 5, have been reported (Li et al., 2000; Maul et al., 1986; Morton et al., 2008). SRV-1, SRV-2 and SRV-3 (Mason-Pfizer monkey virus, MPMV) are commonly found in the genus Macaca and their genomes have been fully sequenced (Marracci et al., 1999; Power et al., 1986; Sonigo et al., 1986; Thayer et al., 1987). SRV-4 was recovered only once from a group of cynomolgus macaques (Macaca fascicularis) in California in 1984 (Voevodin and Marx Jr., 2009). SRV-5 was isolated from rhesus macaques originating in China and a partial gag (encoding capsid protein)-prt/pro (encoding protease) sequence is available (Li et al., 2000). In addition to these five serotypes, new exogenous SRV/Ds have been continuously discovered in various primate species, although their neutralization characteristics have not been determined. SRV-6 was found in wild-caught Hanuman Langurs (Semnopithecus entellus) in India and its env (encoding envelope protein gp70 and gp20) and 3′orf (encoding in an alternate reading frame within env gp70 whose function is still not clear) gene sequences were published (Nandi et al., 2000; Nandi et al., 2003). SRV-7 was isolated from wild rhesus monkeys (Macaca mulatta) in India and its partial env and pol (encoding multiple functional enzymes including reverse transcriptase and integrase) sequences were documented (Nandi et al., 2006). In Japan, a new SRV subtype SRV/D-T was isolated from cynomolgus macaques (Hara et al., 2005b). SRV/D-T antibodies cross-reacted with SRV-2 antigen and SRV/D-T full-length gag and partial env sequences are available (Hara et al., 2005b; Hara et al., 2007).
For about 25 years, SRV-4 (SRV4/CALIF/1984) was only known as a serotype of SRV/Ds that was first isolated at the California National Primate Research Center from a cynomolgus monkey housed at the California Public Health Laboratory in Berkeley, CA in 1984 (Preston A. Marx unpublished results). In the neutralization test, SRV4/CALIF/1984 virus was resistant to antisera against SRV-1, SRV-2 and SRV-3 (Preston A. Marx unpublished results). SRV-4 was therefore named because the next number in SRV series was SRV-4. However, neither viral genome sequences nor any of its characteristics have been documented since 1984. In this paper, we report the identification of SRV-4 from infected cynomolgus macaques with Indochinese and Indonesian/Indochinese mixed ancestry. The virus was isolated, and the entire genome was sequenced and compared to other SRV/D subtype viruses.
Results and discussion
Virus isolation
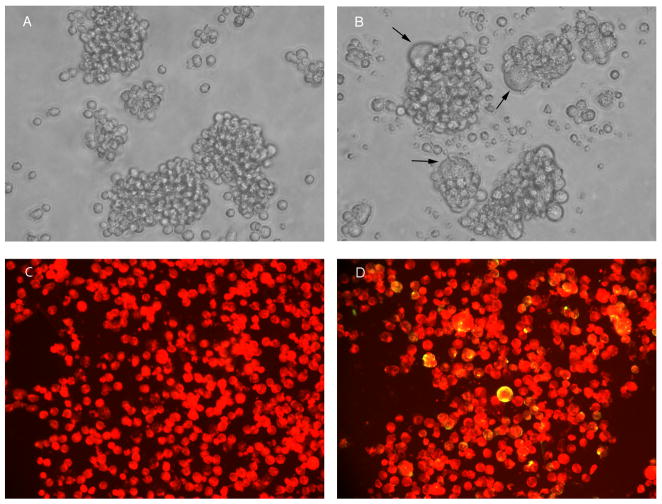
From December 2008 to date, a total of 45 out of 377 cynomolgus monkeys imported from China to the U.S. were found to be SRV/D positive. By analysis of PCR fragments amplified in a routine SRV/D PCR screen, all 45 SRV/D positive animals had the same nucleic acid sequences that differed from published SRV-1, 2, 3, 5 reference strain sequences. This unusual SRV/D prevalence (11.9%) piqued our interest to determine which of the SRV/D serotype was harbored by these animals. A total of 21 virus isolates were obtained from 45 SRV/D positive animals. Syncytia formation of cytopathic effects was observed after 1 to 4 weeks in culture (Figs. 1A and 1B). An immunofluorescent assay using SRV/D positive animal sera staining confirmed the presence of SRV/D (Figs. 1C and 1D). Three virus strains were characterized. The virus strain SRV4/TEX/2009/V1 was isolated from an SRV/D antibody negative animal by using heparined peripheral blood mononuclear cells (PBMCs) co-cultured with Raji cells; virus strain SRV4/TEX/2009/V2 was isolated from an SRV/D antibody positive animal. The attempt to isolate the virus strain SRV4/TEX/2009/V3 from the PBMC sample of an SRV/D antibody negative animal was not successful. However, its complete genome was directly sequenced and assembled from PCR products by using PBMC proviral DNA as templates.
Fig. 1.
SRV4/TEX/2009/V1 virus isolation in Raji cells. Raji cells were co-cultured with cynomolgus monkey PBMCs (400x). A: Uninfected Raji cells. B: Infected Raji cells showing syncytia formation (arrows). C: Uninfected Raji cells stained with sera from SRV/D positive animals and FITC-conjugated anti-human IgG showing red color under fluorescent microscopy. D: Infected cells were observed as yellow-green color with red background for negative cells by immunofluorescent staining.
Mapping of the complete viral genome
PCR fragments of proviral DNA samples prepared from virus isolates and/or their associated PMBCs were sequenced. The total proviral DNA length of both SRV4/TEX/2009/V1 (GenBank accession # FJ971077) and SRV4/TEX/2009/V2 (FJ979638) was 8126 base pairs (bp) long, while that of SRV4/TEX/2009/V3 (FJ979639) was 8127 bp. Although one additional adenosine nucleotide at position 7781 was found in the SRV4/TEX/2009/V3 viral genome as compared to SRV4/TEX/2009/V1 and SRV4/TEX/2009/V2, it was not located in any of the open reading frames. Therefore, the insertion of this nucleotide did not cause an amino acid mutation or frameshift in protein coding regions. Whether this adenosine insertion in the polypurine tract region causes any effect on its role as a primer for plus-strand viral DNA synthesis is unknown.
The genomic organization of these virus strains was similar to that of other SRV/Ds which have long terminal repeat (LTR) flanking sequences at two ends, and gag, prt, pol, env in the middle. The viral genome sequences of proviral DNA extracted from virus isolates and their associated PBMCs were 100% identical for SRV4/TEX/2009/V1 and SRV4/TEX/2009/V2. The identities of viral genome sequences among SRV4/TEX/2009/V1, SRV4/TEX/2009/V2 and SRV4/TEX/2009/V3 (SRV4/TEX/2009/V1-3) were nearly (from 99.7% to 99.9%) identical. However, the viral genome of SRV4/TEX/2009/V1 exhibited only 78.3%, 75.5% and 74.3% identities to SRV-1, 2, and 3, respectively.
Long terminal repeat (LTR) at 5′ and 3′ends
The LTR located at both the 5′end (nucleotide position 1 to 338) and the 3′ end (nucleotide position 7789 to 8126 in SRV4/TEX/2009/V1 and SRV4/TEX/2009/V2; nucleotide position 7790 to 8127 in SRV4/TEX/2009/V3) of the proviral genome was 338 bp long. The LTR sequences of SRV4/TEX/2009/V1 and SRV4/TEX/2009/V2 were 100% identical, and had a 99.7% identity to SRV4/TEX/2009/V3. Although they exhibited only 75.1%, 73.9% and 77% identities to corresponding LTR sequences of SRV-1, SRV-2 and SRV3, the sequence characteristics of LTR were conserved as in all other SRV/Ds (Power et al., 1986; Sonigo et al., 1986; Thayer et al., 1987). The predicted functions of LTR included the integration of viral DNA into the host chromosome, viral DNA synthesis and viral gene expression (Temin, 1981; Varmus, 1982). The inverted repeats U3 (TGTCC, 3′ unique sequence block) and U5 (GGACA, 5′ unique sequence block) were positioned at each end of LTR (Supplemental Fig. 1). Downstream of 5′LTR at position 341 to 354 was the conserved primer binding site (pbs), which allowed cellular tRNA as a primer to bind with and initiate minus-strand DNA synthesis during the reverse transcription process. Upstream of 3′LTR at nucleotide position 7768 to 7786 (7787 in SRV4/TEX/2009/V3) was the polypurine tract (ppt) (AATAAAATAAAAAGGGTGA), which served as a primer for plus-strand viral DNA synthesis. The presumptive “TATA” box (Goldberg-Hogness box), a promoter region for transcription, was at nucleotide position 201 to 207 (TATATAA). A poly-A (ATTAAA) signal was 18 bp downstream of the TATA box and the viral transcript was expected to be polyadenylated at poly-A site of the 3′LTR.
The gag gene
The first open reading frame (ORF) in the viral genome was 1980 bp long and encoded a Gag protein of 659 amino acids (aa). It was genetically related to the gag gene of other SRV/Ds, which encoded a polyprotein precursor and proteolytically processed to six mature virion core structural proteins: p10, pp24/pp18/pp16 (a phosphoprotein), p12, p27 (the major core nucleocapsid protein), p14 (the nucleic acid binding protein) and p4/p6 (Bradac and Hunter, 1984; Henderson et al., 1985).
The amino acid sequences of the Gag protein were 100% identical among our virus strains SRV4/TEX/2009/V1-3. Comparison of full-length gag sequences of SRV4/TEX/2009/V1-3 to the published gag genes of SRV-1, SRV-2, and SRV-3 demonstrated a 78.3% to 80.9% nucleic acid identity (Table 1). The full-length gag nucleotide identity between SRV4/TEX/2009/V1-3 and SRV/D-T (a new SRV/D found in Japan), however, was highly similar ranging from 98.7% to 98.8%. There were 25 nucleotides in the SRV4/TEX/2009/V1-3 gag coding area that were different from the SRV/D-T gag gene, most of them attributed to synonymous substitution with the exception of one non-synonymous amino acid substitution that occurred at amino acid position 271 in the p12 region, where SRV4/TEX/2009/V1-3 had a histidine residue in place of an arginine in SRV/D-T. With only one amino acid difference in the Gag protein between our virus strains and SRV/D-T, the amino acid identity was as high as 99.8%.
Table 1.
The gag nucleotide and amino acid sequences identities comparison among SRVs.
| Virus | V1a | V2b | V3c | SRV-4d | SRV/D-T | SRV-1 | SRV-2 | SRV-3 |
|---|---|---|---|---|---|---|---|---|
| V1a | - | 100 | 100 | 100 | 99.8 | 86.2 | 83.9 | 85.9 |
| V2b | 100 | - | 100 | 100 | 99.8 | 86.2 | 83.9 | 85.9 |
| V3c | 99.9 | 99.9 | - | 100 | 99.8 | 86.2 | 83.9 | 85.9 |
| SRV-4d | 99.3 | 99.3 | 99.4 | - | 99.8 | 86.2 | 83.9 | 85.9 |
| SRV/D-T | 98.7 | 98.7 | 98.8 | 98.8 | - | 86.4 | 84.1 | 86.1 |
| SRV-1 | 80.8 | 80.8 | 80.9 | 81.1 | 81.1 | - | 83.6 | 96.2 |
| SRV-2 | 78.3 | 78.3 | 78.4 | 78.6 | 78.5 | 79.2 | - | 83.6 |
| SRV-3 | 80.4 | 80.4 | 80.5 | 80.5 | 80.6 | 92.8 | 79.6 | - |
The lower half is the comparison of full-length gag nucleotide (1980 bp) identity percentage; the upper half, a comparison of full-length gag amino acid (659 aa) identity percentage.
V1 is the abbreviation of virus full name SRV4/TEX/2009/V1.
V2: SRV4/TEX/2009/V2.
V3: SRV4/TEX/2009/V3.
SRV-4: SRV4/CALIF/1984.
Because no genetic sequences of SRV-4 had been documented, the full-length gag gene was directly sequenced from the viral genome of SRV4/CALIF/1984 (GQ454446) and used as a reference strain to compare with our new virus strains SRV4/TEX/2009/V1-3. Nucleotide sequences identities ranged from 99.3% to 99.4% and amino acid identities were 100% (Table 1). This result confirmed that our virus strains SRV4/TEX/2009/V1-3 were SRV-4 as well.
The prt gene
The second ORF in the viral genome was 945 bp long, encoding a 314 aa protease protein. Because it overlapped a portion of the downstream of gag and the upstream of pol, ribosomal frameshift was suspected to be used in expressing the gag-prt and gag-prt-pol precursor polyproteins (Sonigo et al., 1986). Although the Prt amino acid sequence of SRV4/TEX/2009/V1 strain displayed 84.4%, 80.9% and 84.4% identities to that of SRV-1, 2 and 3, respectively (Supplemental Table 1), five conserved dUTPase motifs in the 5′ end of Prt were found. Therefore, dUTPase activity was believed to be present in the protease protein (Elder et al., 1992; McGeoch, 1990). A potentially active protease site (Asp-Thr-Gly) was located in the 3′ end of Pol (amino acid position 188 to 190), inferring its role as an aspartyl protease.
The pol gene
The third ORF was 2616 bp long, containing a total of 870 aa residues. Two successive frameshifts might have been used to make the long gag-prt-pol fusion protein as proposed in other SRV/Ds (Sonigo et al., 1986; van der Kuyl et al., 1997). The first frameshift (−1) occurred near the 3′ end of the gag ORF and continued into the pro and pol ORFs; near the 3′ end of the prt ORF, the second frameshift (−1) occurred and continued into the pol ORF. The Pol protein of SRV4/TEX/2009/V1 exhibited 84.5%, 80.7%, and 84.2% identities to that of SRV-1, SRV-2, and SRV-3, respectively (Supplemental Table 2). One of the sequence variations was the Pol protein length difference as compared to other exogenous SRV/Ds. The first amino acid residue was the second residue of the corresponding region of SRV-1, 2 and 3, and an additional 4 amino acid residues occurred at the 3′end. Interestingly, this structural feature was the same as the Pol protein structure of simian endogenous retrovirus type D (SERV) (U85505), but the amino acid identity between SRV4/TEX/2009/V1 and SERV was only 78.4%. This suggests that SRV-4 is evolutionally related to endogenous SRV, but still more closely related to exogenous SRV.
Other than these sequence variations, the Pol protein had characteristics typical of other retrovirus Pol proteins, which contained a reverse transcriptase domain (motif A-E of DNA polymerase and RNase H activity) and endonuclease (integrase) domain. A potential proteolytic site at amino acid position 592 (Asn) was proposed to be the boundary between these two domains (Hippenmeyer and Grandgenett, 1984; Power et al., 1986).
The env gene
The fourth ORF was 1749 bp long and encoded a 582 aa envelope glycoprotein. There were 12 Asn-X-Ser/Thr (X can be any amino acid residue) glycosylation sites and 23 cystine residues found in this Env protein. A stretch of hydrophobic amino acids was observed after the start codon Met as was characteristic of signal peptides at the amino termini of many viral and cellular membrane proteins (Wickner and Lodish, 1985). A potential proteolytic cleavage site was present at position 391 (Ala) that yielded a hydrophilic outer member surface gp70 in the N-terminal domain and a hydrophobic transmembrane gp20 in the C-terminal domain. The full-length Env protein of SRV4/TEX/2009/V1 exhibited 74.1%, 70.0%, and 74.1% identities to that of SRV-1, SRV-2, and SRV-3, respectively (Supplemental Table 3).
The induction of protective immunity in SRV/D positive animals is related to the Env glycoprotein (Anderson and Torres, 1999; Hu et al., 1989). SRV/D antibody was detectable in some, but not all, of these SRV-4 infected animals (Chih-Ling Zao unpublished results). Sequence analysis of full-length env genes of SRV4/TEX/2009/V2 isolated from an SRV/D antibody positive animal showed a two amino acid difference (Thr to Ala at position 85 and Gly to Asp at position 148) from SRV4/TEX/2009/V1 and SRV4/TEX/2009/V3 that were both isolated from SRV/D antibody negative animals. It is not clear if these amino acid substitutions in the Env protein are key factors to induce humoral or cellular immunity, but the substitution from Gly to Asp in amino acid residue 148 occurs in the same antigenic determinant region (amino acid residue from147-162) for induction of neutralizing antibodies to SRV-1 (Werner et al., 1990).
The immunosuppressive peptide at Env protein amino acid position 454-485 has been proposed to be associated with inhibiting immunoregulatory functions (Sonigo et al., 1986). This sequence was also conserved in SRV4/TEX/2009/V1-3, but with an amino acid substitution of Ala to Thr at position 482 in the Env protein. More experiments will be needed to explore if this immunosuppressive peptide present in SRV4/TEX/2009/V1-3 also plays a role in inducing immuosuppression in SRV-4 infected monkeys. All SRV-4 infected animals remaining in the facility are healthy except two animals found to have diarrhea. Diarrhea in these two animals has been sporadic and their weights are steadily increasing over time. It could take several years of observation to determine if simian acquired immunodeficiency syndrome would be developed and associated with SRV-4 infection.
Phylogenetic analysis
To study the relationship of our virus strains SRV4/TEX/2009/V1-3 to other SRV/Ds, phylogenetic trees were constructed based on the equivalent regions from published SRV/D sequences. Virus strains SRV4/TEX/2009/V1-3 clustered together with SRV4/CALIF/1984 (GQ454446) and SRV/D-T (AB181392) based on full-length gag gene sequences (Fig. 2A). The same tree pattern was also observed in the phylogeny constructed by comparing SRV-5 partial gag gene (626 bp) (AF252389) to its corresponding position in other SRV/Ds’ genomes (Fig. 2B). Furthermore, phylogenetic analysis based on SRV-6 partial env (701 bp) (AY598468), SRV-7 partial pol gene (452 bp) (AY594212) and their corresponding sequences of other SRV/Ds demonstrated that our virus strains SRV4/TEX/2009/V1-3 were in an independent branch of SRV/Ds (Fig. 2C and 2D). A similar phylogenetic relationship was found by an additional phylogenetic analysis using the Maximum Likelihood method (Nei and Kumar, 2000) (Supplemental Fig. 2). All these data established that our virus strains SRV4/TEX/2009/V1-3 belong to SRV-4. They are closely related to SRV/D-T, and more distantly related to SRV-1, 2, 3, 5, 6, and 7.
Fig. 2.
Phylogenetic analysis of SRV viral genes using the neighbor-joining method. The percentages of 500 replicate trees in which the associated taxa clustered together in the bootstrap test are shown at the branch nodes (only bootstrap values >70% that indicate strong support are shown). The branch lengths of the tree denote the evolutionary distances and are drawn to scale, in which each unit represents the number of base substitutions per site. A: Phylogenetic tree based on full-length gag gene (1980 bp) among SRVs. B: Phylogenetic tree based on SRV-5 partial gag gene (626 bp) (AF252389), and its corresponding position in other SRVs’ genomes. C: Phylogenetic tree based on SRV-6 partial env gene (701 bp) (AY598468), and its corresponding position in other SRVs’ genomes. D: Phylogenetic tree based on SRV-7 partial pol gene (452 bp) (AY594212), and its corresponding position in other SRVs’ genomes.
This is the second identification of SRV-4 documented since the first SRV-4 was isolated 25 years ago. In 2005, Tsukuba Primate Center, Japan also reported a virus isolate SRV/D-T from cynomolgus monkeys (Hara et al., 2005b), which had a high identity with the gag and env genes of SRV4/TEX/2009/V1-3 and SRV4/CALIF/1984 reference strain. Gag gene data has been shown in Table 1 above. Comparison of the partial env gp70 gene (544bp) of SRV/D-T (AB197850) (Hara et al., 2007) to the corresponding position in SRV4/TEX/2009/V1-3 yielded a 95.8 to 96.1% nucleotide identity and a 92.8 to 93.9% amino acid identity. Although not reported as an SRV-4 at the time it was discovered, these high sequence identities between SRV/D-T and our SRV-4 viruses, demonstrate that SRV/D-T is in the SRV-4 group. The SRV-4 complete genome sequences published in this paper (FJ971077, FJ979638, FJ979639) will help identify SRV-4 more easily in the future. The genetic variation observed in SRV4/TEX/2009/V1-3 compared to SRV/D-T might be the evolutional divergence related to different geographic origins of the infected cynomolgus monkeys, but this will only be shown when the origin of SRV/D-T infected monkey is identified.
The origin of SRV-4 infected cynomolgus monkeys
To date, the occurrences of all SRV-4 infections (1984 in California, 2005 in Japan, 2009 in Texas) are restricted only to cynomolgus macaques (Macaca fascicularis) (long-tailed or crab-eating macaque), suggesting that this species is the natural host of SRV-4. No other monkey species have been found to carry SRV-4 thus far, but we cannot rule out that SRV-4 may also infect other monkey species besides cynomolgus macaques. All of our SRV-4 infected cynomolgus monkeys were imported from the same farm in China, but these cynomolgus monkeys did not naturally originate from China. The wide geographic distributions and the difficulties in accessing the importing and breeding history in China’s facilities, make it difficult to trace the exact geographic origin of SRV-4. The origin of SRV-4 infected cynomolgus monkeys might be elsewhere in southeast Asia, where M. fascicularis are naturally distributed from Myanmar (Burma), Thailand, Laos, Cambodia, Vietnam to the Philippines; southward through Malaysia and Indonesia; and northward to the southwest of Myanmar (Wolfheim, 1983). The 835 bp mitochondrial DNA (mtDNA) of 44 SRV-4 positive cynomolgus samples clustered either with haplotypes of the fas1c (21 of the samples) or fas1a (23 of the samples) haplogoups that are restricted to Indonesia/Malaysia and Indochina, respectively (thereby eliminating both Mauritius and the Philippines as ancestral homelands of the 44 animals) in the maximum parsimony tree. The principal components analysis (PCA) based on 14 microsatellite (STR) loci for which the 44 samples were genotyped (data not shown) was able to eliminate Mauritius as a possible region of the samples’ origin, but was unable to differentiate between those animals that belonged to mtDNA haplogroups fas1c and fas1a, suggesting a mixed ancestry in most samples. Two of the three samples sent to the Oregon National Primate Research Center (ONPRC) for single nucleotide polymorphisms (SNP) genotyping, both with Indochinese mtDNA, were assessed as having a greater than 90% probability of being fullblood Indochinese cynomolgus macaques, while the third one with Indonesian/Malaysian mtDNA, was assessed to have a 32% probability of being Indonesian. The STRUCTURE analysis based on 10 of the STR loci provided estimates of Indochinese ancestry ranging from 13%–98%; twenty-three of the 44 samples were assigned greater than 90% Indochinese ancestry while only 4 were assigned less than 25% Indochinese ancestry. As 12 of the 21 animals with Indonesian/Malaysian mtDNA exhibited greater than 90% Indochinese ancestry based on STRs, much of the admixture is likely to have occurred more than one or two generations ago. Thus, while most of the 44 cynomolgus macaques exhibit predominantly Indochinese ancestry, at least half are of mixed Indonesian/Indochinese or Malaysian/Indochinese ancestry. There is a need for SRV-4 epidemiology studies in those areas where cynomolgus macaques are naturally found to help establish its evolutionary relationship among hosts and viruses with different geographic origins.
In conclusion, a new SRV isolated from cynomolgus macaques has been identified as a SRV-4. This is only the second identification of SRV-4 infections documented since the first SRV-4 isolation in California in 1984. The emergence of this virus probably originated from cynomolgus monkeys originating from one or more mainland southeast Asian countries. Because of the limited recognition of SRV-4 in the past, there may have been a large number of undetected presence of SRV-4 circulating in cynomolgus monkeys. The two SRV/D-T prevalence surveys: 22.4% prevalence (11 positive out of 49 cynomolgus monkeys tested) found in 2005 (Hara et al., 2005a) and 13.6% prevalence (57 positive out of 419 cynomolgus monkeys tested) found in 2010 (Fujimoto et al., 2010) at Tsukuba Primate Center in Japan provide two examples to support this hypothesis since these SRV/D-T positive rates can be represented as SRV-4 prevalence as well. Therefore, the complete genome sequence of SRV-4 presented in this paper will be useful for molecular diagnosis of SRV-4 and will be beneficial to control endemic SRV/D in monkey populations.
Materials and methods
Animal history
In December 2008, 200 cynomolgus monkeys were imported from the People’s Republic of China to Covance Research Products in Denver, PA, U.S.A. A total of 24 animals were found to be SRV/D positive through routine PCR and antibody screening by ELISA and/or Western blotting during the quarantine period (manuscript in preparation). In March 2009, 17 animals from a second group of 177 cynomolgus monkeys imported from the same farm in China to Covance Research Products in Alice, TX, were SRV/D positive. By the end of July 2009, 4 more animals also became SRV/D positive. To date, 45 of these imported cynomolgus monkeys are SRV/D positive. Two of these 45 SRV/D animals have experienced diarrhea, but both animals have normal body weight gains. All of the other animals are healthy with no clinical signs of SRV/D infection. The hematocrit values, and lymphocyte, neutrophil, and platelet counts are normal in all animals. The average age of these animals is 3.3 years (age ranges between 2 to 6 years) by March 2009.
Virus isolation
PBMCs were isolated from 5 ml heparin blood using Lymphocyte Separation Medium (Mediatech; Manassas, VA) according to the manufacturer’s instructions. PBMCs were cultured along with Raji cells (Burkitt’s lymphoma B-cell line, ATCC #CCL-86) in RPMI-1640 supplemented with 10% fetal bovine serum (Invitrogen; Grand Island, NY) and antibiotic-antimycotic solution (Sigma; St. Louis, MO). Viral isolations were monitored daily for viral cytopathic effect.
Immunofluorescent assay (IFA)
SRV infected Raji cells were pelleted at 430 ×g for 5 min, and washed twice with cold phosphate-buffered saline (PBS; Invitrogen; Grand Island, NY). Optimal cell numbers were placed in each well of IFA slides. Slides were dried and fixed using ice cold acetone (Fisher Scientific, Pittsburgh, PA). Cells were stained with the primary antibody (SRV/D positive animal sera pool collected from VRL Laboratories) at 37°C for 30 min, followed by incubation with a FITC labeled goat anti-human IgG conjugate (0.1% Evans Blue; Bion, Des Plaines, IL). Slides were mounted using mounting medium (Bion; Des Plaines, IL) and observed under a Nikon ECLIPSE 55i microscope.
PCR of SRV-4 genes for complete genome assembly
The proviral DNA was extracted from whole blood or virus infected Raji cells using the Generation capture column kit (QIAGEN, Valencia, CA). PCR was performed in a 60 μl volume that included DNA template, primers (0.2 μM each), and Platinum PCR SuperMix (Invitrogen, Carlsbad, CA). Primers for 5′LTR, 3′LTR, gag, prt, pol and env gene PCR are listed in Table 2. These primers were designed by primer walking from conserved known SRV/D sequences. PCR cycles used in a 2720 Thermal Cycler (Applied Biosystems, Foster City, CA) were as follows: 94 °C for 2 min, 35 cycles of 94 °C for 1 min, 54 °C for 1 min, 72° C for 4 min and one cycle of 72 °C for 10 min.
Table 2.
Primers list for SRV-4 viral genome mapping.
| Gene | Primer | Sequence (5′-3′) | Amplification size (bp) |
|---|---|---|---|
| 5′LTR | SRV-F3a | TGTCCGGAGCCGTGCWGC | 853 |
| aSRV-R855a | AGGTCTGTTTGGGAAGTACT | ||
| 3′LTR | aSRV-F7370a | GAGAAACGCCGACAACAGT | 802 |
| aSRV-R8171a | TGTCCCGWCCCGCGGGA | ||
| gag | aSRV-F429a | AGCGAAAGTACATTGTCTTA | 2187 |
| aSRV-R2615a | TCATTATCAATDACTCCTGG | ||
| aSRV-F429b | AGCGAAAGTACATTGTCTTA | 427 | |
| aSRV-R855b | AGGTCTGTTTGGGAAGTACT | ||
| aSRV-F731b | GTCACTGCCTTTTCATACTG | 526 | |
| aSRV-R1256b | TTTGCAGTCTGACAATTAGG | ||
| aSRV-F1167b | CAAAGGAGGAACTCAAAGAA | 544 | |
| aSRV-R1710b | CCGGTCAGCATGTCAAAAT | ||
| aSRV-F1974b | CAGCTTGTCAAGCAGCTATT | 642 | |
| aSRV-R2615b | TCATTATCAATDACTCCTGG | ||
| aSRV-F1301 | AAGACTCTTCTCCTGTGCC | ||
| aSRV-R1432 | CATCTACAGTTTCTGTTACTG | ||
| prt | aSRV-F1974a | CAGCTTGTCAAGCAGCTATT | 1628 |
| aSRV-R3601a | GGAATGGTGAAGAAACAGTC | ||
| pol | SRV-F3346a | TTGCTGCTGCCCAACAGT | 2698 |
| aSRV-R6043a | ACAAGAAATGGATGCAACAT | ||
| aSRV-F4284 | ATGTGGATTCATTCACATGCA | ||
| env | SRV-F5714a | GGGCAGAGGTTCAGTCTG | 2017 |
| aSRV-R7730a | GTCATCGGATTAGGCGGTT | ||
| SRV-F6504 | GTGGTAAAGAAAAAATTGATGC |
Denotes primers used for PCR.
Denotes primers used for gag one step RT-PCR.
All primers were also used for sequencing purpose.
RT-PCR of SRV-4 gag gene
SRV-4 genomic RNA was prepared from an SRV-4 reference strain SRV4/CALIF/1984 by QIAamp viral RNA mini kit (QIAGEN, Valencia, CA). The virus strain SRV4/CALIF/1984 was first isolated at the California National Primate Research Center in 1984 from a cynomolgus monkey housed at the Public Health Laboratory in Berkeley, CA (Preston A. Marx unpublished results). RT-PCR was performed by QIAGEN OneStep RT-PCR kit (QIAGEN, Valencia, CA). In 25 μl of each RT-PCR reaction, RNA template, primers (0.6 μM each) (listed in Table 2), dNTP mix (10 μM of each dNTP), RT-PCR enzyme mix and buffer were included. RT-PCR cycles were run in a 2720 Thermal Cycler (Applied Biosystems, Foster City, CA) as follows: 50 °C for 20 min, 95 °C for 15 min, 40 cycles of 95 °C for 30 s, 45 °C for 32 s, 72 °C for 75 s and one cycle of 72 °C for 2 min.
Sequencing
PCR or RT-PCR products were purified using a QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA) and 100 ng per sample was used directly for DNA sequencing on an ABI 3130xl Genetic Analyzer (Foster City, CA) (Nucleic Acids Core Facility, University of Texas Health Science Center at San Antonio). Sequencing primers are listed in Table 2. Sequence identities among different viruses were calculated by global alignment methods in LALIGN (Myers and Miller, 1988).
Phylogenetic analysis and GenBank accession numbers
All sequence data were converted to FASTA format and sequence alignments were done by ClustalW (Larkin et al., 2007) with default settings in MEGA version 4 (Tamura et al., 2007). Phylogenetic analysis was performed by neighbor-joining method in MEGA version 4. The evolutionary distances were computed by Kimura 2-parameter method (Kimura, 1980) and were in the units of the number of base substitutions per site. Condon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated from the dataset. The SERV (baboon endogenous type D retrovirus) sequence was used as an outgroup. Consensus trees were generated using the datasets that were obtained using 500 reshuffles by bootstrapping.
The complete genome of SRV/D strains SRV4/TEX/2009/V1, SRV4/TEX/2009/V2, SRV4/TEX/2009/V3, and full-length gag sequence of SRV4/CALIF/1984 were assembled and deposited into the National Center for Biotechnology Information (NCBI)’s GenBank under the assigned accession number FJ971077, FJ979638, FJ979639 and GQ454446. Reference sequences of SRV-1 (M11841), SRV-2 (AF126467), SRV-3 (M12349), SRV-5 gag (AF252389), SRV-6 env (AY598468), SRV-7 pol (AY594212), SRV/D-T gag (AB181392), SRV/D-T env (AB197850), SERV (U85505) and chimeric type C/type D baboon endogenous virus (BaEV) (D10032) were included for comparison.
Genotyping to identify region of monkey origin
DNA of 44 SRV-4 positive cynomolgus samples (one of 45 SRV-4 positive samples was not available) was genotyped for 14 highly polymorphic microsatellite (STR) loci using primers and methods reported in Kanthaswamy et al. (2006). These genotypes (available upon request) were compared with those of reference samples from three different regional populations of cynomolgus macaques (Malaysia, Vietnam and Mauritius) in a principal components analysis (PCA) constructed as described in Kanthaswamy et al. (2008). An 835 bp fragment of mitochondrial DNA (mtDNA) was also amplified from each sample and sequenced using methods described in Smith et al. (2007) to determine the region of origin of the solely maternally inherited mtDNA genomes of the 44 samples. These samples, together with reference samples (including those from Malaysia, Indonesia, Philippines, Mauritius and Vietnam) representing all mtDNA haplogroups of cynomolgus macaques reported in Smith et al. (2007), were included in a maximum parsimony tree conducted in MEGA version 4 to identify region-specific haplogroups among the 44 samples. The tree was a 50% consensus tree and was rooted with sequences from Macaca sylvanus, M. fuscata and M. cyclopis. In addition, three samples were submitted to the ONPRC for genotyping using a panel of ancestry informative SNPs and methods described by Street et al. (2007) to assess region of origin. Finally, a STRUCTURE analysis (Prichard et al., 2000) was conducted based on 10 of the 14 STR loci for which genotypes were also available for an Indochinese (i.e., Vietnamese) population and four Indonesian/Malaysian populations (Singapore, Bintan Island, Mauritius and Sarawak). For the analysis, the number of ancestral populations (K) was assumed to be 5, the number of comparison populations. The program assigned each of the 44 animals in this study to each of the 5 populations with specific probabilities, QK, whose 5 values sum to 1.0. The probabilities for the four Indonesian/Malaysian populations were summed to estimate a probability of Indonesian/Malaysian ancestry for each of the 44 animals.
Supplementary Material
Acknowledgments
We thank Cecilia Po and Danny Jolly for their assistance in manuscript preparation, Michele Carreon, Susan Gauntt, Sylvia Gonzalez-Arroyo, and Tiffany Rodriguez for SRV/D PCR and antibody screening, Lorna Millen, Amando Elizondo, Daniel Buentello for collecting samples from SRV-4 infected animals, and Debra George for conducting the maximum parsimony and STRUCTURE analyses. We also thank Dr. Betsy Ferguson at ONPRC for genotyping some samples and helpful discussion. This work was funded by VRL Laboratories, Covance Research Products and NIH grants R24RR005090 and R24RR025871 to DGS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson DE, Torres JV. Simian retrovirus receptor and neutralization mechanism by antibodies to the envelope glycoprotein. Viral Immunol. 1999;12:47–56. doi: 10.1089/vim.1999.12.47. [DOI] [PubMed] [Google Scholar]
- Bradac J, Hunter E. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology. 1984;138:260–275. doi: 10.1016/0042-6822(84)90350-7. [DOI] [PubMed] [Google Scholar]
- Elder JH, Lerner DL, Hasselkus-Light CS, Fontenot DJ, Hunter E, Luciw PA, Montelaro RC, Phillips TR. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiomto K, Takano J, Narita T, Hanari K, Shimozawa N, Sankai T, Yosida T, Terao K, Kurata T, Yasutomi Y. Simian betaretrovirus infection in a colony of cynomolgus monkeys (Macaca fascicularis) Comp Med. 2010;60:51–53. [PMC free article] [PubMed] [Google Scholar]
- Giddens WE, Jr, Tsai CC, Morton WR, Ochs HD, Knitter GH, Blakley GA. Retroperitoneal fibromatosis and acquired immunodeficiency syndrome in macaques. Pathologic observations and transmission studies. Am J Pathol. 1985;119:253–263. [PMC free article] [PubMed] [Google Scholar]
- Hara M, Kikuchi T, Ono F, Takano J, Ageyama N, Fujimoto K, Terao K, Baba T, Mukai R. Survey of captive cynomolgus macaque colonies for SRV/D infection using polymerase chain reaction assays. Comp Med. 2005a;55:145–149. [PubMed] [Google Scholar]
- Hara M, Kikuchi T, Sata T, Nakajima N, Ami Y, Sato Y, Tanaka K, Narita T, Ono F, Akari H, Terao K, Mukai R. Detection of SRV/D shedding in body fluids of cynomolgus macaques and comparison of partial gp70 sequences in SRV/D-T isolates. Virus Genes. 2007;35:281–288. doi: 10.1007/s11262-007-0076-1. [DOI] [PubMed] [Google Scholar]
- Hara M, Sata T, Kikuchi T, Nakajima N, Uda A, Fujimoto K, Baba T, Mukai R. Isolation and characterization of a new simian retrovirus type D subtype from monkeys at the Tsukuba Primate Center, Japan. Microbes Infect. 2005b;7:126–131. doi: 10.1016/j.micinf.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Heberling RL, Barker ST, Kalter SS, Smith GC, Helmke RJ. Oncornavirus: isolation from a squirrel monkey (Saimiri sciureus) lung culture. Science. 1977;195:289–292. doi: 10.1126/science.63993. [DOI] [PubMed] [Google Scholar]
- Henderson LE, Sowder R, Smythers G, Benveniste RE, Oroszlan S. Purification and N-terminal amino acid sequence comparisons of structural proteins from retrovirus-D/Washington and Mason-Pfizer monkey virus. J Virol. 1985;55:778–787. doi: 10.1128/jvi.55.3.778-787.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson RV, Maul DH, Lerche NW, Osborn KG, Lowenstine LJ, Prahalada S, Sever JL, Madden DL, Gardner MB. Clinical features of simian acquired immunodeficiency syndrome (SAIDS) in rhesus monkeys. Lab Anim Sci. 1984;34:140–145. [PubMed] [Google Scholar]
- Hippenmeyer PJ, Grandgenett DP. Requirement of the avian retrovirus pp32 DNA binding protein domain for replication. Virology. 1984;137:358–370. doi: 10.1016/0042-6822(84)90228-9. [DOI] [PubMed] [Google Scholar]
- Hu SL, Zarling JM, Chinn J, Travis BM, Moran PA, Sias J, Kuller L, Morton WR, Heidecker G, Benveniste RE. Protection of macaques against simian AIDS by immunization with a recombinant vaccinia virus expressing the envelope glycoproteins of simian type D retrovirus. Proc Natl Acad Sci U S A. 1989;86:7213–7217. doi: 10.1073/pnas.86.18.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthaswamy S, von Dollen A, Kurushima JD, Alminas O, Rogers J, Ferguson B, Lerche NW, Allen PC, Smith DG. Microsatellite markers for standardized genetic management of captive colonies of rhesus macaques (Macaca mulatta) Am J Primatol. 2006;68:73–95. doi: 10.1002/ajp.20207. [DOI] [PubMed] [Google Scholar]
- Kanthaswamy S, Satkoski J, George D, Kou A, Erickson BJ, Smith DG. Hybridization and stratification of nuclear genetic variation in Macaca mulatta and Macaca fascicularis. Int J Primatol. 2008;29:1295–1311. doi: 10.1007/s10764-008-9295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lerche NW, Osborn KG. Simian retrovirus infections: potential confounding variables in primate toxicology studies. Toxicol Pathol. 2003;31(Suppl):103–110. doi: 10.1080/01926230390174977. [DOI] [PubMed] [Google Scholar]
- Lerche NW, Osborn KG, Marx PA, Prahalada S, Maul DH, Lowenstine LJ, Munn RJ, Bryant ML, Henrickson RV, Arthur LO, et al. Inapparent carriers of simian acquired immune deficiency syndrome type D retrovirus and disease transmission with saliva. J Natl Cancer Inst. 1986;77:489–496. [PubMed] [Google Scholar]
- Lerche NW, Switzer WM, Yee JL, Shanmugam V, Rosenthal AN, Chapman LE, Folks TM, Heneine W. Evidence of infection with simian type D retrovirus in persons occupationally exposed to nonhuman primates. J Virol. 2001;75:1783–1789. doi: 10.1128/JVI.75.4.1783-1789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Axthelm MK, Machida CA. Simian retrovirus serogroup 5: partial gag-prt sequence and viral RNA distribution in an infected rhesus macaque. Virus Genes. 2000;21:241–248. doi: 10.1023/a:1008104000920. [DOI] [PubMed] [Google Scholar]
- Marracci GH, Avery NA, Shiigi SM, Couch G, Palmer H, Pilcher KY, Nichols H, Hallick LM, Axthelm MK, Machida CA. Molecular cloning and cell-specific growth characterization of polymorphic variants of type D serogroup 2 simian retroviruses. Virology. 1999;261:43–58. doi: 10.1006/viro.1999.9858. [DOI] [PubMed] [Google Scholar]
- Maul DH, Lerche NW, Osborn KG, Marx PA, Zaiss C, Spinner A, Kluge JD, MacKenzie MR, Lowenstine LJ, Bryant ML, et al. Pathogenesis of simian AIDS in rhesus macaques inoculated with the SRV-1 strain of type D retrovirus. Am J Vet Res. 1986;47:863–868. [PubMed] [Google Scholar]
- McGeoch DJ. Protein sequence comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton WR, Agy MB, Capuano SV, Grant RF. Specific pathogen-free macaques: definition, history, and current production. ILAR J. 2008;49:137–144. doi: 10.1093/ilar.49.2.137. [DOI] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. CABIOS. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Nandi JS, Bhavalkar-Potdar V, Tikute S, Raut CG. A novel type D simian retrovirus naturally infecting the Indian Hanuman langur (Semnopithecus entellus) Virology. 2000;277:6–13. doi: 10.1006/viro.2000.0567. [DOI] [PubMed] [Google Scholar]
- Nandi JS, Tikute SA, Chhangani AK, Potdar VA, Tiwari-Mishra M, Ashtekar RA, Kumari J, Walimbe A, Mohnot SM. Natural infection by simian retrovirus-6 (SRV-6) in Hanuman langurs (Semnopithecus entellus) from two different geographical regions of India. Virology. 2003;311:192–201. doi: 10.1016/s0042-6822(03)00187-9. [DOI] [PubMed] [Google Scholar]
- Nandi JS, Van Dooren S, Chhangani AK, Mohnot SM. New simian beta retroviruses from rhesus monkeys (Macaca mulatta) and langurs (Semnopithecus entellus) from Rajasthan, India. Virus Genes. 2006;33:107–116. doi: 10.1007/s11262-005-0032-x. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press; New York: 2000. [Google Scholar]
- Power MD, Marx PA, Bryant ML, Gardner MB, Barr PJ, Luciw PA. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, McDonough JW, George DA. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol. 2007;69:182–198. doi: 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- Street SL, Kyes RC, Grant R, Ferguson B. Single nucleotide polymorphisms (SNPs) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics. 2007;8:480. doi: 10.1186/1471-2164-8-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–15999. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Temin HM. Structure, variation and synthesis of retrovirus long terminal repeat. Cell. 1981;27:1–3. doi: 10.1016/0092-8674(81)90353-6. [DOI] [PubMed] [Google Scholar]
- Thayer RM, Power MD, Bryant ML, Gardner MB, Barr PJ, Luciw PA. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987;157:317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Benveniste RE, Sherr CJ, Schlom J, Schidlovsky G, Stephenson JR. Isolation and characterization of a new type D retrovirus from the asian primate, Presbytis obscurus (spectacled langur) Virology. 1978;84:189–194. doi: 10.1016/0042-6822(78)90231-3. [DOI] [PubMed] [Google Scholar]
- Tsai CC, Follis KE, Snyder K, Windsor S, Thouless ME, Kuller L, Morton WR. Maternal transmission of type D simian retrovirus (SRV-2) in pigtailed macaques. J Med Primatol. 1990;19:203–216. [PubMed] [Google Scholar]
- van der Kuyl AC, Mang R, Dekker JT, Goudsmit J. Complete nucleotide sequence of simian endogenous type D retrovirus with intact genome organization: evidence for ancestry to simian retrovirus and baboon endogenous virus. J Virol. 1997;71:3666–3676. doi: 10.1128/jvi.71.5.3666-3676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus HE. Form and function of retroviral proviruses. Science. 1982;216:812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Voevodin AF, Marx PA., Jr . Simian Virology. Wiley-Blackwell; Ames, IA: 2009. Betaretroviruses; p. 169. [Google Scholar]
- Werner LL, Malley A, Torres JV, Leung CY, Kwang HS, Benjamini E. Synthetic peptides of envelope proteins of two different strains of simian AIDS retrovirus (SRV-1 and SRV-2) represent unique antigenic determinants for serum neutralizing antibodies. Mol Immunol. 1990;27:1103–1111. doi: 10.1016/0161-5890(90)90098-k. [DOI] [PubMed] [Google Scholar]
- Wickner WT, Lodish HF. Multiple mechanisms of protein insertion into and across membranes. Science. 1985;230:400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Wolfheim JH. Primates of the World: Distribution, Abundance, and Conservation. University of Washington Press; Seattle, WA: 1983. Cercopithecidae Old World Monkeys: Macaca fascicularis Long-tailed or Crab-eating Macaque; pp. 475–483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.