Summary
Small RNAs (sRNA) that act by base pairing with trans-encoded mRNAs modulate metabolism in response to a variety of environmental stimuli. Here, we describe an Hfq-binding sRNA (FnrS) whose expression is induced upon a shift from aerobic to anaerobic conditions and which acts to down regulate the levels of a variety of mRNAs encoding metabolic enzymes. Anaerobic induction in minimal medium depends strongly on FNR but is also affected by ArcA and CRP. Whole genome expression analysis showed that the levels of at least 32 mRNAs are down regulated upon FnrS overexpression, many of which are predicted to base pair with FnrS by TargetRNA. The sRNA is highly conserved across its entire length in numerous enterobacteria, and mutation analysis revealed that two separate regions of FnrS base pair with different sets of target mRNAs. Many of the target genes previously reported to be down regulated in an FNR-dependent manner lack recognizable FNR binding sites. We thus suggest that FnrS extends the FNR regulon and increases the efficiency of anaerobic metabolism by repressing the synthesis of enzymes that are not needed under these conditions.
Keywords: base pairing, FNR, CRP, ArcA, sodB
Introduction
Small RNAs (sRNAs) are now known to be key regulators in all organisms. Approximately 80 sRNAs, generally between 80 and 200 nucleotides in length, have been identified in Escherichia coli. The majority of these sRNAs act either by base pairing with target RNAs or by binding proteins and modifying their activities. Base pairing sRNAs can be encoded in cis or trans with respect to their targets. The cis-encoded sRNAs have extensive complementarity with their targets, while the trans-encoded sRNAs exhibit more limited complementarity with target mRNAs and require the RNA chaperone protein Hfq for base pairing [reviewed in (Waters and Storz, 2009)].
Most of the trans-encoded base pairing sRNAs in E. coli are induced in response to a specific environmental condition and modulate metabolism by regulating the expression of enzymes and transporters. For example, the OmrA and OmrB RNAs, whose transcription is induced by the OmpR-EnvZ two-component regulators in response to high osmolarity, and the MicA and RybB sRNAs, whose expression is regulated by σE in response to perturbations in the cell envelope, all modulate the synthesis of major outer membrane porins [reviewed in (Guillier et al., 2006, Vogel and Papenfort, 2006)]. The Spot42 RNA, whose levels are modulated by CRP (cAMP receptor protein) in response to glucose availability, represses the synthesis of enzymes involved in carbon metabolism (Polayes et al., 1988, Moller et al., 2002). Finally, the RyhB RNA, whose levels increase upon iron depletion when Fur repression is relieved, down regulates a variety of iron-containing enzymes thus making iron available for the most critical enzymes under these iron-limiting conditions (Massé and Gottesman, 2002, Massé et al., 2005).
E. coli is able to grow in both aerobic and anaerobic environments and not surprisingly a wide reprogramming in gene expression, with significant effects on cell metabolism, is observed upon shifts between different low and high oxygen conditions (Salmon et al., 2003, Kang et al., 2005, Constantinidou et al., 2006). Two transcriptional regulators, FNR (fumarate and nitrate reduction) and ArcA (aerobic respiratory control), whose activities are modulated by oxygen availability, impact many of the changes in gene expression associated with a transition from aerobic to anaerobic metabolism. FNR is a direct sensor of oxygen availability. This protein is only active under anaerobic conditions because of the requirement for an [4Fe-4S]2+ cluster which permits FNR dimerization. ArcA is the cytosolic response regulator of a two-component pair, in which ArcB is the transmembrane histidine kinase sensor. Oxygen levels are sensed indirectly by the ArcA/B pair (Georgellis et al., 2001). During aerobiosis, oxidized quinones repress autophosphorylation of ArcB. Under anaerobic conditions, when the levels of oxidized quinone decrease, ArcB becomes autophosphorylated and transfers the phosphate group to ArcA to activate the response regulator. FNR has been shown to repress a number of genes with aerobic functions as well as activate even more genes encoding enzymes of anaerobic pathways (Kang et al., 2005, Salmon et al., 2003, Constantinidou et al., 2006). In contrast, ArcA represses many genes encoding enzymes required for aerobic respiration and acts as a positive regulator of a few genes required for anaerobic metabolism (Iuchi and Lin, 1988, Iuchi et al., 1989) [reviewed in (Gunsalus and Park, 1994)]. There is also significant overlap in the genes modulated by the two regulators. While a subset of the genes induced under anaerobic conditions have been found to have FNR and/or ArcA binding sites, other genes whose expression changes in fnr and arcA mutants do not, raising the question of how these genes are regulated (Constantinidou et al., 2006, Liu and De Wulf, 2004).
In addition to ArcA and FNR, the expression of anaerobic genes is modulated by transcriptional regulators that sense glucose availability or the levels of electron acceptors. For example, CRP activates numerous genes involved in the catabolism of amino acids and sugar when glucose levels are low and cAMP levels are high (Buchet et al., 1999). The amino acid sequence of CRP is similar to that of FNR, and many of the sites recognized by FNR can also be recognized by CRP (Sawers et al., 1997). Furthermore, there are a number of two-component systems that respond to the presence of different electron acceptors. Thus, for example, nitrate, the preferred electron acceptors in the absence of oxygen, and/or nitrite are sensed by the dual two-component systems NarX/L and NarQ/P [reviewed in (Stewart, 2003)].
Here we describe an sRNA, denoted FnrS, whose expression is induced by anaerobic conditions in an FNR- and ArcA-dependent manner. This sRNA down regulates at least 32 mRNAs, many of which encode enzymes directly involved in energy metabolism and previously reported to be part of the FNR regulon. For the five mRNA targets (sodB, maeA, gpmA, folE and folX) examined in more detail, two separate regions of FnrS were shown to base pair with different sets of targets. We suggest that this allows the FnrS RNA, which is conserved across its entire length, to regulate the expression of a large set of genes, thus extending the FNR regulon.
Results
Novel sRNA in the ydaN-dbpA intergenic region
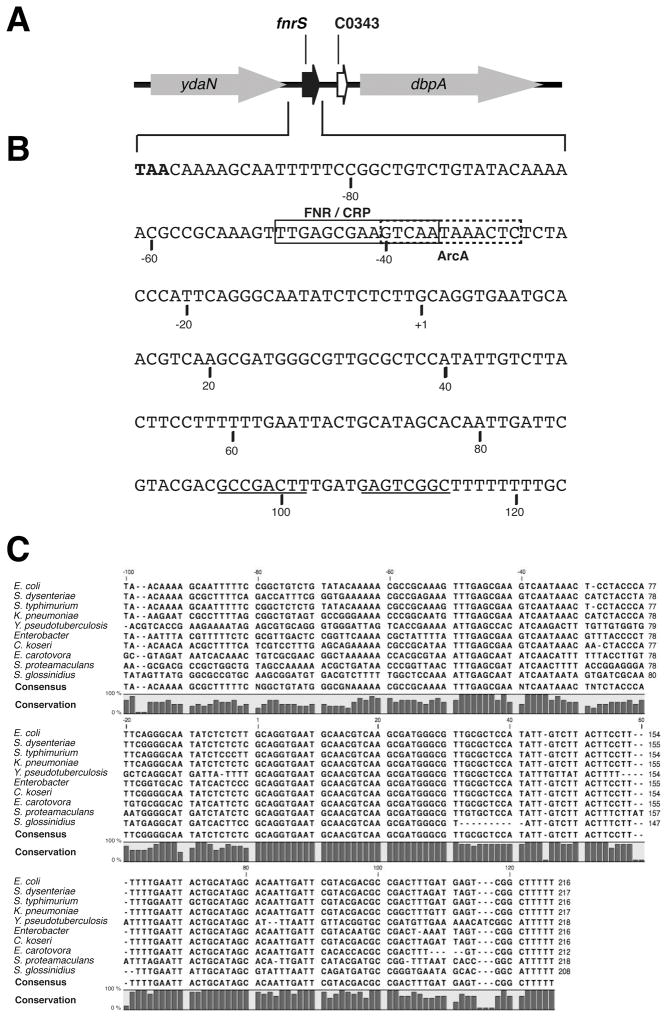
Based on sequence conservation, the 477 base pair intergenic region between E. coli ydaN and dbpA was predicted to encode a sRNA, but no signal was detected by Northern analysis (Wassarman et al., 2001, Carter et al., 2001). However, recent tiling array analysis of RNAs that co-immunoprecipitate with the RNA chaperone protein Hfq in E. coli (Zhang et al., unpublished data) and deep sequencing of Hfq binding sRNAs in Salmonella typhimurium (Sittka et al., 2008) again indicated a sRNA was encoded on the Watson strand of the intergenic region, overlapping the section showing the highest conservation (Fig. 1A). To detect this sRNA by Northern analysis, we used an oligonucleotide probe complementary to the region showing a signal in the tiling array analysis to probe total RNA isolated from wild-type cells as well as RNA that co-immunoprecipated with Hfq (data not shown). A band of slightly longer than 100 nucleotides was barely visible for the Hfq immunoprecipitation sample. The 5′ end of this transcript was mapped to position 1,407,153 by 5′ RACE analysis and a Rho independent terminator was predicted at approximately 100 nt from the +1 (Fig. 1B). An alignment of this region showed that the entire sequence of this sRNA, denoted FnrS as explained below, is conserved among enterobacterial species (Fig. 1C). One flanking gene, ydaN, which is predicted to encode a zinc transporter in Salmonella (Worlock and Smith, 2002), is always found upstream of fnrS. The other flanking gene is more variable. In Escherichia, Salmonella, Shigella, Citrobacter and Klebsiella species, the downstream gene is dpbA, which encodes a 3′ to 5′ RNA helicase (Diges and Uhlenbeck, 2005), while genes encoding a MerR family transcriptional regulator, a tRNA thiolase, a glyoxylase resistance protein and an uncharacterized ORF flank FnrS in Yersinia, Erwinia, Serratia and Sodalis, respectively.
Fig. 1. FnrS RNA encoded in the ydaN-dbpA intergenic region.
A. Map of ydaN-dbpA region.
B. Sequence of the first 217 nucleotides of the ydaN-dbpA intergenic region. +1 indicates the mapped start of the FnrS RNA, the stop codon of ydaN is in bold, the probable FnrS Rho-independent terminator is underlined, and putative binding sites for FNR/CRP and ArcA are boxed in continuous and dotted lines, respectively.
C. Alignment of the region encompassing FnrS created using the CLC sequence viewer (www.clcbio.com). Sequences used for this alignment are from Escherichia coli K12, Shigella dysenteriae Sd197, Salmonella thyphimurium LT2, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Yersinia pseudotuberculosis PB1/+, Enterobacter sp. 368, Citrobacter koseri ATCC BAA-895, Erwinia carotovora subsp. atroseptica SCRI1043, Serratia proteamaculans 568, Sodalis glossinidius str. ‘morsitans’.
Tjaden et al. reported an sRNA microarray signal in the ydaN-dpbA region (C0343), also on the Watson strand but downstream of the region probed above (Fig. 1A) (Tjaden et al., 2002). Possibly this signal corresponds to the leader of the dbpA transcript. However no signal was detected in this region by the tiling array analysis. In contrast, a tiling array signal was noted on the Crick strand. This region was predicted to encode an sRNA by Carter et al. 2001, but we did not detect a transcript in our Northern analysis (data not shown).
FnrS RNA induction during anaerobic growth
Upon examination of the sequence upstream of the FnrS, we noticed a putative FNR (TTGAT-N4-ATCAA) and/or CRP (TGTGA-N6-TCACA) binding site at −41.5 relative to the start of transcription (Eiglmeier et al., 1989). FNR and CRP can activate transcription at two different classes of promoters. Class I promoters have binding sites centered at −61.5, −71.5, −82.5 or −92.5, while class II promoters have binding sites centered at −41.5 relative to the transcriptional start site (Fig. 1B) [reviewed in (Busby and Ebright, 1999)]. In addition, we noticed a putative binding site for ArcA ([A/T]GTTAATTA[A/T]) at approximately −35, overlapping the CRP/FNR site (Lynch and Lin, 1996) and a putative binding site for NarL/NarP (TACYYMT, where Y= C or T and M = A or C) at position −22.5 (Darwin et al., 1997).
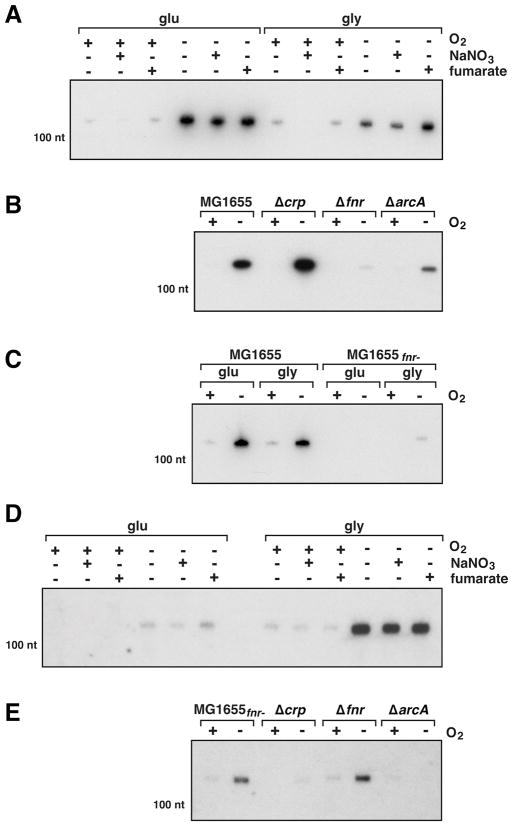
The predicted binding sites prompted us to examine FnrS levels in cells grown with and without oxygen, with different carbon sources (glucose and glycerol), and with different terminal electron acceptors (nitrate and fumarate) (Fig. 2A). This Northern analysis showed that FnrS is barely detectable in cells grown aerobically regardless of carbon source or final electron acceptor. In contrast, FnrS levels are strongly induced when the cells were shifted to anaerobic conditions. The levels of the sRNA are slightly higher in cells grown in glucose medium and appear to be somewhat repressed by nitrate, especially under aerobic conditions. These results show that regulation of FnrS is relatively complex, with maximal expression during anaerobic growth. The available carbon source and final electron acceptor also impact FnrS levels, but to a lesser extent.
Fig. 2. FnrS expression under different growth conditions and in various mutant strains.
A. MG1655 cells were grown in M63 with 0.2% glucose to OD600 ≈ 0.4 under aerobic conditions, the culture was split into multiple aliquots, the cells were collected and resuspended in the indicated medium (M63 with 0.2% glucose or 0.4% glycerol and 20 mM nitrate or 40 mM fumarate) and incubated aerobically or anaerobically for 20 min.
B. MG1655, MG1655Δcrp, MG1655 Δfnr and MG1655 ΔarcA were grown in M63 with 0.2% glucose to OD600 ≈ 0.4 under aerobic conditions, the cultures were split into two aliquots, the cells were collected and resuspended in M63 with 0.4% glycerol and 40 mM fumarate and incubated aerobically or anaerobically for 20 min.
C. MG1655 and MG1655fnr- strain were grown in M63 with 0.2% glucose to OD600 ≈ 0.4 under aerobic conditions, the cultures were split into four aliquots, cells were collected and resuspended in M63 with either 0.2% glucose or 0.4% glycerol and incubated aerobically or anaerobically for 20 min.
D. MG1655fnr- cells were grown and treated as for (A).
E. MG1655fnr-, MG1655fnr- Δcrp, MG1655fnr- Δfnr or MG1655fnr- ΔarcA were grown and treated as for (B). For all samples, total RNA (5 μg) was separated on an acrylamide gels, transferred to nitrocellulose and probed with a 32P-labelled oligonucleotide specific to FnrS. For all panels the position of the band corresponding to a 100-nucleotide marker RNA is indicated on the left. The Northern blots in (A), (B) and (C) were exposed overnight, while the Northern blots in (D) and (E) were exposed for one week.
FNR, ArcA and CRP-dependent transcription of FnrS RNA
To determine what transcriptional regulators are required for FnrS induction, we deleted the genes encoding the CRP, FNR and ArcA regulators and examined FnrS levels in these strains in glycerol medium with fumarate during aerobic and anaerobic growth. In the presence of oxygen, the sRNA is barely detectable in all strains (Fig. 2B). After a shift from aerobic to anaerobic conditions, FnrS levels are slightly higher in the Δcrp strain than in wild-type cells and barely detectable in the Δfnr strain. The anaerobic induction is also significantly reduced in the ΔarcA strain. Thus FNR (hence the name FnrS), and to a lesser extent ArcA, act together to activate FnrS transcription during anaerobic growth. Furthermore, CRP has a negative impact on anaerobic expression of FnrS.
FnrS activation by CRP and ArcA in an fnr mutant strain
In the course of this study, we found that one laboratory stock of MG1655 (hereafter referred to as MG1655fnr-) had significantly lower levels of FnrS compared to another laboratory stock of MG1655 (Fig. 2C). Since FNR had the strongest effect on FnrS expression, we sequenced the fnr gene in the MG1655fnr- strain. The sequencing revealed an insertion of six amino acids between amino acids 21 and 22 of FNR (Fig. S1). The arginine at position 10 and the serine at position 13 were also mutated to glycine and phenylalanine, respectively (Fig. S1). Previous studies showed that the [4Fe-4S]2+ cluster required to form the transcriptionally-active FNR dimer is ligated by cysteines 20, 23, 29, 122 (Sharrocks et al., 1990 1990, Melville and Gunsalus, 1990). Thus the mutations in the MG1655fnr- probably disrupt the binding of the [4Fe-4S]2+ cluster, explaining the lower FnrS expression in this strain.
Nevertheless, FnrS expression is still induced by a shift to anaerobic conditions in MG1655fnr-, though the induction is approximately 10 fold higher in glycerol than in glucose (Fig. 2D). In contrast to what was found for MG1655, no effect of the final electron acceptor was observed for MG1655fnr-. To identify the transcription factors responsible for FnrS induction, we deleted the arcA and crp genes in MG1655fnr-. Northern analysis showed that ArcA and CRP are both required for full FnrS expression in anaerobic growth (Fig. 2E). We also deleted the fnr gene; not surprisingly, the deletion does not affect FnrS expression in the MG1655fnr- strain, showing that the six amino acid insertion and point mutations completely abolish FNR activity. We conclude that FnrS expression during anaerobic growth is completely ArcA- and CRP-dependent in MG1655fnr-. Furthermore, in MG1655fnr- grown aerobically in minimal medium containing glycerol, FnrS expression is essentially CRP-dependent.
FnrS secondary structure
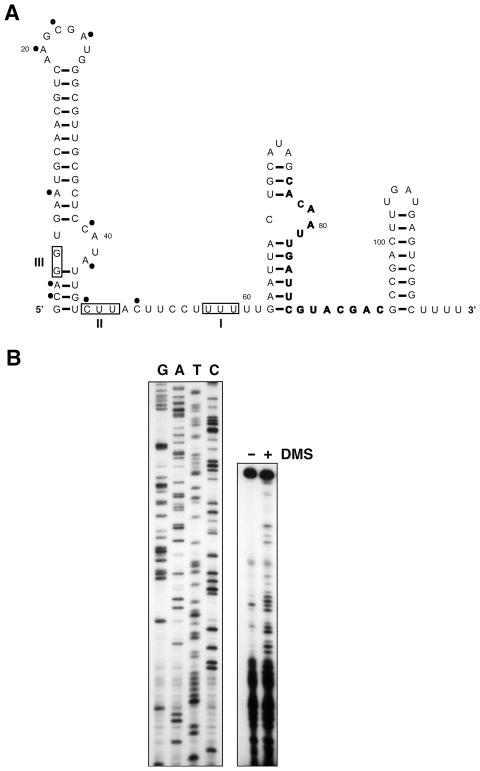
Several related secondary structures are predicted by the Mfold program (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) (Zuker, 2003) for the E. coli, Serratia, Shigella, Yersinia, Erwinia and Citrobacter FnrS RNAs. All structures contain the same Rho independent terminator. However, the first seven nucleotides at the 5′ end are alternatively predicted to be unstructured or in a small stem followed by a long stem-loop structure. The most stable predicted structure for E. coli FnrS is shown in Fig. 3A. To directly examine the FnrS structure, we carried out in vivo probing with 50 mM dimethylsulfate (DMS), which methylates unpaired adenosines and cytidines in RNA, and then carried out primer extension reactions, which terminate at the methylated nucleosides. The results of the primer extension analysis on the DMS treated samples (Fig. 3B) and the positions of the methylated residues (Fig. 3A) are consistent with the structure in Fig. 3A, with some breathing of the bottom of the first stem-loop. This structure with three stem-loops and an extended single-stranded region is typical of other Hfq-binding sRNAs (Wassarman et al., 1999).
Fig. 3. FnrS structure.
A. FnrS structure predicted by Mfold and supported by dimethylsulfate (DMS) modification data. The sequence complementary to the oligonucleotide used in the reverse transcription reaction is in bold. Dots indicate residues that reacted with DMS and boxes denote residues modified in mutants I, II and III (see Fig. 5A).
B. In vivo probing of the FnrS RNA structure. Cells were grown anaerobically in M63 with 0.2% glucose and 40 mM fumarate and half of the culture was treated with dimethylsulfate for four min. Total RNA extracted from these cultures was analyzed by primer extension reactions.
FnrS down regulation of mRNAs encoding a variety of metabolic enzymes
Given that the FnrS RNA co-immunoprecipitated with the Hfq protein, we surmised that FnrS would act by base pairing with trans-encoded mRNAs as is the case for other Hfq-bound sRNAs. To identify potential base pairing targets of FnrS, we transiently overexpressed FnrS and examined the genome-wide changes in transcript levels by microarray analysis, an approach that has led to the successful identification of other sRNA targets (Tjaden et al., 2006, Guillier and Gottesman, 2006, De Lay and Gottesman, 2009). FnrS was cloned behind the arabinose-inducible PBAD promoter of pAZ3, and expression was induced by the addition of 0.2% arabinose for 15 min. As a control, strains carrying the parental plasmid (pAZ3) were also treated with 0.2% arabinose. Microarray analysis revealed that 32 genes were repressed more than two fold after FnrS over-expression in three independent experiments (Table 1 and Table S1).
Table 1.
Genes repressed by FnrS overexpression.
| gene/ORF | Descriptiona | Ratiob | FnrS base-pairing corec | FNR regulond | FNR sitee |
|---|---|---|---|---|---|
| Cytochrome | |||||
| cydD | ATP-binding component of cytochrome-related transport, Zn sensitive; NarL and ArcA regulated | 3.8 | SS | N | |
| cydC | ATP-binding component of cytochrome-related transport; NarL and ArcA regulated | 6.3 | N | ||
| yceI | hypothetical protein, homology with cytochrome b561 of Caulobacter | 4.5 | SS | Y | |
| Central intermediary metabolism/energy metabolism | |||||
| sfcA/maeA | NAD-linked malate dehydrogenase (malic enzyme) | 7.3 | SS | N | |
| mqo | malate:quinone oxidoreductase | 4.3 | SS | Y | 11.5 |
| adhPf | alcohol dehydrogenase, propanol-preferring | 3.7 | SS | Y | |
| gpmA | phosphoglycerolmutase 1; Fur regulated | 3.4 | SS | Y | |
| dld | D-lactate dehydrogenase, FAD protein, NADH independent | 2.5 | Y | ||
| nfsA | modulator of drug activity; SoxS regulated | 2.3 | SS | N | |
| Folate biosynthesis | |||||
| folX | D-erythro-7,8-dihydroneopterin tri-phosphate epimerase | 3.7 | 5′ | Y | |
| folEf | GTP cyclohydrolase I | 2.8 | 5′ | Y | |
| Amino acid biosynthesis | |||||
| metEf | tetrahydropteroyltriglutamate methyltransferase; MetR, MetJ regulated | 2.4 | SS/5′ | ||
| tyrB | tyrosine aminotransferase, tyrosine repressible; TyrR regulated | 2.3 | Y | ||
| Metalloprotease | |||||
| dcpf | dipeptidyl carboxypeptidase II | 3.6 | N | ||
| yggGf | hypothetical protein, Putative metalloprotease lipoprotein | 2.6 | N | ||
| ybjC | hypothetical protein, predicted metal-dependent membrane protease; SoxS regulated | 2.5 | N | ||
| Stress resistance proteins | |||||
| sodB | superoxide dismutase; Fur, CRP, HNS and IHF regulated | 4.8 | 5′ | Y | |
| ydhD/grxD | glutaredoxin 4 | 3.4 | SS | N | |
| marA | multiple antibiotic resistance, transcriptional activator of defense system; CRP and Fis regulated | 2.1 | SS/5′ | N | |
| Transporters/outer membrane proteins | |||||
| ygiW | hypothetical outer membrane protein | 5.2 | Y | ||
| yobA | hypothetical protein, homolog to copper resistance protein, putative cation transporter | 4.2 | SS | Y | −26.5 |
| yebZ | putative resistance protein, copper ion homeostasis | 4.3 | Y | −26.5 | |
| yebY | hypothetical protein | 3.8 | Y | −26.5 | |
| bhsA/ycfR | hypothetical protein involved in stress resistance and biofilm formation, putative outer membrane | 3.8 | 5′ | N | |
| chaA | calcium protein antiporter | 3.1 | N | ||
| Others | |||||
| azuC | 28 aa ORF; CRP regulated | 4.4 | N | ||
| yncE | putative receptor, possible ATP-binding protein | 3.9 | N | ||
| yfcL | hypothetical protein | 2.7 | N | ||
| eco | serine protease inhibitor convergent to mqo | 2.5 | N | ||
| yoaB | hypothetical protein, putative translation initiation inhibitor | 2.4 | N | ||
| ycaO | hypothetical protein | 2.3 | N | ||
Categories of gene function based on http://www.ecocyc.org/.
Average ratio of signal for pAZ3 control:pAZ3-FnrS for three experiments.
Region of FnrS predicted to base pair with target mRNA by TargetRNA (SS = single strand region between the first and second stem loops, 5′ = 5′ end).
Genes suggested to part of FNR regulon based on microarray data from (Constantinidou et al., 2006) (Y = yes, N = No).
Position of known or predicted FNR binding sites relative to transcriptional start sites based on (Constantinidou et al., 2006).
Targets known or predicted to bind zinc.
The mRNAs whose levels were repressed by FnrS encode proteins required for a variety of processes. Several of the mRNAs whose levels are down regulated encoded dehydrogenases. Two of the dehydrogenases are NAD-dependent; malate dehydrogenase (maeA) which catalyzes the conversion of malate to pyruvate (Yamaguchi, 1979), and the ethanol dehydrogenase/reductase (adhP), which can convert ethanol to an acetaldehyde or ketone and can also catalyze the reverse reaction of acetaldehyde reduction to ethanol. A third dehydrogenase, D-lactate dehydrogenase (dld), which converts lactate to pyruvate and is required for aerobic growth on lactate, is a membrane bound flavoprotein (Haugaard, 1959). Another down regulated mRNA encodes the most abundant phosphoglycerate mutase isozyme (gpmA), which converts 3-phosphoglycerate into 2-phosphoglycerate (Fraser et al., 1999). The sodB mRNA, already known to be down regulated by the RyhB RNA (Massé and Gottesman, 2002) and encoding one of the three E. coli superoxide dismutase enzymes which protect against superoxide radicals generated during aerobic growth, is also repressed (Carlioz and Touati, 1986, Farr et al., 1986). Finally, two mRNAs encoding enzymes involved in folate metabolism, dihydroneopterin triphosphate epimerase (folX) and a GTP cyclohydrolase I (folE), are down regulated by FnrS overexpression.
Half of the genes whose expression is down regulated by FnrS overproduction were reported to be in the FNR regulon based on microarray analysis of a wild-type MG1655 strain and the corresponding Δfnr mutant (Constantinidou et al., 2006). However, most of these genes do not have a predicted FNR binding site indicating that the effects of FNR could be indirect, possibly mediated by FnrS (Table 1).
To confirm FnrS regulation of putative targets encoding a broad range of enzymes, we specifically examined the levels of the maeA and gpmA mRNAs (central metabolism), the sodB mRNA (oxidative stress) and the folE and folX mRNAs, (folate metabolism) upon FnrS overexpression. Total RNA was isolated from wild-type cells carrying the pAZ3 control plasmid or pAZ3-FnrS plasmid and treated with 0.2% arabinose to induce the PBAD promoter. After 15 min, cells were washed two times in LB + 0.2% glucose, to repress the PBAD promoter. The cultures were then incubated an additional 15 min. The RNA isolated from the samples at different time points after induction was subjected to Northern analysis with oligonucleotide probes specific to maeA, gpmA, sodB, folE, folX as well as to FnrS (Fig. 4). We detected transcripts of the size expected for the monocistronic mRNA for all of the genes. The levels of all the mRNAs decreased upon FnrS induction and then increased after the arabinose was removed.
Fig. 4. FnrS repression of maeA, gpmA, sodB, folE and folX.
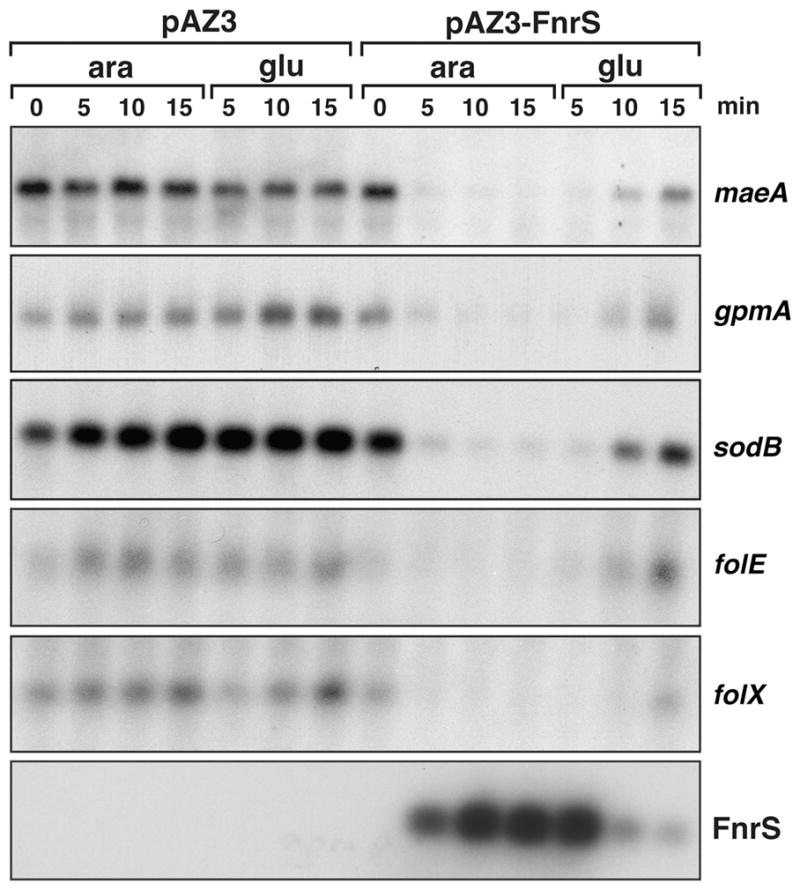
Cultures of MG1655 carrying pAZ3 or pAZ3-FnrS were grown in LB to OD600 ≈ 0.4 and treated with 0.2% arabinose. After 15 min, cells were washed two times in LB + 0.2% glucose and grown an additional 15 min. The time of incubation (min) with arabinose (ara) and glucose (glu) before RNA extraction are indicated on top. For all samples, total RNA (5 μg) was separated on an agarose gels, transferred to nitrocellulose and probed with a 32P-labelled oligonucleotides specific the genes indicated on the right.
FnrS base pairing with target mRNAs
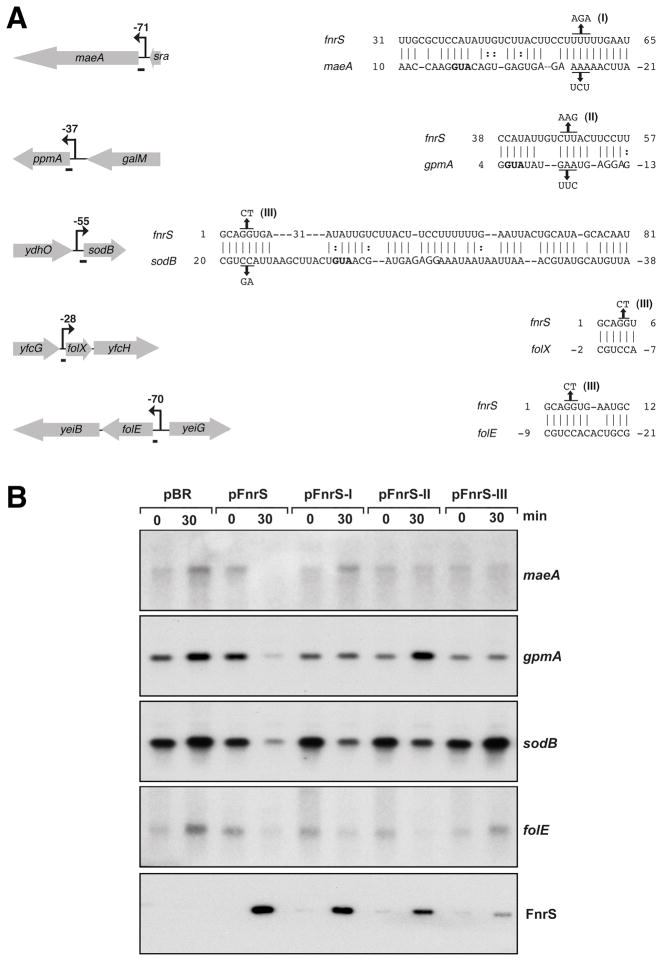
The maeA, gpmA, sodB, folE and folX mRNAs are all predicted to be base pairing targets of FnrS by the TargetRNA program (http://snowwhite.wellesley.edu/targetRNA) (Tjaden et al., 2006), and given that 59% of the genes identified by the microarray analysis are predicted by TargetRNA, we assume that many base pair directly with FnrS (Table 1). Interestingly, both TargetRNA and Mfold analysis predict that different regions of FnrS base pair with the maeA and gpmA mRNAs compared to the sodB, folE and folX mRNAs (Fig. 5A). The first region comprises the single-stranded region between the first and second stem-loops and the second region is the 5′ end of the partially open stem of the first stem-loop (Fig. 3A).
Fig. 5. Base pairing between FnrS and target mRNAs.
A. Predicted base pairing interactions. Black arrows indicate the promoters mapped by 5′ RACE PCR, and the numbers correspond to the number of nucleotides between the transcriptional and the translational start sites. The regions of base pairing between FnrS and its targets as predicted by the TargetRNA program are symbolized by short bars on the left and are given on the right. FnrS mutations I, II and III are also indicated. The ribosome binding sites are italicized and the start codons are in bold. The sequences of the compensatory mutations are also given.
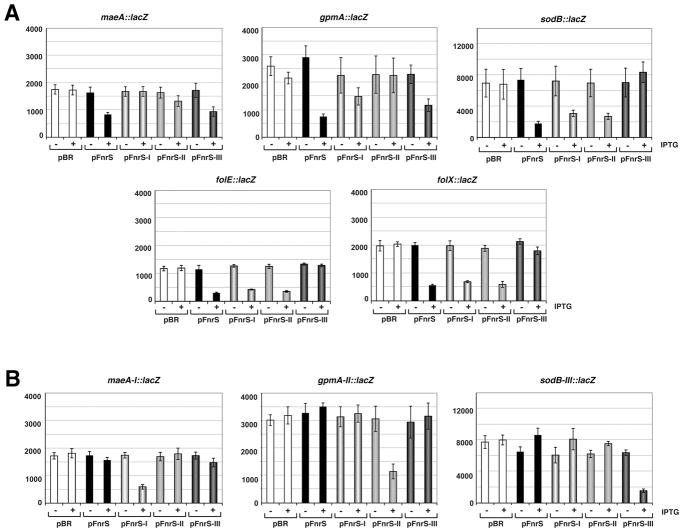
B. Repression of maeA, gpmA, sodB, folE expression by FnrS and FnrS mutants. Total RNA was extracted from MG1655 before and 30 min after the induction of FnrS (from pBR-FnrS) or FnrS mutant I, II or III (pBR-FnrS-I, II or III) with 100 μM IPTG. Genes probed are indicated on the left. The last panel shows the levels of the wild-type and mutant FnrS transcripts. The Northern blots were carried out as in Fig. 2 and 4.
To test whether pairing was direct or indirect and whether the predicted regions were required, we constructed three mutants of FnrS (I, II, III, Fig. 3A and 5A) by directed mutagenesis of pBR-FnrS. This plasmid contains an IPTG inducible promoter Plac (Guillier and Gottesman, 2006). Upon induction, the levels of FnrS-I are comparable to wild-type FnrS levels expressed from the same vector. FnrS-II levels are about 50% relative to wild type, while FnrS-III levels are somewhat lower (Fig. 5B). Northern analysis showed that none of the mutant forms of FnrS were as effective as the wild-type sRNA in repressing the sodB, gpmA, maeA and folE mRNA levels. However, FnrS-I is particularly defective at down regulating maeA, FnrS-II is very defective are repressing gpmA, and FnrS-III was most defective with respect to sodB and folE. In general the mutations support the conclusion that different regions of FnrS base pair with different sets of targets.
We also constructed chromosomal fusions between the PBAD promoter, the 5′ untranslated regions and first codons of the maeA, gpmA, sodB, folE and folX transcripts and lacZ such that these translational fusions are inducible by arabinose as described in (Mandin and Gottesman, 2009). We then monitored the effects of FnrS, FnrS-I, FnrS-II and FnrS-III overexpression in these strains. Expression of the translational fusions was first induced by the addition of 0.2% arabinose. After 5 min, expression of the FnrS wild-type or FnrS mutant RNAs from the pBR plasmids was induced with 100 μM IPTG, and the levels of β-galactosidase activity were assayed after 30 min. All fusions were repressed from 2- to 4-fold by wild-type FnrS overexpression and were unaffected by the empty vector (Fig. 6A). The maeA-lacZ fusion was also repressed by FnrS-III, despite the decreased levels of this sRNA, but was barely repressed by FnrS-II and not repressed by FnrS-I. Similarly, the gpmA-lacZ fusion was repressed by FnrS-III, but was repressed less well by FnrS-I and was unaffected by FnrS-II. The sodB-lacZ, folE-lacZ and folX-lacZ fusions were repressed by all mutants except FnrS-III. These assays substantiate the results obtained by Northern analysis and support the conclusion that the central single-stranded region of FnrS is required for base pairing with maeA and gpmA while the 5′ end of FnrS base pairs with sodB, folE and folX (Fig. 5A).
Fig. 6. Mutational analysis of FnrS base pairing.
A. β-galactosidase assays of FnrS target mRNA-lacZ fusions in presence of pBR (empty plasmid), pBR-FnrS, pBR-FnrS-I, pBR-FnrS-II and pBR-FnrS-III.
B. β-galactosidase assays of FnrS target mRNA-lacZ fusions carrying complementary mutations. For both (A) and (B), expression of the lacZ fusions was pre-induced for 5 min by the addition of 0.2% arabinose, after which cells were treated with 100 μM IPTG to induce the Plac promoter on the pBR plasmids. The levels of β-galactosidase activity were assayed 30 min later. The averages for the activity in Miller units determined in three independent experiments are shown together with the standard deviation.
We further constructed compensatory mutations in three of the target fusions denoted maeA-I, gpmA-II and sodB-III based on the corresponding FnrS mutations (Fig. 6B). Assays of the mutant lacZ fusion strains expressing the mutant FnrS RNAs showed that only FnrS-I down regulates maeA-I, only FnrS-II down regulates gpmA-II and only FnrS-III down regulates sodB-III. These assays confirm direct base pairing between FnrS and these targets and again support the conclusion that different regions of FnrS are involved in base pairing with different targets.
Decreased target mRNA repression in an ΔfnrS mutant strain
We examined FnrS RNA levels after shifts between aerobic and anaerobic growth. Cells were grown aerobically in M63 medium containing glucose and fumarate until mid-exponential phase. The cultures were then placed into an anaerobic chamber and total RNA was extracted after 10 and 20 min of anaerobic growth. Subsequently, the cultures were shifted back to aerobic conditions, and total RNA was extracted after 10, 20 and 60 min. As shown in Fig. 7, FnrS levels were maximally induced around 20 min after cells were shifted to anaerobic conditions. In addition, the sRNA is almost completely absent 10 min after the culture was re-aerated. These results show that both induction and presumably also degradation of FnrS are rapid.
Fig. 7. Effect of RydD on gpmA and sodB expression upon shifts between aerobic and anaerobic conditions.
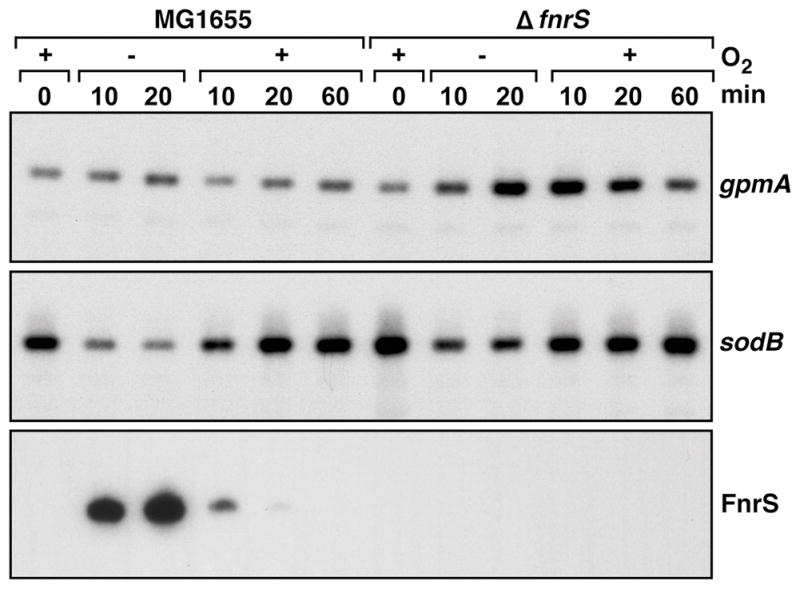
MG1655 and MG1655 ΔfnrS strains were grown in M63 with 0.2% glucose and 40 mM fumarate to OD600 ≈ 0.4 under aerobic conditions. Cells were collected and resuspended in the same medium and grown anaerobically. After 20 min, cells were again harvested, resuspended in the same medium and incubated aerobically for 60 min. Total RNA was extracted at several times (min) during this cycle. Northern blots were probed as in Fig. 2 and 4 for the genes indicated on the right.
We also monitored the levels of the sodB and gpmA mRNAs in the samples taken above as well as in the samples taken from ΔfnrS cells exposed to the same treatment (Fig. 7). The levels of the gpmA mRNA were stable between aerobic and anaerobic growth in the wild-type strain. In contrast, the gpmA RNA levels increased after the anaerobic shift in the ΔfnrS mutant strain. These elevated levels persisted upon the shift back to aerobic growth. In contrast to the gpmA transcript, the levels of the sodB mRNA decreased under anaerobic conditions, particularly in the wild-type strain. These results show that FnrS expressed from the chromosome impacts the levels of the target mRNAs, presumably to fine tune metabolism as cells are shifted between environments with different oxygen levels.
Discussion
An increasing number of Hfq-binding sRNAs are being found to remodel metabolism and transport in response to particular environmental stimuli. Here we describe a newly-identified sRNA that is induced upon a shift to anaerobic conditions and that represses the expression of enzymes that are dispensable during growth in low oxygen.
Regulation of FnrS expression
The finding that the strong anaerobic induction of FnrS depends on FNR is in agreement with the prediction of an FNR binding site at −41.5 and the FNR signal detected by chromatin immunoprecipitation experiments (Grainger et al., 2007). ArcA had a more limited effect on FnrS transcription, but was still required for optimal anaerobic induction. Dual regulation by both FNR and ArcA has been described for other anaerobically-induced genes (reviewed in (Sawers and Nakano, 2004)). These regulators can act in conjunction to either increase or decrease expression of the target gene or they can act in an opposing manner; one regulator activating transcription and the other repressing transcription. Only a few genes whose expression is activated by both FNR and ArcA, as observed for FnrS, have been described. These include fumB gene (encoding fumarase) and the focA-pflB operon (encoding a formate transporter and pyruvate formate-lyase, respectively) (Tseng, 1997, Sawers, 2006). In general, dual regulation by FNR and ArcA may allow the fine-tuning of gene expression during the shift from aerobic to anaerobic growth. An examination of transcriptomic data shows that FNR responds very rapidly while ArcA responds more slowly to a shift (Partridge et al., 2007).
Interestingly, in an MG1655fnr- strain lacking functional FNR, FnrS levels were still induced during anaerobic growth, though at a 20-fold lower level and more so in glycerol than in glucose medium (Fig. 2D). In the MG1655fnr- background, FnrS expression is dependent on CRP and ArcA. These results indicate that CRP can recognize the FNR binding site as demonstrated previously for the focA-pflB operon as well as the hlyE gene encoding hemolysin E (Kaiser and Sawers, 1997, Sawers et al., 1997, Green and Baldwin, 1997, Westermark et al., 2000) and can even partially replace FNR under anaerobic conditions. FnrS activation by CRP and ArcA is less efficient than the activation by FNR and ArcA possibly because CRP has a lower affinity than FNR for the binding site and/or is less efficient at stimulating FnrS transcription together with ArcA. The lower activation of FnrS by CRP could explain its negative impact on FnrS anaerobic expression in wild type strain where FNR and CRP compete for the same binding site (Fig. 2A and 2B). The dual regulation at the hlyE promoter allows induction of this gene under several different growth conditions, during anaerobic growth and upon glucose starvation, respectively. However, unlike the case for hlyE, expression of focA-pflB and FnrS is only slightly induced by CRP in a wild type strain growing aerobically. We proposed that at these promoters, CRP acts more as a negative regulator to limit focA-pfl and FnrS expression during anaerobic growth (Fig. 2A and 2B).
Finally, we found that nitrate slightly represses FnrS expression during both aerobic and anaerobic growth (Fig. 2A and 2D). The putative NarL/NarQ binding site centered at −22.5 in the FnrS promoter might explain this effect. The NarXL and NarPQ two-component systems program cell metabolism for nitrate respiration when nitrate is available (Unden and Bongaerts, 1997). It is unclear why these regulators might repress fnrS, but the expression of other anaerobically-induced genes has also been found to be modulated by a plethora of transcriptional regulators.
Two base pairing domains in FnrS
Microarray analysis indicated that the levels of at least 32 mRNA species are down regulated two-fold or more by FnrS overexpression. Five of these targets (maeA, gpmA, sodB, folE and folX) were confirmed to be strongly repressed by FnrS by Northern blot analysis and β-galactosidase assays of translational fusions to the 5′ untranslated regions of these mRNAs. The TargetRNA program predicts that FnrS base pairs with these targets, as well as most of the other targets identified by microarray experiments, at or near the Shine-Dalgarno sequences of the target mRNAs (Fig. 5A). The program also predicts that two different single stranded regions of FnrS are involved in base pairing with the targets. Mutations introduced into these regions together with the corresponding compensatory mutations into targets confirmed the TargetRNA predictions. We observed that pairing with a central single stranded region of FnrS is important for regulation of maeA and gpmA, while the 5′ end of FnrS is required for sodB, folE and folX repression. Both mutations in the central single stranded region affect gpmA and maeA repression, probably because base pairing between FnrS and the maeA or gpmA mRNAs can be extended through the whole region affected by the mutations (Fig. 5A). We also predicted that FnrS could use both the 5′ and the central regions to regulate sodB mRNA (Fig. 5A) but only the FnrS-III mutation at the 5′ end strongly affected sodB regulation.
For most base pairing sRNAs described thus far, only one region of the sRNA has been found to be involved in base pairing. This single-stranded region can be localized at the 5′ end of the sRNA as described for OmrA and OmrB in E. coli and RybB in Salmonella (Guillier and Gottesman, 2006, Bouvier et al., 2008) or in a central part such as for E. coli RyhB (Massé and Gottesman, 2002. Results presented here show that two regions of FnrS are involved in base pairing, both the 5′ end and a more central region. The results presented here are the first to clearly demonstrate that an E. coli sRNA can use two separate regions to target two different sets of targets. DsrA was proposed to use two different stem loop structures to regulate rpoS and hns, but these regions are adjacent and probably even overlap since some mutations affect both rpoS and hns regulation (Lease et al., 1998, Majdalani et al., 1998). GcvB has also been predicted to use different regions to base pair with the cycA, oppA and dppA mRNAs, but mutational analyses have not confirmed this prediction (Pulvermacher et al., 2009). In addition, two different regions of the OxyS RNA have been shown to be involved in base pairing, but, in this case, the different regions base pair with the same mRNA target (Argaman and Altuvia, 2000).
The involvement of two different regions of FnrS in base pairing could allow for the regulation of a larger number of targets and could explain the extensive conservation of FnrS through out its entire length. Based on predictions by TargetRNA and the results presented above, we propose that different sets of targets are regulated by the two regions, with genes linked to oxidative stress as well as folate and methionine metabolism regulated by the 5′ end and genes of central metabolism regulated by the central region.
Physiological role of FnrS RNA
Many of the mRNAs whose expression is repressed by FnrS encode enzymes involved in central and energy metabolism while a few encode enzymes linked to aerobic respiration, folate and amino acid metabolism and protection against stress (Table 2). Down regulation of these genes by FnrS during anaerobic growth reduces the expression of proteins that are not required under these conditions, such as enzymes used for aerobic respiration (dld) or cytochrome assembly (cydDC), or redundant enzymes like the malate dehydrogenase (mqo). Mqo, which has the same properties as Mdh, seems to be unnecessary since the TCA cycle is blocked before malate formation during anaerobic growth. Moreover, FnrS repression of sodB, which encodes an abundant superoxide dismutase, can also be explained in that oxidative stress is limited under anaerobic conditions.
Table 2.
Strains and plasmids used in this study
| Strains | Relevant features | References |
|---|---|---|
| MG1655 | MG1655 mal::lacIq (NM525) | Laboratory stock |
| CV600 | MG1655 lacX74 crp::cat | De Lay and Gottesman, 2009 |
| GSO388 | MG1655 Δcrp::cat | This study |
| GSO389 | MG1655 Δfnr::kan | This study |
| GSO390 | MG1655 ΔarcA::kan | This study |
| MG1655fnr- | MG1655 with duplication of amino acids 22 to 27 and mutations R10G and S13F in fnr gene | Laboratory stock |
| GSO391 | MG1655fnr- Δcrp::cat | This study |
| GSO392 | MG1655fnr- Δfnr::kan | This study |
| GSO393 | MG1655fnr- ΔarcA::kan | This study |
| PM1205 | lacI::PBAD-cat-sacB-lacZ, mini lambda tetR | Mandin et al., 2009 |
| GSO394 | PM1205 lacI′::PBAD-gpmA–lacZ | This study |
| GSO395 | PM1205 lacI′::PBAD-sodB–lacZ | This study |
| GSO396 | PM1205 lacI′::PBAD-maeA–lacZ | This study |
| GSO397 | PM1205 lacI′::PBAD-folE–lacZ | This study |
| GSO398 | PM1205 lacI′::PBAD-folX–lacZ | This study |
| GSO399 | PM1205 lacI′::PBAD-maeA-I–lacZ | This study |
| GSO400 | PM1205 lacI′::PBAD-gpmAII–lacZ | This study |
| GSO401 | PM1205 lacI′::PBAD-sodBIII–lacZ | This study |
| GSO402 | MG1655 ΔfnrS::kan | This study |
| Plasmids | ||
| pAZ3 | pBAD promoter based expression, kanR | Kawano et al., 2007 |
| pAZ3-FnrS | EcoRI-HindIII FnrS containing fragment cloned into pBR-Plac | This study |
| pBR-lac | Plac promoter based expression vector, ampR | Guillier et al., 2006 |
| pBRlac-FnrS | AatII-EcoRI FnrS containing fragment cloned into pBR-Plac | This study |
| pBRlac-FnrSI | U57A U58G U59A site-directed mutation in pBRlac-FnrS | This study |
| pBRlac-FnrSII | C47A U48A U49G site-directed mutation in pBRlac-FnrS | This study |
| pBRlac-FnrSIII | G4C G5T site-directed mutation in pBRlac-FnrS | This study |
Several targets down regulated by FnrS encode proteins known or predicted to bind zinc. For example, the FolE and MetE proteins require zinc for their enzymatic activities (Auerbach et al., 2000, Gonzalez et al., 1996), and, based on sequence comparisons, YggG, AdhP and Dcp are inferred to bind zinc (Henrich et al., 1993). It is also noteworthy that FnrS is always encoded adjacent to zntB (ydaN in E. coli), a zinc transporter in Salmonella. The ZntB protein is an efflux pump (Caldwell & Smith, 2003) and its expression is decreased in an fnr mutant strain (Salmon et al., 2003). Since the anaerobic growth of facultative anaerobes is absolutely dependent on ribonucleotide reductase III, which requires zinc to be active (Luttringer et al., 2009), we propose that FnrS may also contribute to the optimization of zinc utilization under anaerobic conditions, analogous to how RyhB optimizes iron utilization under aerobic conditions.
Like many commensal and pathogenic microorganisms, E. coli thrives in the gastrointestinal tract of humans and animals. In this environment, oxygen is limited and the cell must produce energy from anaerobic respiration with alternative electron acceptors or by the fermentation of simple sugars. The cell responds to decreases in oxygen tension by modulating pathways for carbon and energy flow. Thus far, FNR was considered as the main regulator of these adaptations. We now described an FNR-regulated sRNA whose expression is induced by anaerobic conditions and regulates numerous genes. We propose that FnrS expands the FNR regulon since many of the FnrS-repressed genes were reported to show altered expression in an fnr mutant, yet did not have upstream FNR binding sites (Constantinidou et al., 2006). This layered regulation allows the cells to precisely adjust energy consumption during anaerobic growth.
Experimental procedures
Bacterial strains
The bacterial strains used in this study are listed in Table 2, and the oligonucleotides used to generate the strains are listed in Table S2. The mini-λ-Red recombination system was used to create the arcA and fnr deletion strains (Datsenko and Wanner, 2000, Yu et al., 2000, Court et al., 2003). In all cases, pKD13 (Datsenko and Wanner, 2000), which encodes kanamycin resistance, was used as a template in PCR reactions together with oligonucleotide primers containing 20 bases of pKD13 sequence and approximately 40 nucleotides of homology to the chromosomal region being replaced. Subsequently, the kanamycin cassette was removed using pCP20 (Cherepanov and Wackernagel, 1995). The ΔfnrS strain was constructed by replacing fnrS with a barcoded sequence as described in (Hobbs et al., 2009). Each deletion mutation was moved into MG1655 by P1 transduction and was confirmed by PCR. The Δcrp::cat allele from CV600 (De Lay and Gottesman, 2009) was also moved into MG1655 by P1 transduction.
Translational fusions between the 5′ ends of the maeA, gpmA, sodB, folE and folX genes and lacZ were generated in PM1205 as previously described (Mandin and Gottesman, 2009). Briefly, the products from 5′ RACE-PCR were used to transform PM1205 to place each fusion under the arabinose-inducible PBAD promoter. Directed mutagenesis on maeA, gpmA and sodB was carried out by PCR in using the parental strain as the template. PCR products were used to transform PM1205. The lacZ fusions were all confirmed by sequencing.
Plasmid construction
The plasmids used in this study are listed in Table 2, and the oligonucleotides used for cloning are listed in Table S2. Plasmid DNA was always isolated using the Qiagen Mini Plasmid Kit. Plasmid pAZ3 (Kawano et al., 2007), a derivative of pBAD18 with an EcoRI site at +1, was used to overproduce FnrS from the PBAD promoter. For cloning into pAZ3, the fnrS gene along with 20 bases pairs downstream of the 3′ end were amplified from MG1655 genomic DNA by PCR. The products were purified using a Qiagen PCR Purification Kit, digested with EcoRI and HindIII and cloned into the corresponding sites of pAZ3. Wild-type fnrS was cloned into pBR-lac (Guillier and Gottesman, 2006) using the same strategy except both the plasmid and PCR fragment were digested with AatII and EcoRI. The fnrS mutants were generated by PCR-directed mutagenesis using pBRlac-FnrS as a template. For each mutation, two oligonucleotides of approximately 40 bases complementary to each other and carrying the mutation in the middle of the oligonucleotide were used in PCR reaction with the primers used to clone the wild-type fragment. The fragments again were digested with AatII and EcoRI and cloned into pBR-lac. The sequences of all inserts were confirmed by sequencing.
Growth conditions
E. coli K-12 MG1655 was grown in Luria–Bertani (LB, 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl per l) at 37°C. When needed kanamycin (30 μg/ml) and chloramphenicol (25 μg/ml) were added. To overexpress FnrS from pAZ3-FnrS, arabinose was added at OD600 ≈ 0.4 at a final concentration of 0.2%. Wild-type FnrS and the FnrS mutants were induced from the pBR-lac plasmid by the addition of 100 μM IPTG for 30 min. For anaerobic experiments, cells were first grown aerobically in M63 minimal medium (KD Medical) with 0.001% vitamin B1, and 0.2% glucose. At OD600 ≈ 0.4, 5 ml of cells were collected by centrifugation at 4°C and then resuspended in M63 medium pre-incubated in anaerobic chamber (Coy Laboratory) and containing 0.001% vitamin B1, 10 μM ammonium molybdate, 0.2% glucose or 0.4% glycerol and 40 mM fumarate or 20 mM nitrate as indicated. When anaerobically-growing cells were shifted back to aerobic conditions, the cells again were collected by centrifugation and then resuspended in aerobic M63 glucose fumarate medium.
RNA extraction
For cultures grown in LB, total RNA was extracted by using a modified version of the hot phenol technique (Massé et al., 2003). Briefly, 750 μl of cell culture were mixed with 102 μl of lysis solution (320 mM Na acetate at pH 4.6, 8% SDS, 16 mM EDTA). The lysed cells were then mixed with 500 μl of acid phenol (Ambion) at 65°C for 10 min. After centrifugation, the supernatant was extracted twice with acidic phenol and precipitated with 700 μl of 100% ethanol. For cultures grown in M63 medium, total RNA was extracted by using a modified version of the hot phenol technique as described in (Kawano et al., 2002). Briefly, 5 ml of cells were collected in 50 ml Falcon tubes by centrifugation at 5000 g for 5 min at 4°C. Cells were then resuspended in 500 μl of solution A (0.5% SDS, 20 mM sodium acetate, 10 mM EDTA, pH 5.5) and mixed thoroughly. Thereafter the mixture was transferred to a 1.5 ml eppendorf tube containing 500 μl of acid phenol and incubated at 65°C for 10 min. After vortexing, cells were extracted exactly as described for the LB samples. In all cases, the resulting RNA pellets were resuspended in H2O treated with diethyl pyrocarbonate (DEPC) and stored at −80°C. RNA concentrations were determined based on OD260.
Northern analysis
For the detection of FnrS transcript, total RNA (5 μg) was separated on a denaturing 6% polyacrylamide-8 M urea gel and transferred to a Zeta-Probe Membrane (Bio-Rad) for 5 h at 55 V in 0.5X TBE. Oligonucleotide probes, specific for the FnrS RNA, were labelled with 32P using T4 polynucleotide kinase (New England Biolab). Hybridization and wash steps were as described previously (Opdyke et al., 2004). For the detection of the sodB, maeA, gpmA, folE and folX mRNAs, total RNA (5 μg) was separated on a 1X TBE-1% agarose gel, transferred to a Zeta-Probe Membrane (Bio-Rad) by a gravity blotting for 3 h in a 0.01 N NaOH, 5X SSC buffer. Membranes were hybridized and washed as described previously (Opdyke et al., 2004). The RNA century marker (Ambion) and RNA millennium marker (Ambion) are used with acrylamide and agarose northerns, respectively, and visualized on the membrane by U.V. (254 nm).
5′ RACE
5′ RACE analysis was carried out as described (Argaman et al., 2001). The sequences of the oligonucleotides used to generate FnrS or the mRNA target cDNAs are given in Table S1. The amplified cDNA fragments were then cloned into vector pCRII Topo (Invitrogen) for FnrS or used to transform PM1205 (Mandin and Gottesman, 2009) to generate the target fusions. All 5′ ends were mapped by sequencing.
In vivo RNA structure probing
To probe the FnrS structure in vivo, a culture of MG1655 grown aerobically in M63 with glucose to OD600 ≈ 0.4 was split, the cells were harvested and then resuspended in M63 medium containing glucose, fumarate, vitamin B1 and ammonium molybdate (pre-incubated in anaerobic chamber). One of the cultures was left untreated while the other was treated immediately with 50 mM dimethylsulfate (DMS). After 4 min, total RNA was extracted as described above, and primer extension reactions using end-labeled oligo FnrS-ext were carried out with AMV reverse transcriptase (Life Sciences). The extension products together with sequencing reactions primed with the same end-labeled FnrS-ext primer were separated on an 8% sequencing gel.
Microarray analysis
MG1655 cells harboring pAZ3 or pAZ3-FnrS were grown to OD600 ≈ 0.5 in LB and were induced with arabinose at a final concentration of 0.2%. Cells were harvested after 15 min and total RNA was prepared as described previously (Kawano et al., 2002). The preparation of the cDNA and hybridization to the Affymetrix E. coli Genome 2.0 array were performed as described in Affymetrix manual Section 3: Prokaryotic Sample and array Processing (www.affymetrix.com/support/downloads/manuals/expression_s3_manual.pdf).
β-galactosidase assays
All strains were grown until OD600 ≈ 0.3 and induced with 0.2% arabinose. After 5 min of induction, 100 μM IPTG was added to induce expression from the pBR-lac plasmid, pBR-FnrS, pBR-FnrS-I, pBR-FnrS-II and pBR-FnrS-III. After 30 min of induction with IPTG, cells were lysed in 800 μl of Z-buffer with 5 μl of SDS 0.1% and 10 μl of chloroform. β-galactosidase assays were carried out as described by Miller (1972).
Supplementary Material
Acknowledgments
We thank P. Valentin-Hansen and P. Kiley for sharing unpublished information, S. Gottesman, P. Mandin and P. Kiley for advice and strains and F. Barras for helpful discussions. We also are grateful to C. Beisel, S. Gottesman, E. Hobbs, and P. Kiley for comments on the manuscript. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
References
- Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner E, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Buchet A, Nasser W, Eichler K, Mandrand-Berthelot MA. Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol Microbiol. 1999;34:562–575. doi: 10.1046/j.1365-2958.1999.01622.x. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH. Transcription activation by catabolite activator protein (CAP) J Mol Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life ? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Dubchak I, Holbrook S. A computational approach to identify genes for functional RNAs in genomic sequences. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Constantinidou C, Hobman J, Griffiths L, Patel M, Penn C, Cole J, Overton T. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem. 2006;281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- Court D, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan S. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- Darwin A, Tyson K, Busby S, Stewart V. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol Microbiol. 1997;25:583–595. doi: 10.1046/j.1365-2958.1997.4971855.x. [DOI] [PubMed] [Google Scholar]
- Datsenko K, Wanner B. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol. 2009;191:461–476. doi: 10.1128/JB.01157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diges C, Uhlenbeck O. Escherichia coli DbpA is a 3′ --> 5′ RNA helicase. Biochemistry. 2005;44:7903–7911. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- Eiglmeier K, Honoré N, Iuchi S, Lin E, Cole S. Molecular genetic analysis of FNR-dependent promoters. Mol Microbiol. 1989;3:869–878. doi: 10.1111/j.1365-2958.1989.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Farr SB, D’Ari R, Touati D. Oxygen-dependent mutagenesis in Escherichia coli lacking superoxide dismutase. Proc Natl Acad Sci USA. 1986;83:8268–8272. doi: 10.1073/pnas.83.21.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H, Kvaratskhelia M, White M. The two analogous phosphoglycerate mutases of Escherichia coli. FEBS Lett. 1999;455:344–348. doi: 10.1016/s0014-5793(99)00910-2. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin E. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Grainger D, Aiba H, Hurd D, Browning D, Busby S. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res. 2007;35:269–278. doi: 10.1093/nar/gkl1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Baldwin ML. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology. 1997;143:3785–3793. doi: 10.1099/00221287-143-12-3785. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- Guillier M, Gottesman S, Storz G. Modulating the outer membrane with small RNAs. Genes Dev. 2006;20:2338–2348. doi: 10.1101/gad.1457506. [DOI] [PubMed] [Google Scholar]
- Gunsalus RP, Park SJ. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- Henrich B, Becker S, Schroeder U, Plapp R. dcp gene of Escherichia coli: cloning, sequencing, transcript mapping, and characterization of the gene product. J Bacteriol. 1993;175:7290–7300. doi: 10.1128/jb.175.22.7290-7300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs EC, Astarita JL, Storz G. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: Analysis of a Bar-coded Mutant Collection. J Bacteriol. 2009 doi: 10.1128/JB.00873-09. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Cameron DC, Lin EC. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989;171:868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser M, Sawers G. Overlapping promoters modulate Fnr- and ArcA-dependent anaerobic transcriptional activation of the focApfl operon in Escherichia coli. Microbiology. 1997;143:775–783. doi: 10.1099/00221287-143-3-775. [DOI] [PubMed] [Google Scholar]
- Kang Y, Weber K, Qiu Y, Kiley P, Blattner F. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Aravind L, Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Oshima T, Kasai H, Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci U S A. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, De Wulf P. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004;279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- Luttringer F, Mulliez E, Dublet B, Lemaire D, Fontecave M. The Zn center of the anaerobic ribonucleotide reductase from E. coli. J Biol Inorg Chem. 2009;14:923–933. doi: 10.1007/s00775-009-0505-9. [DOI] [PubMed] [Google Scholar]
- Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia F, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Vanderpool C, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville SB, Gunsalus RP. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opdyke J, Kang J, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Partridge JD, Sanguinetti G, Dibden DP, Roberts RE, Poole RK, Green J. Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J Biol Chem. 2007;282:11230–11237. doi: 10.1074/jbc.M700728200. [DOI] [PubMed] [Google Scholar]
- Polayes DA, Rice PW, Garner MM, Dahlberg JE. Cyclic AMP–cyclic AMP receptor protein as a repressor of transcription of the spf gene of Escherichia coli. J Bacteriol. 1988;170:3110–3114. doi: 10.1128/jb.170.7.3110-3114.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvermacher SC, Stauffer LT, Stauffer GV. Role of the sRNA GcvB in regulation of cycA in Escherichia coli. Microbiology. 2009;155:106–114. doi: 10.1099/mic.0.023598-0. [DOI] [PubMed] [Google Scholar]
- Salmon K, Hung S, Mekjian K, Baldi P, Hatfield G, Gunsalus R. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem. 2003;278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- Sawers G, Kaiser M, Sirko A, Freundlich M. Transcriptional activation by FNR and CRP: reciprocity of binding-site recognition. Mol Microbiol. 1997;23:835–845. doi: 10.1046/j.1365-2958.1997.2811637.x. [DOI] [PubMed] [Google Scholar]
- Sawers RG. Differential turnover of the multiple processed transcripts of the Escherichia coli focA-pflB operon. Microbiology. 2006;152:2197–2205. doi: 10.1099/mic.0.28951-0. [DOI] [PubMed] [Google Scholar]
- Sawers RG, Nakano MM. Strict and facultative anaerobes: Medical and Environmental Aspects. In Redox(Oxygen)-dependent gene regulation in facultative anaerobes. In: Nakano MM, Zuber P, Sonnenschein AL, editors. Horizon Bioscience. 2004. pp. 67–86. [Google Scholar]
- Sharrocks AD, Green J, Guest JR. In vivo and in vitro mutants of FNR the anaerobic transcriptional regulator of E. coli. FEBS Lett. 1990;270:119–122. doi: 10.1016/0014-5793(90)81248-m. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem Soc Trans. 2003;31:1–10. doi: 10.1042/bst0310001. [DOI] [PubMed] [Google Scholar]
- Tjaden B, Goodwin S, Opdyke J, Guillier M, Fu D, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CP. Regulation of fumarase (fumB) gene expression in Escherichia coli in response to oxygen, iron and heme availability: role of the arcA, fur, and hemA gene products. FEMS Microbiol Lett. 1997;157:67–72. doi: 10.1111/j.1574-6968.1997.tb12754.x. [DOI] [PubMed] [Google Scholar]
- Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta. 1997;1320:217–234. doi: 10.1016/s0005-2728(97)00034-0. [DOI] [PubMed] [Google Scholar]
- Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol. 2006;9:605–611. doi: 10.1016/j.mib.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Wassarman K, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Zhang A, Storz G. Small RNAs in Escherichia coli. Trends in microbiology. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark M, Oscarsson J, Mizunoe Y, Urbonaviciene J, Uhlin BE. Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J Bacteriol. 2000;182:6347–6357. doi: 10.1128/jb.182.22.6347-6357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlock AJ, Smith RL. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:4369–4373. doi: 10.1128/JB.184.16.4369-4373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M. Studies on regulatory functions of malic enzymes. IV. Effects of sulfhydryl group modification on the catalytic function of NAD-linked malic enzyme from Escherichia coli. J Biochem. 1979;86:325–333. doi: 10.1093/oxfordjournals.jbchem.a132530. [DOI] [PubMed] [Google Scholar]
- Yu D, Ellis H, Lee E, Jenkins N, Copeland N, Court D. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.