Abstract
During the last step of αβ T cell development, thymocytes that have rearranged genes encoding TCR chains and express CD4 and CD8 coreceptor are selected on the basis of their TCR reactivity to escape programmed cell death and become mature CD4 or CD8 T cells. This process is triggered by intrathymic TCR signaling, that activates ‘sensor’ transcription factors ‘constitutively’ expressed in DP thymocytes. Eventually, TCR-signaled thymocytes evolve effector transcriptional circuits that control basal metabolism, migration, survival and initiation of lineage-specific gene expression. This review examines how components of the ‘sensing’ transcription apparatus responds to positive selection signals and highlights important differences with mature T cell responses. In a second part, we evaluate current observations and hypotheses on the connections between sensing transcription factors and effector circuitries.
Introduction: sensors and effectors of positive selection signals
The development of αβ T cells in the thymus can be schematically divided into two phases that (i) generate a large repertoire of precursors carrying clonotypic TCR specificities [1,2] and (ii) select from this pool the subset of specificities matching self antigen presenting molecules. At the end of the first phase, αβ-lineage thymocytes (which we will consider in this review) have evolved into a large population of cells that express TCRαβ complexes and both CD4 and CD8 coreceptors (double positive, DP). These ‘preselection’ cells serve as input for the second phase of αβ T cell intrathymic development, that for the sake of simplicity we will refer to as ‘thymocyte selection’, during which those cells expressing TCRs of appropriate avidity for self MHC peptides are ‘positively selected’ to differentiate into mature T cells [3]. In parallel, thymocytes carrying TCRs with high avidity for self antigens are actively deleted, a process called negative selection [4] that we will not address in this review. Positively selecting TCR signals not only rescue thymocytes from programmed cell death (this is positive selection per se) but also trigger a series of accompanying developmental events [3], including lineage choice and differentiation into single-positive (SP) CD4+CD8− or CD4−CD8+ cells [5-7], termination of TCR gene rearrangement [8,9], migration of thymocytes from the thymic cortex to the medulla and egress from the thymus [10,11].
These events are ultimately caused by modifications in gene transcription, either of conventional protein encoding genes, e.g. the termination of Rag1 and Rag2 expression, the up-regulation of genes encoding IL-7Rα or the chemokine receptor CCR7 [8,9,12-14], or of non-protein coding genes (the best example so far being the micro-RNA mir181a [15]). These events occur in an orderly fashion, so that cells terminate Rag gene expression before undergoing CD4-CD8 differentiation [9], and some changes, including the termination of Cd4 or Cd8 gene expression [6], are mutually exclusive.
Even though these developmental events require TCR signaling, many are not directly triggered by TCR signals. TCR engagement on DP thymocytes in vitro terminates Rag gene expression [8] and up-regulates the anti-apoptotic gene Bcl2 [16], but does not induce expression of IL-7Rα or of Thpok and Runx3, two transcription factors critical for commitment to the CD4 or CD8 lineage, respectively, and which are not expressed by pre-selection DP thymocytes [17-21]. These observations suggest that there are multiple intermediates between the transcription factors that ‘sense’ TCR signals, which are expressed in pre-selection DP thymocytes and whose activity is modulated by TCR signals, and the circuitry that controls effector gene expression.
Sensing
Some membrane receptors trigger gene expression through ‘messenger’ transcription factors that translocate from the receptor to their nuclear targets, including Notch family receptors, whose engagement generates a cleavage product that directly activates Notch target genes [22], and cytokine receptors, which activate Stat molecules that trigger cytokine-induced gene expression [23]. However, no such direct messenger has been found associated with TCR complexes. Rather, TCR engagement initially assembles membrane proximal signaling complexes through sequential tyrosine phosphorylations [24]; this activates cytosolic signal transduction cascades which in turn trigger transcriptional changes by affecting the intra-cellular localization or the activity of pre-existing transcription factors [25-27]. In the first part of this review, we will discuss four such signal transduction pathways: the Erk MAP kinase cascade, the calcineurin-NFAT axis, the NF-κB pathway, and the PI-3 kinase-mediated deactivation of Foxo molecules.
Erk kinases and Elk activation
TCR engagement activates a cascade of kinases, collectively known as MAP kinases, whose downstream members translocate to the nucleus and control transcription factor activity by serine-threonine phosphorylation [25]. The ‘classical’ MAP kinase cascade transduces TCR signals from Ras GTPases, activated as an end-point of membrane-proximal signaling events, to the closely related and partly redundant kinases Erk1 and Erk2. The Erk pathway is required for positive selection, but appears dispensable for negative selection, even though positively and negatively selecting signals markedly differ in the kinetics and geography of Erk activation [28-31]. Neither of the two ‘non classical’ JNK and p38 MAP kinase cascades, that share a similar organization, seems to be involved in the transduction of positive selection TCR signals; the implication of these pathways in thymocyte apoptosis and negative selection, notably for p38 kinases, was reviewed recently and will not be discussed further [32,33].
In addition to a variety of cytosolic substrates, multiple transcription factor targets have been identified for Erk kinases in vitro. This includes factors important for T cell selection, including Runx1, Ets1 and Gata3 [34-36]. However, in most cases, the physiological relevance of such phosphorylations has yet to be established. In fact, ‘knockin’ studies have found that the major Erk phosphorylation sites in Runx1 are dispensable for T cell development [37], thereby questioning the importance of Erk-mediated phosphorylation for Runx1 function.
In contrast, strong evidence points to the involvement of the Elk subfamily of Ets transcription factors as primary targets of Erk kinases [38]. Elk factors include Elk1, Elk3 (or Net) and Elk4 (or SAP1, unrelated to the cytosolic adaptor SAP), of which Elk4 is the predominantly expressed in thymocytes [39]. Interest in these factors initially came from their role in the transcription of cfos, one of the few ‘immediate’ genes transcribed without de novo protein synthesis in response to serum stimulation in fibroblasts, or to TCR stimulation in mature T cells. The current model is that Erk-mediated phosphorylation of Elk proteins activates a transcription factor complex constitutively bound to DNA and causes cfos transcription [38]. Specifically, Elk proteins associate with the unrelated Serum Response Factor (SRF) on the cfos promoter Serum Response Element, forming a complex that is transcriptionally inactive unless its Elk component is phosphorylated on specific serine residues. The resulting cfos transcription results in the synthesis of Fos proteins that associate with Jun molecules to form Fos-Jun heterodimers; these are the canonical version of the transcription factor AP1 [40], which in mature T cells is involved in expression of IL-2 [41]. The Erk-Elk-AP1 sequence is one of best characterized avenues of cell activation.
How does this apply to thymocyte selection? Genetic analyses demonstrate that Elk4 and SRF are required for positive selection [39,42]. This supports the possibility that Elk4 acts as a target of Erk during in positive selection, although it is worth noting that Elk proteins are potentially controled by additional signals [43]. However, it is unclear whether Elk4 promotes positive selection by inducing cfos expression and AP1 activation. Indeed, two sets of results challenge the notion that AP1 is involved in positive selection. First, AP1 activity seems resistant to induction in preselection DP thymocytes. Unlike in post-selection thymocytes and mature T cells, TCR stimulation, or even pharmacologic stimuli that typically bypass proximal receptor signaling (e.g. phorbol esters), fail to activate AP1 in preselection thymocytes [39,44,45]. Second, none of the genes contributing to AP1 activity, when considered individually, is essential for positive selection [40]. Enforced expression of the AP1 inhibitor BATF or of a dominant-negative version of Jun did not appear to affect the generation of SP thymocytes and T cells [46,47], suggesting that functional redundancy among Fos and Jun family members is not the only explanation behind these results. Thus, the current evidence does not favor a role of AP1 as an Erk target during positive selection; however, additional genetic analyses will be required for a definitive assessment of the role of AP1, and we will return to possible functions of AP1 in thymocyte development in the second part of this review.
Two lines of evidence offer clues to possible Elk targets in thymocytes. First, Elk4 promotes the transcription of Egr1 (Early Growth Response 1) [39], which belongs to a small family of transcription factors induced in response to TCR signaling [48]. Second, recent ‘ChIP-chip’ analyses of binding sites for Elk1 in the HeLa cell line suggest that Elk proteins control expression of genes encoding basal transcription factors [49]. Thymocyte selection involves increased protein and RNA synthesis, which at late stages of selection is sustained by IL-7 signaling [50]; however, cytokine signaling does not operate during the initial stages of selection, notably because DP thymocytes do not express IL-7 receptors, and it is therefore possible that TCR-induced Elk4 activation contributes to reactivate basal cell metabolism. Of note, analyses of Elk1 binding sites in HeLa cells showed that a large fraction of them are not associated with an SRF site [49]. Thus, even though Elk4 and SRF are both required for positive selection, their target sets may not be entirely overlapping.
Calcium and NFAT signaling
Intracellular calcium entry, which in lymphocytes mainly occurs as a result of depletion of endoplasmic reticulum calcium stores and the ensuing activation of calcium-release activated calcium channels, is a critical consequence of TCR engagement in mature T cells. Similar to Erk activation, calcium entry has multiple targets effects, both in the cytosol and the nucleus. The best characterized pathway transducing calcium signals to the nucleus involves calcineurin, a calcium-inducible protein phosphatase, and transcription factors of the NFAT family [26,51]. In unsignaled cells, NFAT proteins are phosphorylated on several sites in their amino-terminal half, and as a result are retained in the cytosol. The rise in intra-cellular calcium concentration activates calcineurin, which dephosphorylates NFAT molecules, thereby allowing their nuclear entry (possibly in part by unmasking a nuclear localization signal) and their transcriptional function.
Initial hints at the role of this pathway in positive selection came from observations that cyclosporin or FK506 (two inhibitors of calcineurin) inhibited T cell selection [52-55]. Genetic inactivation of calcineurin was difficult to achieve, owing to the functional redundancy of its two catalytic subunits [56,57]. However, T cell targeted disruption of the gene encoding its B1 regulatory subunit (CnB1) [58] demonstrated that calcineurin is required for positive selection. In contrast, CnB1 disruption did not seem to affect negative selection, in agreement with some [55] but not other [52-54] studies analyzing the effects of calcineurin inhibitors on T cell development in vivo, suggesting that the effect of these inhibitors is mediated by their action on the thymic stroma [58].
The genetic complexity of the NFAT family, and the requirement of these molecules for multiple developmental processes, has so far limited our understanding of their role in T cell development. Three calcineurin-sensitive NFAT family members, NFATc1, NFATc2 and NFATc3 (also called NFAT4), are expressed in DP thymocytes [59]. NFATc3 expression peaks in DP thymocytes, whereas expression of NFATc1 and NFATc2 becomes more prominent in SP thymocytes and mature T cells [60,61]. Accordingly, NFATc3 disruption has the most prominent effect on positive selection [59,60]. Combinatorial gene disruption further suggest that NFATc2 minimally contributes to positive selection, and that c3 and c1 are the main contributors to NFAT activity during T cell selection [59].
Despite its importance in mature T cells, the role of the calcineurin-NFAT axis as an actual sensor of positive selection signals was questioned by the observation that CnB1-deficient thymocytes, or thymocytes lacking NFATc2/c3, have defective Erk activation in response to TCR engagement [58]. Because CnB1-deficient thymocytes normally phosphorylate Erk in response to phorbol ester stimulation [58,62], these observations raise the possibility that the calcineurin-NFAT axis ‘sensitizes’ the Erk pathway, presumably in pre-DP thymocytes [62], rather than acting as a direct sensor of TCR signals. It is difficult at present to evaluate the respective share of these direct and indirect contributions of calcineurin-NFAT to positive selection. The observation that a constitutively active form of the Raf serine-threonine kinase, the upstream member of the Erk cascade, partially rescues the positive selection block caused by CsA treatment or CnB1 disruption raises the possibility that the calcineurin-NFAT axis is not actually involved in the transduction of positively selecting TCR signals [62]. Countering that perspective, short term treatment with CsA impairs positive selection before the appearance of Erk ‘desensitization’. In view of this finding, and of the major importance of this pathway in mature T cells, the tentative conclusion that calcineurin affects positive selection independently from its effect on Erk activity seems reasonable, even though it will have to be re-evaluated.
Although the calcineurin-NFAT axis is an essential relay of calcium signaling into the nucleus, it is not the only one. Other transcription factors are phosphorylated as a result of calcium entry, in a manner independent from calcineurin or NFAT. These include CREB, phosphorylated by CaMK kinases [63] and whose role in positive selection has yet to be fully evaluated [64], and Ets1, the prototype of the Ets family that also includes Elk proteins [65]. Ets1 is phosphorylated on multiple serine residues in a region, encoded by Ets1 exon 7, which lies immediately upstream of its DNA binding domain [34,66]. Biochemical and structural analyses indicate that such phosphorylation impairs Ets1 binding to DNA [34,66,67]. Ets1 is important for multiple aspects of T cell development, including TCRβ allelic exclusion in DN thymocytes and CD8 cell differentiation [68-70]. Supporting the possibility that ‘Exon 7 phosphorylations’ are important for Ets1 function during positive selection, ‘knockin’ deletion of Exon 7-encoded sequences resulted in increased numbers of CD8-lineage thymocytes [71].
Additional questions about calcium signaling in thymocytes have emerged from the recent identification of components of the store-operated calcium entry machinery, including ‘channel’ proteins referred to as CRACM or Orai, and STIM1 and STIM2 regulatory proteins [72,73]. While the function of CRACM/Orai proteins in T cell selection has yet to be analyzed, it was unexpectedly found that conditional disruption of the genes encoding STIM1 and STIM2 in DP thymocytes did not substantially disrupt T cell development [74]. While this result may reflect the delayed loss of STIM proteins after conditional gene deletion, this would not explain that germline disruption of STIM1 only modestly affects T cell development despite massively impairing calcium entry in DP cells [72,75].
Altogether, these few examples illustrate how fragmentary is our current picture of calcium signaling in thymocytes, and underscore the need for future studies both of the control of calcium entry and of its nuclear targets.
NF-κ B and activating kinases
In mature T cells, NF-κB is an important sensor of TCR signaling. NF-κB factors control transcription as homo or heterodimers associating five distinct gene products (NF-κB1, NF-κB2, Rel, RelA, and RelB) [76,77]. NF-κB activation requires the serine-phosphorylation and proteolytic degradation of inhibitory proteins (inhibitors of NF-κB, IκB) that bind NF-κB and Rel molecules and sequester them in the cytosol. Two IκB phosphorylating activities have been identified: a ‘canonical’ (IκB kinase, IKK) complex made of IKKα, β and γ molecules (the last one also called Nemo), and a ‘non-canonical’ pathway involving the activation of IKKα by the unrelated kinase NIK. There is no known connection between TCR signaling and ‘non canonical’ IKK activity. In contrast, TCR engagement in mature T cells induces ‘canonical’ IKK complexes through the activation of protein kinase C θ(PKCθ) and the three adaptor molecules Carma1, Bcl10 and Malt1 [78].
While NF-κB activity is essential in mature T cells for survival and function, and notably for the generation of normal populations of regulatory T cells [79], its role during T cell selection has been more difficult to evaluate [80-82]. Nonetheless, a few salient results have emerged. None of the NF-κB or Rel genes is individually required for positive selection [83]; as for AP1 and NFAT proteins, the significance of these observations is limited by the potential for functional redundancy and the pleiotropic functions of NF-κB. In contrast, IKK activity promotes in a cell-intrinsic manner the generation of mature T cells from DP thymocytes, as shown by T cell-targeted deletion of Nemo or ‘knockin’ expression of a catalytically inactive version of IKKβ [79]. However, IKK-deficient thymocytes are not arrested at an ‘unsignaled’ preselection stage (as are their Erk-deficient counterparts), but instead during their late maturation as SP thymocytes. It is possible that NF-κB is important throughout thymocyte selection, but that the kinetics of conditional gene deletion and protein degradation would mask this requirement until a late developmental stage. Alternatively, the late developmental arrest of IKK-deficient thymocytes may indicate a specific requirement for IKK activity for the survival of mature thymocytes. If correct, the latter perspective raises the question of whether TCR engagement is responsible for IKK activation, as TCR signaling is not thought to operate during the latest stages of thymic maturation.
In fact, independent genetic analyses question whether TCR-mediated NF-κB activation is important for thymocyte selection. Similar to mature T cells, TCR stimulation of DP or SP thymocytes activates NF-κB in a Bcl10- and Malt1-dependent manner [84], and reporter analyses indicate that NF-κB is activated in cells signaled to undergo positive selection in vivo [85]; however, careful studies of mice deficient for Bcl10 or Malt1 have failed to reveal any effect on positive selection [84]. While other pathways have been proposed to contribute to NF-κB activation in response to TCR engagement [86], their contribution to thymocyte selection remains to be established. Studies of Tak1, a kinase required for canonical NF-κB activation in thymocytes, offer possible clues to this apparent paradox [87-89]. IKK- and Tak1-deficient thymocytes are arrested at a similar developmental stage, and in both cases the requirement is particularly stringent for CD8 cells, consistent with other observations and with a greater NF-κB activity in CD8 than in CD4 SP thymocytes [81,85]. However, Tak1-deficient SP thymocytes seemed unable to respond to IL-7, a cytokine that is specifically required during the selection of CD8 but not CD4 T cells [87]. Thus, it is possible that cytokine signals, rather than those emanating from the TCR, contribute to NF-κB activation during thymocyte selection.
The parallel with Tak1 raises another possible solution to the conflicting phenotypes caused by IKK and Bcl10 or Malt1 deficiencies. Tak1 is a multifunctional kinase at the cross-roads of multiple activation pathways and downstream target cascades [90]. Similarly, it is conceivable that the requirement for IKK during selection is unrelated to NF-κB, notably because IKKα has a nuclear localization signal and has been proposed to phosphorylate a variety of transcription-associated substrates [91]. Further studies will evaluate such NF-κB-independent IKK functions in thymocytes, including whether they would also involve Nemo.
Foxo’s: targets of Erk and PI-3 kinases
Foxo transcription factors have emerged within the last few year as important targets of surface receptors [92]. Foxo proteins form a subgroup of the large family of ‘forkhead box’ or ‘winged helix’ transcription factors (http://biology.pomona.edu/fox/). Of the three Foxo factors expressed in immune cells (Foxo1, Foxo3 and Foxo4), Foxo1 is the only one shown so far to be important in T cells [93,94], although it has been reported that it is in part redundant with Foxo3 [93]. Unlike transcription factors considered in the previous sections, Foxo1 expression itself is induced after cells have initiated positive selection [93,95]. However, Foxo proteins deserve discussion in this review because their activity is directly controled by at least two pathways activated by TCR engagement [96]. They are substrates for the serine-threonine kinase Akt, a PI-3 kinase target, and they are phosphorylated (at least Foxo3) by activated Erk. Both phosphorylations contribute to Foxo inactivation: Akt-dependent phosphorylation excludes Foxo proteins from the nucleus, whereas Erk-mediated phosphorylation triggers their degradation.
What is the function, if any, of the PI-3 kinase-Foxo axis in sensing TCR signaling? Genetic analyses so far have not found a role for any individual Foxo gene as target of TCR signals in DP cells [93,95]. While in developing B cells Foxo1 controls the expression of Rag genes [97], there is no evidence for such a function in DP thymocytes where Foxo1 expression is low. Furthermore, whereas PI-3 kinase activity is required for early T cell development [98], notably in conjunction with Notch signals [99], its potential contribution to the sensing of TCR signals remains to be evaluated. Thus, the role of Foxo factors as sensors of TCR signals during selection remains hypothetical at present; we will discuss in the last section of this review their potential implication in the late stages of thymocyte selection.
Connecting sensing and effecting circuits
Tracking signaling progression in thymocytes undergoing selection
In a simple perspective, the output of the sensing circuitry described above is a new state of gene expression that reflects attributes of TCR signals. At the other end of the developmental sequence are effector circuitries that that control lineage choice [20,21], the terminal maturation of thymocytes and their exit from the thymus (which both require the transcription factor Klf2 [100]), and the acquisition of the IL-7-dependent survival pattern that characterizes mature T cells [101,102]. Our current knowledge stops somewhere in the chain(s) of events connecting sensors and effectors. In some cases, e.g. NFAT or NF-κB, there is detailed connection to TCR signals, but little if any information about potential targets in thymocytes undergoing selection. Reciprocally, there is information on the biological function and molecular targets of Runx1 or Ets1, but their connection to TCR signaling pathways remains somewhat tenuous [20,37,70,71,103]. The following three examples illustrate emerging connections and remaining unexplored territory between sensing and effector circuitries; they underscore the scope of the challenges we face in building a dynamic transcriptional network of thymocyte selection.
Downstream of Elk
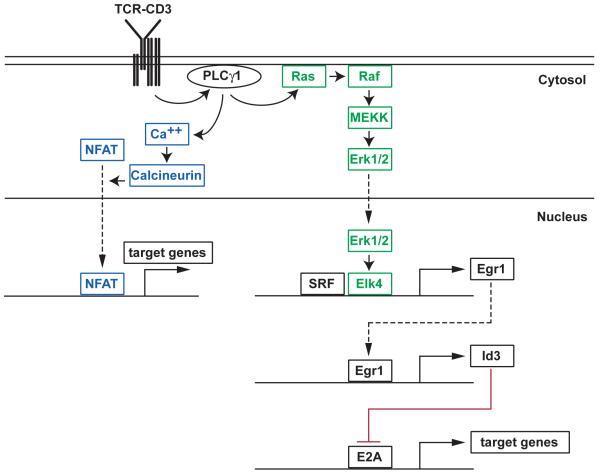
The best transcriptional sequence characterized so far implicates Elk4, Egr1 and E2A E-box binding proteins. Egr1 (Early Growth factor Response-1) is part of a four member family of zinc finger proteins (Egr1-4, of which the first three are expressed in thymocytes) [48]. They are expressed in response to environmental cues in a variety of cell types, and in thymocytes appear to serve as intermediates between sensing and effector circuitries. Both Egr1 and Egr2 contribute to positive selection and mature T cell development [104-106], and their role is most likely underestimated owing to functional redundancy between family members [107]. Initial analyses in a DP thymocyte cell line had indicated that expression of all Egr proteins is Ras dependent [48]. More recently, Egr1 has emerged as a link between the Erk signaling pathway and E2A (Fig. 1). ChIP analyses have identified Egr1 as a direct target of Elk4, and Elk4 promotes Egr1 expression in thymocytes [39]. In turn, Egr1 promotes expression of Id3 [108, 108a], an inhibitor of E2A proteins; Egr1 binds the Id3 promoter in vivo, suggesting that the control is at least in part direct [39].
Figure 1.
Schematic representation of the Erk-Elk (green) and calcineurin-NFAT (blue) sensor modules in thymocytes. Plain arrows indicate activation, dashed arrows nuclear import. The membrane-proximal signaling event that lead to PLCγ 1 activation are not depicted. Active PLCγ 1 releases inositol triphosphate and diacyl glycerol, which cause calcium release from intra-cellular stores (and the ensuing opening of plasma membrane CRAC channels) and activation of Ras via the exchange factor RasGRP1 [157,158], respectively.
The connection to E2A proteins, which bind DNA sites known as ‘E-boxes’, is significant for several reasons. E2A activity is high in preselection DP cells, where it promotes Cd4 expression notably through its HEB component [109]. E2A promotes expression of multiple genes characteristic of DP thymocytes, including the chemokine receptor CXCR4 and RORγ t, a transcription factor essential for DP cell survival and shown to promote Rag2 expression in a DP cell line [110-113]; conversely, E2A disruption promotes the expression by DP cells of genes characteristic of mature thymocytes, including IL-7Rα, the chemokine receptor CCR7 and the transcription factor Klf2 [112]. Genetic analyses show that Id3 is important for positive selection, supporting the importance of the Elk-Egr-Id3 sequence [108]. Perhaps most spectacularly, disruption of E2A activity in DP thymocytes allows the development of mature T cells even in the absence of TCR signaling [112].
How oversimplified is this simple scheme? As might be expected, additional links between TCR stimulation, Egr activity and effector mechanisms add complexity to this linear cascade. NFATc1 cooperates with Egr1 to induce Id3 after pre-TCR signaling [114], and the same may be true in TCR signaled DP cells. Other Egr factors affect selection; notably, Egr2 promotes expression of Bcl-2 and has been proposed to promote IL-7 receptor expression and signaling [105,106]. The up-regulation of Bcl-2 and IL-7Rα are characteristic of positive selection, and both genes are required for the long-term survival of mature T cells [115,116]. Thus, the connection between Egr proteins and Bcl-2 or IL-7Rα is biologically significant; whether it is direct or occurs though antagonism of E2A, as suggested for IL-7Rα by analyses of E2A-deficient thymocytes [112], remains to be determined. But there is reason to think that the overall picture will be more complex. Egr binding sites are frequent in a variety of gene promoters, suggesting a wide array of molecular targets; indeed, microarray gene expression analyses in DN thymocytes found that Egr genes promoted expression of multiple genes involved in cell metabolism [107]. It is not known if such effects are direct, nor do we appreciate the extent to which Egr proteins are functionally interchangeable. Additional stimuli feed into Egr activity. Expression of Egr2 and Egr3 is NFAT-dependent [48,117]. Egr1 is a target of AP1 in other developmental systems [118], and it is conceivable that AP1 further enhances Egr1 expression in late thymocyte differentiation. In addition, it has been reported that Fos promotes expression of Bcl2, suggesting a cooperation of two distinct Elk targets on the same gene [119]. In mature T cells, the activity of Egr factors is controled by the two proteins Nab1 and Nab2 whose expression is controled by TCR signaling [120].
Thpok expression and CD4 lineage differentiation
A typical example of the challenges in linking sensing and effector pathways is to connect TCR signals to the up-regulation of the CD4-committing gene Thpok. The research on CD4-CD8 differentiation was reviewed recently [7,20,21,121,122], and we will simply mention that Thpok is required for CD4-lineage commitment during positive selection [17,123,124], that it is not expressed in preselection DP cells, and that it is selectively up-regulated in MHC II-restricted thymocytes [17,18]. Because of its expression pattern and requirement for CD4-lineage commitment, it is important to understand how Thpok is induced by TCR signaling. Assuming that TCR-induced Thpok up-regulation is cell-intrinsic (i.e. that this is not mediated through a secondary ligand-receptor system), the question is equivalent to connecting the sensing circuitry to Thpok cis-regulatory elements.
A simple hypothesis is that Thpok up-regulation involves a linear cascade of transcription factors, similar to that connecting Elk and E2A. A prime candidate for such a function would be the zinc finger transcription factor Gata3, which is induced by TCR signaling [125], is required for Thpok expression in thymocytes and is recruited in vivo to the Thpok locus [123]. However, enforced Gata3 expression in DP thymocytes is not sufficient to induce substantial Thpok expression [126, and L. W., K.F. Wildt and R.B, unpublished observations], challenging the idea of a simple linear cascade. Adding to this caveat, in vitro TCR engagement in DP thymocytes up-regulates Gata3 but not (at least not significantly) Thpok [18, 125], suggesting that other factors are required. Such additional levels of controls could include post-translational modifications, or combinatorial effects, and previous studies suggest a few speculations. NFAT proteins and Gata3 cooperate for IL-4 gene expression during Th2 effector differentiation [26,127], and a similar cooperation could operate at the Thpok locus. NFAT also induces expression of the HMG-binding protein Tox [128], which is required for CD4-lineage differentiation, presumably by inducing chromatin modifications [129,130]. While there is at present no evidence supporting a specific role of NFAT in lineage differentiation, the potential for an NFAT-Gata3 cooperation remains to be evaluated. In addition, studies in Th2-differentiating T cells indicate that Erk-mediated phosphorylation stabilizes Gata3 proteins by preventing their ubiquitin-proteasome mediated degradation [36]. However, the control of Gata3 expression and activity by Erk kinases is complex [131] and it remains unclear which effect, if any, Erk has on Gata3 in thymocytes.
None of these mechanisms, however, would readily explain why TCR engagement on DP thymocytes fails to up-regulate Thpok. A possible, although at present highly speculative, explanation comes from the parallel kinetics of Thpok expression [124,132,133] and restoration of AP1 inducibility [44,45] during thymocyte selection. In MHC II-signaled thymocytes, both are contemporaneous with TCR up-regulation, raising the possibility that AP1 contributes to Thpok expression. Such an idea would fit with the repeated observation that Erk activity is more important for CD4 than CD8 T cell development [29,134,135]. AP1 would connect the Erk pathway, of which it is a target, to the calcineurin-NFAT axis, as exemplified in mature T cells by the cooperative binding of NFAT and AP1 factors to cytokine gene promoters [26,136]. In such complexes, NFAT factors are recruited to composite elements in association with an AP1 Fos-Jun dimer [137], unlike their binding as homodimers to ‘NFAT-only’ sites. While the idea that AP1 contributes to Thpok expression remains highly speculative, it has great appeal in light of the low AP1 activity in TCRlo preselection cells. A requirement for AP1 for Thpok expression would fit with the ‘kinetic signaling’ perspective of lineage commitment, and notably with the concept that TCRlo DP thymocytes are not competent for lineage choice [138,139]. Additional genetic studies of AP1, using gene disruption analyses in addition to dominant negative approaches [47], would be needed to ascertain its function in lineage choice.
Foxo1 and IL-7Rα expression
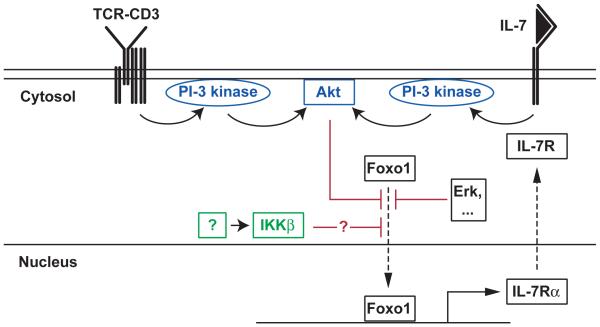
Four recent studies put the spotlight on the role of Foxo1 in T cell homeostasis [93,94,140,141]. Foxo1 promotes the expression of IL-7Rα and of the zinc finger transcription factor Klf2. The effect of Foxo1 on IL-7Rα expression is at least in part direct, as Foxo1 proteins are recruited in vivo to a cis-regulatory element 3.6 kb upstream of the IL-7Rα gene promoter [93]. In post-thymic cells, it has been proposed that Foxo1 contributes to a self-regulatory loop that limits cell sensitivity to IL-7 [93,142]. That is, IL-7-mediated activation of PI-3 kinase would promote Foxo1 phosphorylation and its exclusion from the nucleus, thereby reducing IL-7Rα gene expression.
CD4 and CD8 SP thymocytes express Foxo1 and IL-7Rα, and such IL-7Rα expression depends in part on Foxo1, suggesting that similar mechanisms may operate in the thymus [12,93,95]. However, TCR signaling also activates PI-3 kinase [143], adding an extra twist to the control of Foxo1 activity (Fig. 2). Indeed, TCR signaling has a dual role in IL-7Rα expression in post-DN thymocytes. While only positively selected thymocytes express IL-7Rα [12], indicating a requirement for TCR engagement, IL-7Rα expression is highest on the most mature thymocytes, that have ceased TCR signaling as indicated by their down-regulation of the TCR signaling marker CD69. Accordingly, in vitro signaled DP thymocytes up-regulate IL-7Rα only after signaling has been terminated [138]. A related pattern was reported for Klf2, another Foxo1 target in T cells [93,140]: Klf2 is expressed in mature but not immature thymocytes, and its expression, at least in mature T cells, is down-regulated by TCR signaling [144]. These observations suggest a two step model, whereby TCR signaling would promote Foxo1 expression, through a yet unidentified pathway, but prevent Foxo1-mediated IL-7Rα transcription by excluding Foxo1 molecules from the nucleus through activation of PI3 kinase (Fig. 2). As a result, actual IL-7Rα expression would be restrained in cells until the cessation of TCR signaling. While future studies will evaluate this possibility, it would illustrate how signals from distinct receptor could target the same sensing factor and contribute to effector gene expression. Of note, IKKβ was shown to promote Foxo3 phosphorylation and nuclear exclusion in tumor cells [145]. While it is not yet known if IKKβ phosphorylates Foxo1 in T cells, this observation is interesting to correlate with the finding that Tak1, an IKK activator, promotes IL-7 signaling (although not IL-7Rα expression) in SP thymocytes [87]. As the consequences of Tak1, IKK and Foxo1 deficiencies all become apparent in mature SP thymocytes [79,93], it is possible that Foxo factors are a relevant physiological target of IKK in SP thymocytes.
Figure 2.
Foxo1 as a sensor for TCR and IL-7 signals in thymocytes. In mature T cells, IL-7 activates PI-3 kinase and Akt, thereby causing Foxo1 phosphorylation. As Foxo1 promotes IL-7Rα expression, IL-7-mediated Foxo1 phosphorylation contributes to repress IL-7Rα expression and thereby IL-7 signaling. In SP thymocytes, where Foxo1 also promotes IL-7Rα expression, both TCR signaling and IL-7 signaling potentially contribute to PI-3 kinase activation. This mechanism could have a ‘licensing’ function by preventing the terminal maturation of SP thymocytes unable to cease TCR signaling. In tumor cells, IKKβ has been shown to phosphorylate Foxo3 and prevent its nuclear translocation [145]; it is not yet known which stimulus activates IKKβ, and whether it also acts on Foxo1 in thymocytes or T cells.
As an aside, if Foxo1 control of Klf2 expression mirrored that of IL-7Rα, continuous TCR-induced PI-3 kinase activation could also contribute to prevent the terminal maturation and thymic egress of thymocytes with excessive affinity for self-ligands, thereby enabling a simple ‘licensing’ mechanism to prevent the thymic egress of self-reactive T cells.
Conclusions and Future challenges
The last few years have seen much progress in our understanding of thymocyte signaling. As in other biological systems, multiple layers of control, at the level of gene expression and post-translational modifications, contribute to transduce signals from cytosolic intermediates to sensing transcription factors, and to the emergence of effector expression networks. Emerging evidence also points to micro-RNAs as important contributors to signaling and potentially transcriptional circuits [15,146-148]. Many pieces are missing from the sketch drawn in this review. Other TCR-induced kinases have potential targets in the nucleus [149], including protein kinase D, which activates histone deacetylases and thereby has pleitropic effects on gene expression, whereas other ligand-receptor systems directly or indirectly affect TCR signaling [150]. Conversely, several ‘orphan’ transcription factors, including Bcl11b [151] or Schnurri2 [152], are essential for positive selection, but what induces them or controls their activity remains obscure.
Future studies will have to tackle broader challenges. The first is to define target genes for transcription factors and assemble sensing and regulatory networks. New massively parallel sequencing approaches coupled to transcription factor immunoprecipitation (‘Chipseq’ analyses) have transformed into a realistic perspective what appeared as an impossible task only a few years ago [153]. However, networks assembled on binding sites give only a partial picture, and need to be refined to take into account the combinatorial and dynamic components of transcriptional responses, as in vivo recruitment observed by ChIP analyses does not equate to transcriptional regulation [154,155]. In this context, new tools will be necessary to decipher the logic of transcription factor combinatorial binding [156]. The multiple post-translational modifications, including, beyond phosphorylation, acetylation methylation, ubiquitination and sumoylation, are as many dimensions to these analyses.
Acknowledgments
We thank Jon Ashwell, Dietz Conze, Ellen Rothenberg and Murty Shrinivasula for stimulating discussions, and Jon Ashwell and Paul Love for critical reading of the manuscript. Research work in the authors’ laboratory is supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 4.Sohn SJ, Thompson J, Winoto A. Apoptosis during negative selection of autoreactive thymocytes. Curr Opin Immunol. 2007;19:510–515. doi: 10.1016/j.coi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Germain RN. T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol. 2002;2:309–322. doi: 10.1038/nri798. [DOI] [PubMed] [Google Scholar]
- 6.Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- 7.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turka LA, Schatz DG, Oettinger MA, Chun JJ, Gorka C, Lee K, McCormack WT, Thompson CB. Thymocyte expression of RAG-1 and RAG-2: termination by T cell receptor cross-linking. Science. 1991;253:778–781. doi: 10.1126/science.1831564. [DOI] [PubMed] [Google Scholar]
- 9.Bhandoola A, Cibotti R, Punt JA, Granger L, Adams AJ, Sharrow SO, Singer A. Positive selection as a developmental progression initiated by alpha beta TCR signals that fix TCR specificity prior to lineage commitment. Immunity. 1999;10:301–311. doi: 10.1016/s1074-7613(00)80030-8. [DOI] [PubMed] [Google Scholar]
- 10.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 11.Schwab SR, Cyster JG. Finding a way out: lymphocyte egress from lymphoid organs. Nat Immunol. 2007;8:1295–1301. doi: 10.1038/ni1545. [DOI] [PubMed] [Google Scholar]
- 12.Sudo T, Nishikawa S, Ohno N, Akiyama N, Tamakoshi M, Yoshida H, Nishikawa S. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci U S A. 1993;90:9125–9129. doi: 10.1073/pnas.90.19.9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwan J, Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 14.Ueno T, Saito F, Gray DH, Kuse S, Hieshima K, Nakano H, Kakiuchi T, Lipp M, Boyd RL, Takahama Y. CCR7 Signals Are Essential for Cortex-Medulla Migration of Developing Thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Cibotti R, Punt JA, Dash KS, Sharrow SO, Singer A. Surface molecules that drive T cell development in vitro in the absence of thymic epithelium and in the absence of lineage-specific signals. Immunity. 1997;6:245–255. doi: 10.1016/s1074-7613(00)80327-1. [DOI] [PubMed] [Google Scholar]
- 17.He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- 18.Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- 19.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol. 2009;9:106–115. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Bosselut R. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J Immunol. 2009;183:2903–2910. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu Rev Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 23.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 24.Samelson LE. Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu Rev Immunol. 2002;20:371–394. doi: 10.1146/annurev.immunol.20.092601.111357. [DOI] [PubMed] [Google Scholar]
- 25.Rincon M, Flavell RA, Davis RJ. Signal transduction by MAP kinases in T lymphocytes. Oncogene. 2001;20:2490–2497. doi: 10.1038/sj.onc.1204382. [DOI] [PubMed] [Google Scholar]
- 26.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 27.Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-kappaB. Nat Rev Immunol. 2005;5:435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- 28.Alberola-Ila J, Forbush KA, Seger R, Krebs EG, Perlmutter RM. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 1995;373:620–623. doi: 10.1038/373620a0. [DOI] [PubMed] [Google Scholar]
- 29.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 31.McGargill MA, Ch’en IL, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. Cutting edge: Extracellular signal-related kinase is not required for negative selection of developing T cells. J Immunol. 2009;183:4838–4842. doi: 10.4049/jimmunol.0902208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugawara T, Moriguchi T, Nishida E, Takahama Y. Differential roles of ERK and p38 MAP kinase pathways in positive and negative selection of T lymphocytes. Immunity. 1998;9:565–574. doi: 10.1016/s1074-7613(00)80639-1. [DOI] [PubMed] [Google Scholar]
- 33.Rincon M, Davis RJ. Regulation of the immune response by stress-activated protein kinases. Immunol Rev. 2009;228:212–224. doi: 10.1111/j.1600-065X.2008.00744.x. [DOI] [PubMed] [Google Scholar]
- 34.Rabault B, Roussel MF, Quang CT, Ghysdael J. Phosphorylation of Ets1 regulates the complementation of a CSF-1 receptor impaired in mitogenesis. Oncogene. 1996;13:877–881. [PubMed] [Google Scholar]
- 35.Tanaka T, Kurokawa M, Ueki K, Tanaka K, Imai Y, Mitani K, Okazaki K, Sagata N, Yazaki Y, Shibata Y, Kadowaki T, Hirai H. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol Cell Biol. 1996;16:3967–3979. doi: 10.1128/mcb.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamashita M, Shinnakasu R, Asou H, Kimura M, Hasegawa A, Hashimoto K, Hatano N, Ogata M, Nakayama T. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J Biol Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- 37.Tachibana M, Tezuka C, Muroi S, Nishimoto S, Katsumoto T, Nakajima A, Kitabayashi I, Taniuchi I. Phosphorylation of Runx1 at Ser249, Ser266, and Ser276 is dispensable for bone marrow hematopoiesis and thymocyte differentiation. Biochem Biophys Res Commun. 2008;368:536–542. doi: 10.1016/j.bbrc.2008.01.124. [DOI] [PubMed] [Google Scholar]
- 38.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 39.Costello PS, Nicolas RH, Watanabe Y, Rosewell I, Treisman R. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat Immunol. 2004;5:289–298. doi: 10.1038/ni1038. [DOI] [PubMed] [Google Scholar]
- 40.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim HP, Imbert J, Leonard WJ. Both integrated and differential regulation of components of the IL-2/IL-2 receptor system. Cytokine Growth Factor Rev. 2006;17:349–366. doi: 10.1016/j.cytogfr.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Fleige A, Alberti S, Grobe L, Frischmann U, Geffers R, Muller W, Nordheim A, Schippers A. Serum response factor contributes selectively to lymphocyte development. J Biol Chem. 2007;282:24320–24328. doi: 10.1074/jbc.M703119200. [DOI] [PubMed] [Google Scholar]
- 43.Yang SH, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rincon M, Flavell RA. Regulation of AP-1 and NFAT transcription factors during thymic selection of T cells. Mol Cell Biol. 1996;16:1074–1084. doi: 10.1128/mcb.16.3.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen F, Chen D, Rothenberg EV. Specific regulation of fos family transcription factors in thymocytes at two developmental checkpoints. Int Immunol. 1999;11:677–688. doi: 10.1093/intimm/11.5.677. [DOI] [PubMed] [Google Scholar]
- 46.King LB, Tolosa E, Lenczowski JM, Lu F, Lind EF, Hunziker R, Petrie HT, Ashwell JD. A dominant-negative mutant of c-Jun inhibits cell cycle progression during the transition of CD4(−)CD8(−) to CD4(+)CD8(+) thymocytes. Int Immunol. 1999;11:1203–1216. doi: 10.1093/intimm/11.8.1203. [DOI] [PubMed] [Google Scholar]
- 47.Williams KL, Zullo AJ, Kaplan MH, Brutkiewicz RR, Deppmann CD, Vinson C, Taparowsky EJ. BATF transgenic mice reveal a role for activator protein-1 in NKT cell development. J Immunol. 2003;170:2417–2426. doi: 10.4049/jimmunol.170.5.2417. [DOI] [PubMed] [Google Scholar]
- 48.Shao H, Kono DH, Chen LY, Rubin EM, Kaye J. Induction of the early growth response (Egr) family of transcription factors during thymic selection. J Exp Med. 1997;185:731–744. doi: 10.1084/jem.185.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boros J, Donaldson IJ, O’Donnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;19:1963–1973. doi: 10.1101/gr.093047.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 52.Gao EK, Lo D, Cheney R, Kanagawa O, Sprent J. Abnormal differentiation of thymocytes in mice treated with cyclosporin A. Nature. 1988;336:176–179. doi: 10.1038/336176a0. [DOI] [PubMed] [Google Scholar]
- 53.Jenkins MK, Schwartz RH, Pardoll DM. Effects of cyclosporine A on T cell development and clonal deletion. Science. 1988;241:1655–1658. doi: 10.1126/science.241.4873.1655. [DOI] [PubMed] [Google Scholar]
- 54.Shi YF, Sahai BM, Green DR. Cyclosporin A inhibits activation-induced cell death in T-cell hybridomas and thymocytes. Nature. 1989;339:625–626. doi: 10.1038/339625a0. [DOI] [PubMed] [Google Scholar]
- 55.Wang CR, Hashimoto K, Kubo S, Yokochi T, Kubo M, Suzuki M, Suzuki K, Tada T, Nakayama T. T cell receptor-mediated signaling events in CD4+CD8+ thymocytes undergoing thymic selection: requirement of calcineurin activation for thymic positive selection but not negative selection. J Exp Med. 1995;181:927–941. doi: 10.1084/jem.181.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bueno OF, Brandt EB, Rothenberg ME, Molkentin JD. Defective T cell development and function in calcineurin A beta -deficient mice. Proc Natl Acad Sci U S A. 2002;99:9398–9403. doi: 10.1073/pnas.152665399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang BW, Zimmer G, Chen J, Ladd D, Li E, Alt FW, Wiederrecht G, Cryan J, O’Neill EA, Seidman CE, Abbas AK, Seidman JG. T cell responses in calcineurin A alpha-deficient mice. J Exp Med. 1996;183:413–420. doi: 10.1084/jem.183.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neilson JR, Winslow MM, Hur EM, Crabtree GR. Calcineurin B1 is essential for positive but not negative selection during thymocyte development. Immunity. 2004;20:255–266. doi: 10.1016/s1074-7613(04)00052-4. [DOI] [PubMed] [Google Scholar]
- 59.Cante-Barrett K, Winslow MM, Crabtree GR. Selective role of NFATc3 in positive selection of thymocytes. J Immunol. 2007;179:103–110. doi: 10.4049/jimmunol.179.1.103. [DOI] [PubMed] [Google Scholar]
- 60.Oukka M, Ho IC, de la Brousse FC, Hoey T, Grusby MJ, Glimcher LH. The transcription factor NFAT4 is involved in the generation and survival of T cells. Immunity. 1998;9:295–304. doi: 10.1016/s1074-7613(00)80612-3. [DOI] [PubMed] [Google Scholar]
- 61.Amasaki Y, Masuda ES, Imamura R, Arai K, Arai N. Distinct NFAT family proteins are involved in the nuclear NFAT-DNA binding complexes from human thymocyte subsets. J Immunol. 1998;160:2324–2333. [PubMed] [Google Scholar]
- 62.Gallo EM, Winslow MM, Cante-Barrett K, Radermacher AN, Ho L, McGinnis L, Iritani B, Neilson JR, Crabtree GR. Calcineurin sets the bandwidth for discrimination of signals during thymocyte development. Nature. 2007;450:731–735. doi: 10.1038/nature06305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Baumann S, Kyewski B, Bleckmann SC, Greiner E, Rudolph D, Schmid W, Ramsay RG, Krammer PH, Schutz G, Mantamadiotis T. CREB function is required for normal thymic cellularity and post-irradiation recovery. Eur J Immunol. 2004;34:1961–1971. doi: 10.1002/eji.200324826. [DOI] [PubMed] [Google Scholar]
- 65.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 66.Pognonec P, Boulukos KE, Bosselut R, Boyer C, Schmitt-Verhulst AM, Ghysdael J. Identification of a Ets1 variant protein unaffected in its chromatin and in vitro DNA binding capacities by T cell antigen receptor triggering and intracellular calcium rises. Oncogene. 1990;5:603–610. [PubMed] [Google Scholar]
- 67.Pufall MA, Lee GM, Nelson ML, Kang HS, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 68.Eyquem S, Chemin K, Fasseu M, Bories JC. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc Natl Acad Sci U S A. 2004;101:15712–15717. doi: 10.1073/pnas.0405546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clements JL, John SA, Garrett-Sinha LA. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J Immunol. 2006;177:905–912. doi: 10.4049/jimmunol.177.2.905. [DOI] [PubMed] [Google Scholar]
- 70.Zamisch M, Tian L, Grenningloh R, Xiong Y, Wildt KF, Ehlers M, Ho IC, Bosselut R. The transcription factor Ets1 is important for CD4 repression and Runx3 up-regulation during CD8 T cell differentiation in the thymus. J Exp Med. 2009;206:2685–2699. doi: 10.1084/jem.20092024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Higuchi T, Bartel FO, Masuya M, Deguchi T, Henderson KW, Li R, Muise-Helmericks RC, Kern MJ, Watson DK, Spyropoulos DD. Thymomegaly, microsplenia, and defective homeostatic proliferation of peripheral lymphocytes in p51-Ets1 isoform-specific null mice. Mol Cell Biol. 2007;27:3353–3366. doi: 10.1128/MCB.01871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oh-hora M. Calcium signaling in the development and function of T-lineage cells. Immunol Rev. 2009;231:210–224. doi: 10.1111/j.1600-065X.2009.00819.x. [DOI] [PubMed] [Google Scholar]
- 73.Vig M, Kinet JP. Calcium signaling in immune cells. Nat Immunol. 2009;10:21–27. doi: 10.1038/ni.f.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oh-Hora M, Yamashita M, Hogan PG, Sharma S, Lamperti E, Chung W, Prakriya M, Feske S, Rao A. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beyersdorf N, Braun A, Vogtle T, Varga-Szabo D, Galdos RR, Kissler S, Kerkau T, Nieswandt B. STIM1-independent T cell development and effector function in vivo. J Immunol. 2009;182:3390–3397. doi: 10.4049/jimmunol.0802888. [DOI] [PubMed] [Google Scholar]
- 76.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 77.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 78.Lin X, Wang D. The roles of CARMA1, Bcl10, and MALT1 in antigen receptor signaling. Semin Immunol. 2004;16:429–435. doi: 10.1016/j.smim.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 79.Schmidt-Supprian M, Courtois G, Tian J, Coyle AJ, Israel A, Rajewsky K, Pasparakis M. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 80.Boothby MR, Mora AL, Scherer DC, Brockman JA, Ballard DW. Perturbation of the T lymphocyte lineage in transgenic mice expressing a constitutive repressor of nuclear factor (NF)-kappaB. J Exp Med. 1997;185:1897–1907. doi: 10.1084/jem.185.11.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hettmann T, Leiden JM. NF-kappa B is required for the positive selection of CD8+ thymocytes. J Immunol. 2000;165:5004–5010. doi: 10.4049/jimmunol.165.9.5004. [DOI] [PubMed] [Google Scholar]
- 82.Mora AL, Stanley S, Armistead W, Chan AC, Boothby M. Inefficient ZAP-70 phosphorylation and decreased thymic selection in vivo result from inhibition of NF-kappaB/Rel. J Immunol. 2001;167:5628–5635. doi: 10.4049/jimmunol.167.10.5628. [DOI] [PubMed] [Google Scholar]
- 83.Pasparakis M, Luedde T, Schmidt-Supprian M. Dissection of the NF-kappaB signalling cascade in transgenic and knockout mice. Cell Death Differ. 2006;13:861–872. doi: 10.1038/sj.cdd.4401870. [DOI] [PubMed] [Google Scholar]
- 84.Jost PJ, Weiss S, Ferch U, Gross O, Mak TW, Peschel C, Ruland J. Bcl10/Malt1 signaling is essential for TCR-induced NF-kappaB activation in thymocytes but dispensable for positive or negative selection. J Immunol. 2007;178:953–960. doi: 10.4049/jimmunol.178.2.953. [DOI] [PubMed] [Google Scholar]
- 85.Jimi E, Strickland I, Voll RE, Long M, Ghosh S. Differential role of the transcription factor NF-kappaB in selection and survival of CD4+ and CD8+ thymocytes. Immunity. 2008;29:523–537. doi: 10.1016/j.immuni.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M. Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science. 2005;307:1465–1468. doi: 10.1126/science.1104765. [DOI] [PubMed] [Google Scholar]
- 87.Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 88.Sato S, Sanjo H, Tsujimura T, Ninomiya-Tsuji J, Yamamoto M, Kawai T, Takeuchi O, Akira S. TAK1 is indispensable for development of T cells and prevention of colitis by the generation of regulatory T cells. Int Immunol. 2006;18:1405–1411. doi: 10.1093/intimm/dxl082. [DOI] [PubMed] [Google Scholar]
- 89.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delaney JR, Mlodzik M. TGF-beta activated kinase-1: new insights into the diverse roles of TAK1 in development and immunity. Cell Cycle. 2006;5:2852–2855. doi: 10.4161/cc.5.24.3558. [DOI] [PubMed] [Google Scholar]
- 91.Chariot A. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 2009;19:404–413. doi: 10.1016/j.tcb.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 92.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10:1057–1063. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kerdiles YM, Beisner DR, Tinoco R, Dejean AS, Castrillon DH, DePinho RA, Hedrick SM. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ouyang W, Beckett O, Flavell RA, Li MO. An essential role of the Forkheadbox transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–71. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leenders H, Whiffield S, Benoist C, Mathis D. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur J Immunol. 2000;30:2980–2990. doi: 10.1002/1521-4141(200010)30:10<2980::AID-IMMU2980>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 96.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 97.Kuo TC, Schlissel MS. Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol. 2009;21:173–178. doi: 10.1016/j.coi.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Webb LM, Vigorito E, Wymann MP, Hirsch E, Turner M. Cutting edge: T cell development requires the combined activities of the p110gamma and p110delta catalytic isoforms of phosphatidylinositol 3-kinase. J Immunol. 2005;175:2783–2787. doi: 10.4049/jimmunol.175.5.2783. [DOI] [PubMed] [Google Scholar]
- 99.Ciofani M, Zuniga-Pflucker JC. Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 100.Carlson CM, Endrizzi BT, Wu J, Ding X, Weinreich MA, Walsh ER, Wani MA, Lingrel JB, Hogquist KA, Jameson SC. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 101.Schluns KS, Kieper WC, Jameson SC, Lefrancois L. Interleukin-7 mediates the homeostasis of naive and memory CD8 T cells in vivo. Nat Immunol. 2000;1:426–432. doi: 10.1038/80868. [DOI] [PubMed] [Google Scholar]
- 102.Tan JT, Dudl E, LeRoy E, Murray R, Sprent J, Weinberg KI, Surh CD. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc Natl Acad Sci U S A. 2001;98:8732–8737. doi: 10.1073/pnas.161126098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grenningloh R, Miaw SC, Moisan J, Graves BJ, Ho IC. Role of Ets-1 phosphorylation in the effector function of Th cells. Eur J Immunol. 2008;38:1700–1705. doi: 10.1002/eji.200738112. [DOI] [PubMed] [Google Scholar]
- 104.Bettini M, Xi H, Milbrandt J, Kersh GJ. Thymocyte development in early growth response gene 1-deficient mice. J Immunol. 2002;169:1713–1720. doi: 10.4049/jimmunol.169.4.1713. [DOI] [PubMed] [Google Scholar]
- 105.Lauritsen JP, Kurella S, Lee SY, Lefebvre JM, Rhodes M, Alberola-Ila J, Wiest DL. Egr2 is required for Bcl-2 induction during positive selection. J Immunol. 2008;181:7778–7785. doi: 10.4049/jimmunol.181.11.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lawson VJ, Weston K, Maurice D. Early growth response 2 regulates the survival of thymocytes during positive selection. Eur J Immunol. 2009 doi: 10.1002/eji.200939567. [DOI] [PubMed] [Google Scholar]
- 107.Carter JH, Lefebvre JM, Wiest DL, Tourtellotte WG. Redundant role for early growth response transcriptional regulators in thymocyte differentiation and survival. J Immunol. 2007;178:6796–6805. doi: 10.4049/jimmunol.178.11.6796. [DOI] [PubMed] [Google Scholar]
- 108.Rivera RR, Johns CP, Quan J, Johnson RS, Murre C. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 2000;12:17–26. doi: 10.1016/s1074-7613(00)80155-7. [DOI] [PubMed] [Google Scholar]; 108a Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–171. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 109.Sawada S, Littman DR. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993;13:5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, Littman DR. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 111.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 112.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–870. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xi H, Schwartz R, Engel I, Murre C, Kersh GJ. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–826. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 114.Koltsova EK, Ciofani M, Benezra R, Miyazaki T, Clipstone N, Zuniga-Pflucker JC, Wiest DL. Early growth response 1 and NF-ATc1 act in concert to promote thymocyte development beyond the beta-selection checkpoint. J Immunol. 2007;179:4694–4703. doi: 10.4049/jimmunol.179.7.4694. [DOI] [PubMed] [Google Scholar]
- 115.Marsden VS, Strasser A. Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu Rev Immunol. 2003;21:71–105. doi: 10.1146/annurev.immunol.21.120601.141029. [DOI] [PubMed] [Google Scholar]
- 116.Fry TJ, Mackall CL. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J Immunol. 2005;174:6571–6576. doi: 10.4049/jimmunol.174.11.6571. [DOI] [PubMed] [Google Scholar]
- 117.Rengarajan J, Mittelstadt PR, Mages HW, Gerth AJ, Kroczek RA, Ashwell JD, Glimcher LH. Sequential involvement of NFAT and Egr transcription factors in FasL regulation. Immunity. 2000;12:293–300. doi: 10.1016/s1074-7613(00)80182-x. [DOI] [PubMed] [Google Scholar]
- 118.Hoffmann E, Ashouri J, Wolter S, Doerrie A, Dittrich-Breiholz O, Schneider H, Wagner EF, Troppmair J, Mackman N, Kracht M. Transcriptional regulation of EGR-1 by the interleukin-1-JNK-MKK7-c-Jun pathway. J Biol Chem. 2008;283:12120–12128. doi: 10.1074/jbc.M800583200. [DOI] [PubMed] [Google Scholar]
- 119.Wang X, Zhang Y, Xiao G, Gao X, Liu X. c-Fos enhances the survival of thymocytes during positive selection by upregulating Bcl-2. Cell Res. 2009;19:340–347. doi: 10.1038/cr.2008.322. [DOI] [PubMed] [Google Scholar]
- 120.Collins S, Wolfraim LA, Drake CG, Horton MR, Powell JD. Cutting Edge: TCR-induced NAB2 enhances T cell function by coactivating IL-2 transcription. J Immunol. 2006;177:8301–8305. doi: 10.4049/jimmunol.177.12.8301. [DOI] [PubMed] [Google Scholar]
- 121.Laky K, Fowlkes B. Receptor signals and nuclear events in CD4 and CD8 T cell lineage commitment. Curr Opin Immunol. 2005;17:116–121. doi: 10.1016/j.coi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 122.Hedrick SM. Thymus lineage commitment: a single switch. Immunity. 2008;28:297–299. doi: 10.1016/j.immuni.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 123.Wang L, Wildt KF, Zhu J, Zhang X, Feigenbaum L, Tessarollo L, Paul WE, Fowlkes BJ, Bosselut R. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol. 2008;9:1122–1130. doi: 10.1038/ni.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 126.van Hamburg JP, de Bruijn MJ, Ribeiro de Almeida C, Dingjan GM, Hendriks RW. Gene expression profiling in mice with enforced Gata3 expression reveals putative targets of Gata3 in double positive thymocytes. Mol Immunol. 2009;46:3251–3260. doi: 10.1016/j.molimm.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 127.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 128.Aliahmad P, O’Flaherty E, Han P, Goularte OD, Wilkinson B, Satake M, Molkentin JD, Kaye J. TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med. 2004;199:1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aliahmad P, Kaye J. Commitment issues: linking positive selection signals and lineage diversification in the thymus. Immunol Rev. 2006;209:253–273. doi: 10.1111/j.0105-2896.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 130.Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu X, Taylor BJ, Sun G, Bosselut R. Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol. 2005;175:4465–4474. doi: 10.4049/jimmunol.175.7.4465. [DOI] [PubMed] [Google Scholar]
- 133.Muroi S, Naoe Y, Miyamoto C, Akiyama K, Ikawa T, Masuda K, Kawamoto H, Taniuchi I. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol. 2008;9:1113–1121. doi: 10.1038/ni.1650. [DOI] [PubMed] [Google Scholar]
- 134.Sharp LL, Schwarz DA, Bott CM, Marshall CJ, Hedrick SM. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 1997;7:609–618. doi: 10.1016/s1074-7613(00)80382-9. [DOI] [PubMed] [Google Scholar]
- 135.Bommhardt U, Basson MA, Krummrei U, Zamoyska R. Activation of the extracellular signal-related kinase/mitogen-activated protein kinase pathway discriminates CD4 versus CD8 lineage commitment in the thymus. J Immunol. 1999;163:715–722. [PubMed] [Google Scholar]
- 136.Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- 137.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 138.Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, Singer A. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. [DOI] [PubMed] [Google Scholar]
- 139.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 140.Fabre S, Carrette F, Chen J, Lang V, Semichon M, Denoyelle C, Lazar V, Cagnard N, Dubart-Kupperschmitt A, Mangeney M, Fruman DA, Bismuth G. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181:2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 141.Gubbels Bupp MR, Edwards B, Guo C, Wei D, Chen G, Wong B, Masteller E, Peng SL. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur J Immunol. 2009;39:2991–2999. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Park JH, Yu Q, Erman B, Appelbaum JS, Montoya-Durango D, Grimes HL, Singer A. Suppression of IL7Ralpha transcription by IL-7 and other prosurvival cytokines: a novel mechanism for maximizing IL-7-dependent T cell survival. Immunity. 2004;21:289–302. doi: 10.1016/j.immuni.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 143.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110delta isoform of PI3K. Trends Immunol. 2007;28:80–87. doi: 10.1016/j.it.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kuo CT, Veselits ML, Leiden JM. LKLF: A transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 145.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 146.Cobb BS, Hertweck A, Smith J, O’Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O’Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matthews SA, Cantrell DA. New insights into the regulation and function of serine/threonine kinases in T lymphocytes. Immunol Rev. 2009;228:241–252. doi: 10.1111/j.1600-065X.2008.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Laky K, Fowlkes BJ. Presenilins regulate alphabeta T cell development by modulating TCR signaling. J Exp Med. 2007;204:2115–2129. doi: 10.1084/jem.20070550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Takagi T, Harada J, Ishii S. Murine Schnurri-2 is required for positive selection of thymocytes. Nat Immunol. 2001;2:1048–1053. doi: 10.1038/ni728. [DOI] [PubMed] [Google Scholar]
- 153.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 155.Yu M, Wan M, Zhang J, Wu J, Khatri R, Chi T. Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci U S A. 2008;105:3873–3878. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc Natl Acad Sci U S A. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- 158.Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS. RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation. Immunity. 2002;17:617–627. doi: 10.1016/s1074-7613(02)00451-x. [DOI] [PubMed] [Google Scholar]