Abstract
Background
All four Francisella tularensis subspecies possess gene clusters with potential to express type IV pili (Tfp). These clusters include putative pilin genes, as well as pilB, pilC and pilQ, required for secretion and assembly of Tfp. A hallmark of Tfp is the ability to retract the pilus upon surface contact, a property mediated by the ATPase PilT. Interestingly, out of the two major human pathogenic subspecies only the highly virulent type A strains have a functional pilT gene.
Results
In a previous study, we were able to show that one pilin gene, pilA, was essential for virulence of a type B strain in a mouse infection model. In this work we have examined the role of several Tfp genes in the virulence of the pathogenic type A strain SCHU S4. pilA, pilC, pilQ, and pilT were mutated by in-frame deletion mutagenesis. Interestingly, when mice were infected with a mixture of each mutant strain and the wild-type strain, the pilA, pilC and pilQ mutants were out-competed, while the pilT mutant was equally competitive as the wild-type.
Conclusions
This suggests that expression and surface localisation of PilA contribute to virulence in the highly virulent type A strain, while PilT was dispensable for virulence in the mouse infection model.
Background
Francisella tularensis is a Gram-negative facultative intracellular bacterial pathogen and the causative agent of tularemia. Infections have been reported in a range of vertebrates as well as invertebrates [1]. Natural infections can occur via ingestion of contaminated food or water, handling of infected animals, bites by infected arthropods, including mosquitoes and ticks, or via inhalation [2]. F. tularensis is divided into four subspecies, where ssp. holarctica (type B) is most widely spread and found in the major part of Europe, Asia, and North America. F. tularensis ssp. tularensis (type A) is found exclusively in North America and ssp. mediasiatica in Central Asia. Finally, ssp. novicida has been isolated in several locations in North America, as well as in Australia [3,4]. Human infections are mainly caused by type A or type B strains, where type A strains are significantly more virulent than type B strains. Our knowledge regarding virulence determinants in F. tularensis is rather limited. However, available genome information [5,6] together with development of genetic tools [7], has resulted in increased understanding of the molecular mechanisms of F. tularensis infections.
The genome of F. tularensis encodes gene clusters involved in secretion and assembly of type IV pili (Tfp) [5]. Tfp are complex adhesins involved in important host cell interactions for human pathogens like Neisseria spp., Pseudomonas aeruginosa and Vibrio cholerae [8-11]. The pilus fiber is composed of one major pilin subunit and several additional minor pilins required for function and/or assembly of the pilus [12,13]. However, the exact roles of the minor pilins are still not completely understood. The pilus is translocated to the cell surface via the secretin, PilQ, which forms a pore in the outer membrane through which the pilus is transported and extended [14]. PilD is a peptidase cleaving the prepilin subunits [11] and PilC is a transmembrane protein spanning across the plasma membrane [15]. Furthermore, two ATPases, PilB and PilT, are involved in extension and retraction, of the pilus [16,17]. In some bacteria Tfp can mediate twitching motility, an activity that is PilT dependent [18].
There is evidence that F. tularensis expresses Tfp-like surface structures on the bacterial surface [19-21], and the putative pilin, PilA, has been shown to be required for virulence of type B strains in a mouse infection model [22]. Interestingly, due to direct repeat mediated deletion, the pilA gene has been lost in the attenuated live vaccine strain LVS [22,23], supporting the significance of PilA for virulence [24]. There are also other potentially significant differences between different F. tularensis subspecies. In ssp. novicida that is non-pathogenic for humans, PilA differs in the amino acid sequence compared to the virulent type A strain SCHU S4 [25]. On the contrary, pilA of virulent type B strains is essentially identical to the corresponding gene in type A strains, however, several other differences are apparent between the two subspecies. Two predicted pilin genes, pilE and pilV, and the ATPase encoding gene, pilT, are pseudogenes in type B strains [19,21,22,26]. These differences are likely to be significant since pilA has been linked to virulence of type B strains and is missing in the attenuated vaccine strain LVS. Furthermore, PilA of ssp. novicida was recently shown to be involved in protein secretion that was coupled to Tfp [20,25]. Interestingly, mutation of pilA and loss of protein secretion resulted in increased virulence in a mouse infection model [25]. As the human pathogenic type A and type B strains do not secrete detectable levels of proteins in vitro, it is possible that one step in the evolution of human pathogenic variants of F. tularensis from ssp. novicida has involved loss of protein secretion as a consequence of changes in PilA structure and function.
In this work we wanted to address the question if PilA is involved in virulence of the highly pathogenic type A strain SCHU S4, similarly to what we have previously shown for type B strains, and if Tfp secretion and assembly genes are required for virulence.
Results
Construction of non-polar pilin gene mutants
In a recent study, we were able to demonstrate that the pilA gene can be lost by a deletion event mediated by direct repeats flanking the gene [22]. Type B strains lacking pilA were found to be attenuated for virulence in a mouse infection model. In this study we wanted to extend this work to the highly pathogenic type A strain SCHU S4, and therefore we constructed a specific pilA deletion mutant using our previously described allelic exchange technique [7]. In addition, to address the significance of secretion and assembly of PilA, we also engineered in-frame deletions in pilC and pilQ, encoding a transmembrane protein and a secretin, respectively. For some pathogens, Tfp expression is associated with a unique ability to retract the pili, a phenotype depending on the ATPase PilT. Interestingly, pilT appears to be functional in type A strains, while it is a pseudogene in the less pathogenic type B strains. In order to elucidate if the expression of PilT could be correlated to the higher virulence of type A strains, we also constructed an in-frame deletion in the pilT gene.
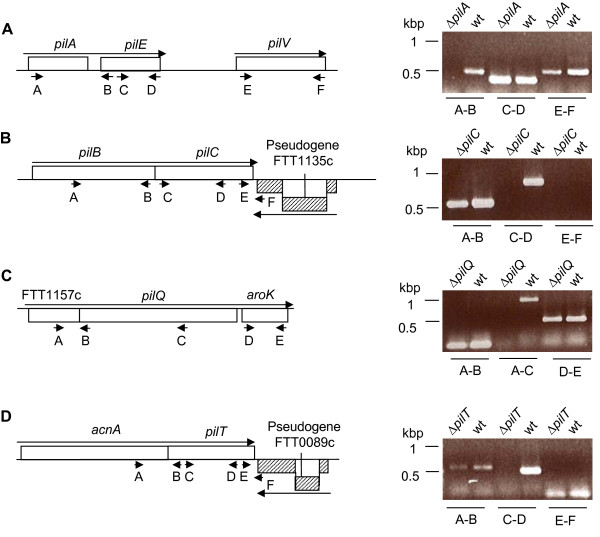
In order to verify that the mutations did not have a major impact on neighboring gene transcription, each region was analysed by RT-PCR on mRNA extracted from the mutant strains and compared to the isogenic wild-type strain (Fig. 1). Thereby we could confirm that none of the deletion events caused any polar effects on transcription. Both pilC and pilT are flanked by pseudogenes situated directly downstream of each gene that were found not to be transcribed neither in the wild-type nor in the pilC or pilT mutant strains. The upstream genes of pilC and pilT were readily transcribed at similar levels in the wild-type and mutant strains. In the case of the pilQ mutant, we could verify non-polarity on the downstream aroK gene.
Figure 1.
A-D. Analysis of gene transcription in wild-type and mutant strains of F. tularensis using RT-PCR of mRNA. The location of the different primers used is indicated by small arrows. Large arrows illustrate direction of transcription. Control reactions where reverse transcriptase was omitted were all negative (data not shown).
We also attempted to make an in-frame deletion of the pilA gene, but in spite of several attempts we were unable to generate an unmarked deletion. It is possible that this is linked to the fact that there are two direct repeats flanking pilA and that this somehow affects the recombination in this region [22]. We therefore chose a different strategy where we introduced a chloramphenicol resistance gene to allow for direct selection of the mutational event. In order to lower the risk of polar effects on the downstream pilE gene the resistance gene was inserted in the same orientation as the pilAE genes. We could also verify that the levels of pilE transcription were similar in the pilA mutant and wild-type strain, suggesting that there were no major polar effects on downstream genes. We have previously shown that pilV is transcribed from a promoter downstream of pilE gene in type strains [22] and also in this case pilV transcription levels were similar in the pilA and the wild-type strain. From this we conclude that none of the mutations generated any major polar effect on transcription of neighboring genes.
PilA expression in Tfp mutant strains
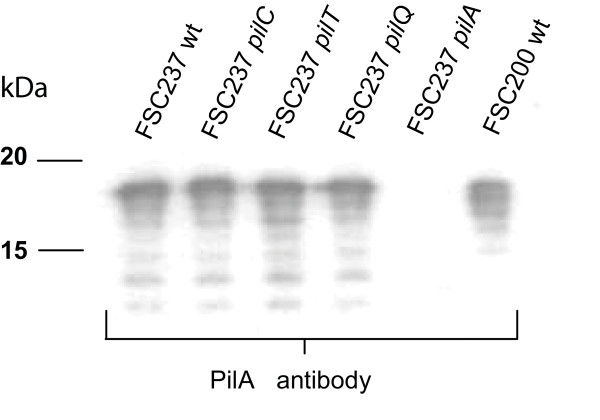
Next we wanted to address if any of the mutations influenced PilA expression. Therefore the expression of PilA in the different mutant strains was analysed by Western blot analysis. All mutants, except for pilA, expressed PilA at levels similar to the isogenic wild-type strain SCHU S4 (Fig. 2). The apparent molecular mass of PilA was similar to what has previously been shown for type B strains, 4-5 kDa larger than expected from their calculated molecular masses, indicating PilA to be post-transcriptionally modified, presumably by glycosylation [22]. Thus, with the exception of the pilA mutant, all the mutants expressed PilA at similar levels as the wild-type strain SCHU S4.
Figure 2.
PilA is expressed at wild-type levels in all strains, except for the pilA deletion mutant. Different pili mutants in the F. tularensis strain SCHU S4, analysed by Western blot using an anti-PilA antiserum. Lane; 1, FSC237 (SCHU S4, Type A); 2, FSC237 pilC deletion mutant; 3, FSC237 pilT deletion mutant; 4, FSC237 pilQ deletion mutant; 5, FSC237 pilA deletion mutant; 6, FSC200 (Type B).
pilA, pilC and pilQ contribute to virulence of SCHU S4
When we studied the role of pilA in LVS we could establish that the pilin had a major impact on virulence [24]. More recently, we have also made a specific pilA mutant in a recent clinical type B isolate. In this highly virulent type B strain, the attenuation seen for the pilA mutant was less marked, but still the lethal infection dose for the mutant was about 40-fold higher compared to the isogenic wild-type strain (unpublished data). Here, we wanted to address the role of pilA, the assembly/secretion genes, pilC and pilQ, and the ATPase encoding gene pilT, in the virulence of the highly pathogenic type A strain SCHU S4. First we verified that the growth kinetics in vitro was not affected in the mutant strains by measuring growth in liquid medium (data not shown). In order to evaluate the importance of the pili genes for in vivo virulence, mice were infected via the subcutaneous (s.c.) route with the mutant strains, as well as the isogenic wild-type strain SCHU S4. We used the s.c. route of infection as it can be more discriminative than the intraperitoneal (i.p.) route of infection. For instance, the attenuated vaccine strain LVS is still virulent by the i.p. route but highly attenuated by the s.c. route of infection in mice. Two different infection doses were used; approximately 10 and 100 bacteria respectively. Groups of six mice were infected with each dose and the progress of the infection was monitored over time. Small differences in infection kinetics were observed for the pilA, pilC and pilQ mutants, and mice infected with these strains showed a slightly delayed time to death compared to mice infected with the wild-type strain. Still, as SCHU S4 is very virulent in mice, even at the lowest doses (5 - 10 bacteria), all animals had succumbed to the infection after six to eight days post infection, making it difficult to establish the significance of the result (Fig. 3, Table 1). Therefore, we decided to perform competitive infections between the wild-type strain and the different isogenic mutants. In this case all mutants, except pilT, were out-competed by the wild-type strain SCHU S4. For the pilA, pilC and pilQ mutant strains, the ratios were 0.14, 0.34 and 0.16 (Table 1), respectively, suggesting PilA to be a virulence determinant also in the type A strain SCHU S4. The fact that the ratio was similar for the pilC and pilQ mutants indicate that assembly/surface localisation of the pilin PilA is required for full virulence in the mouse infection model. Statistical analysis verified that the differences in ratios for these three mutants were significant at P < 0.05 (data not shown). Somewhat surprisingly, the pilT mutant was not out-competed by the wild-type strain in the mixed infection experiment. The ratio (1.98) suggests that PilT is dispensable for virulence in the subcutaneous mouse infection model (Table 1). In this case the higher ratio for the pilT mutant was not statistically significant (data not shown).
Figure 3.
Infection kinetics are slightly delayed for mice infected with the pilA, pilC, and pilQ deletion strains. Groups of six mice were infected via the subcutaneous route and the survival followed over time. The exact dose for each strain was determined by retrospective viable count.
Table 1.
Mice infection data.
| SCHU S4 | Infection dose (cfu) |
CI value |
|---|---|---|
| wt | 11 | |
| pilA | 4.8 | 0.14 |
| pilC | 8.5 | 0.34 |
| pilT | 6.0 | 1.98 |
| pilQ | 10 | 0.16 |
Infection dose (cfu) in a standard infection study, and CI value in a competitive index assay.
Discussion
Recently, we have provided evidence that PilA, one of the type IV pilins encoded by the human pathogenic subspecies of F. tularensis, is an important virulence determinant for type B strains [22]. In addition, we have established that loss of the pilA gene is one of two major genetic events, responsible for the attenuation of the live vaccine strain, LVS [6,24]. Even though we have been able to demonstrate PilA to be both surface located in F. tularensis [22] and able to form functional Tfp in the heterologous system in Neisseria gonorrhoeae [27], we have still not managed to verify PilA to be an actual structural component of Tfp expressed by F. tularensis.
In this study, we present evidence that PilA and the Tfp assembly/secretion proteins, PilC and PilQ, are required for full virulence of the type A strain, SCHU S4, the most virulent subspecies of F. tularensis. In infections with individual mutants, we were unable to show that mutations of the putative Tfp genes resulted in a significant attenuation. However, when we conducted mixed infections, where the ability of the mutants to compete with the wild-type strain was assessed, it became more obvious that Tfp encoding genes may play a role in the virulence of SCHU S4. This is in line with our observation that pilA mutants in highly virulent clinical isolates of type B strains are less attenuated compared to type B strains with weaker virulence, like LVS [22,24]. A general problem with the mouse infection model is that mice are highly susceptible to Francisella and do not discriminate between the virulence properties of different F. tularensis subspecies in the same way as the human infection. The emerging picture is that pilA mutants show less attenuation in the most pathogenic subspecies. Still, we believe that PilA, and potentially also Tfp, may play an important role in virulence. This theory is supported by the fact that LVS has lost pilA, and that this is one of the causes of its attenuation [24]. When genomes of different subspecies are compared, one striking difference is that the pilT gene is a pseudogene in type B strains, due to a point mutation introducing a stop codon in the middle of the gene [26]. Interestingly, in a study involving the attenuated type B strain LVS the pilT gene was demonstrated to be involved in pili assembly, adherence and virulence [19]. Chakraborty with colleagues have suggested the possibility that the truncated PilT protein somehow has retained function in LVS [19]. Their findings are somewhat surprising since in other Tfp expressing pathogens the PilT protein is only involved in pilus retraction and not in pilus assembly. The pilT mutant in SCHU S4 did not have any impact on the virulence in the subcutaneous mouse infection model. However, the fact that pilT is intact in most pathogenic type A strains suggests that PilT might, at least partly, contribute to the higher virulence of type A strains. As mentioned above, this infection model has limitations and may not reflect virulence in humans. Another possibility is that PilT may rather play a role in the environment and/or in transmission of tularemia than in the animal/human infection. With the genetic tools and the availability of specific mutants in the Tfp encoding gene clusters of SCHU S4, it will now be possible to address the role of the Tfp system in other infection models, for survival in the environment, and perchance for vector-borne transmission.
Conclusions
We have shown that pilA is required for full virulence of SCHU S4 in mice - a result in line with our earlier findings in type B strains. In addition, we have also demonstrated that the pilin assembly genes, pilC and pilQ, are needed for full virulence of SCHU S4. An unexpectedly finding is that PilT, even though it is functional only in type A strains, did not contribute to virulence in the mouse subcutaneous infection model.
Methods
Bacterial strains, plasmids, growth conditions, and DNA methods
The bacterial strains and plasmids used in this study are listed in Table 2. F. tularensis strains were grown on modified Thayer-Martin agar or Blood Cystine Glucose agar (BCGA) at 37ºC in 5% CO2. Escherichia coli strains were grown on blood agar base (BAB; Merck) plates or in Luria Bertani broth (LB). Antibiotics were used at the following concentrations: kanamycin 50 μg/ml and chloramphenicol 2.5 μg/ml (F. tularensis), or 25 μg/ml (E. coli). Preparation of plasmid DNA, restriction enzyme digests, ligations and transformations into E. coli were performed essentially as described [28]. Generally, the primers (Table 3) were constructed based on the genomic information from the FSC237 (SCHU S4) and FSC155 (LVS) genomes. The amplified PCR fragments were first cloned into the pCR®4.0-TOPO cloning vector (Invitrogen AB, Stockholm, Sweden), sequenced by Eurofins MWG Operon, and subsequently cloned into the suicide vectors pSMP22 [29] or pDM4 [30].
Table 2.
Strains and plasmids used in this study
| Strains | Genotype/phenotype | Source |
|---|---|---|
| F. tularensis | ||
| FSC237 | tularensis; SCHU S4 | Human ulcer 1941, Ohio |
| FSC237; ΔpilA; deletion of codons 1-135 | This study | |
| FSC237; ΔpilC (FTT1134); in frame deletion of codons 5-405 | This study | |
| FSC237; ΔpilQ (FTT1156); in frame deletion of codons 13-593 | This study | |
| FSC237; ΔpilT (FTT0088); in frame deletion of codons 7-336 | This study | |
| E. coli | ||
| Top10 | F- mcrA Δ(mrr-hsdRMS-mcrBC), Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 (Δara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| S17-1Λpir | recA, thi, pro, hsdR-M+,<RP4:2-Tc:Mu:Km:Tn7> TpR, SmR | [32] |
| Plasmids | ||
| pCR®4.0 | TOPO-cloning vector. AmpR, KmR | Invitrogen |
| pDM4 | Suicide plasmid. sacB; mobRP4; oriR6K; CmR | [30] |
| pSMP22 | Suicide plasmid. groESL promoter, ori T, bla, sacB | [29] |
| pSMP50CAM | 432 bp fragment of pilA including a chlorampenicol resistance cassette cloned into pSMP22. CmR | This study |
| pAL12 | 2072 bp fragment of approximately 1 kb upstream and 1 kb downstream of pilC cloned in XbaI and SalI site of pDM4. CmR | This study |
| pAL16 | 2122 bp fragment of approximately 1 kb upstream and 1 kb downstream of pilQ cloned in XbaI and SalI site of pDM4. CmR | This study |
| pAL18 | 2133 bp fragment of approximately 1 kb upstream and 1 kb downstream of pilT cloned in XbaI and SalI site of pDM4. CmR | This study |
Table 3.
Primers used in this study
| Primer | Primer sequence 5'-3' | RE site |
|---|---|---|
| pilA LFF | GAGCTCACGCGT-CTTACTTGCCGGATCATTACCAAC | SphI |
| pilA LFR | CTGCAG-CCTTCTTTATAGTTTAGTTTAC | PstI |
| pilA RFF | CTGCAGGTAGATAAACTAAGCCACTTTCATGTG | PstI |
| pilA RFR | GGATCCGCATGCTCAAGGCTTCTGTCAATCTTGTTC | MluI |
| CAM PstIF | GCCTGCAGGTAAGAGGTTCCAACTTTCAC | PstI |
| CAM PstIR | TGATCTGCAGTTACGCCCCGCCCTGCCACTCATC | PstI |
| PilC-A | GCATGTCCTAGGGTCAAGCTTAGATATTGCTGAA | AvrII |
| PilC-B | TATATCGCATCGCCAAATAGCATATTTTTTATTCC | |
| PilC-C | GCTATTTGGCGATGCGATATAATATACTTTTAAAAA | |
| PilC-D | GCATGTGTCGACGTCCTGAGAAAATATCTAGACA | SalI |
| PilT-A | CATTATGTCGACTATGCAACAGTTCTTGATGGT | SalI |
| PilT-B | TACTACAATGTATAGTAATTTTCTTATCATATCAAG | |
| PilT-C | AGAAAATTACTATACATTGTAGTAAGGTAATCA | |
| PilT-D | CATTATTCTAGACAGGATTAACGGCAGCTAAAA | XbaI |
| PilQ-A3 | GCATGTCCTAGG TCAGTCAATGGAAGCACAGAT | AvrII |
| PilQ-B3 | TATCTGCTATCATGTTAGAACAACTAATAACTTCTT | |
| PilQ-C3 | TTGT TCTAACATGATAGCAGATAATAGTTGCAAA | |
| PilQ-D3 | GCATGTGTCGACAGAAAGTAATGTTGTTGGTATTT | SalI |
|
RT-PCR primers |
||
| PilA_A | GATCCCGATGTACTCTAACTA | |
| PilA_B | CCATTAGCTCAACTAGTGAGAA | |
| PilA_C | ATCTTAGCAGCTGTAGCAATA | |
| PilA_D | GGGGTAGTACTTTAAATCCT | |
| PilA_E | CTTACTGAGTTACTTGTTGTTAT | |
| PilA_F | GTCTTTCTGATCTATATGCTTC | |
| PilC_A | GTCAAGCTTAGATATTGCTGAA | |
| PilC_B | GTCTCTGGAGCACTGTTTGTAT | |
| PilC_C | AAGGTAGTATTGATGCTGACAC | |
| PilC_D | CCGTTGCTAAAGACACCATA | |
| PilC_E | GATGCGATATAATATACTTTTAAAAA | |
| PilC_F | CGAATTGGTATTGGCCAGAT | |
| PilQ_A | TATGGTCAGGTAGAAGATGTAA | |
| PilQ_B | CATCAATTTACCTTACTAATGTAT | |
| PilQ_C | GCCTGAGCAGTAGTATAGTTT | |
| PilQ_D | AGTTGGTGCTGGAAAATCTAC | |
| PilQ_E | CAGGATAGTTTCTTCTACTAAA | |
| PilT_A | CTATTAGGCGTGAAAGCAGTT | |
| PilT_B | TAGTAATTTTCTTATCATATCAAG | |
| PilT_C | ATGATGCGAGATTTAGGGTA | |
| PilT_D | CAGCAGGTGGAAATACAGAT | |
| PilT_E | TACATTGTAGTAAGGTAATCA | |
| PilT_F | GGTAGAGTTGAATCAGCGTTTA |
Construction of deletion mutants of pilA, pilC, pilQ, and pilT in FSC237
Left and right flanking regions of pilA (FTT0890c, SCHU S4 nomenclature) were PCR amplified using the primer pairs pilA_LFF/pilA_LFR and pilA_RFF/pilA_RFR, and cloned into pGEMT-easy (Promega). The left flank was excised with EcoRI and PstI and the right flank was excised with BamHI and PstI. The fragments were ligated into an EcoRI/BamHI digested pBluescript KS+ vector (Stratagene), giving rise to pSMP47. A chloramphenicol resistance gene was PCR amplified from pDM4 with the primer pair CAM_PstIF/CAM_PstIR, digested with PstI, and cloned into pSMP47, generating pSMP48 containing the left and right flanks of pilA disrupted by a chloramphenicol cassette. The mutated allele of pilA was excised from pSMP48 with SphI and MluI, cloned into pSMP22, and the resulting plasmid pSMP50CAM (Table 2) was introduced into strain FSC237 by conjugal mating as previously described [7].
The deletion constructs for pilC, pilQ and pilT were obtained by overlapping PCR using the following primer pairs PilX_A, PilX_B, PilX_C and PilX_D (Table 3). For the deletion constructs of pilC and pilQ strain FSC237 was used as template and for the pilT deletion the strain FSC155 was used as a template. The sequence for the pilT construct is almost identical between FSC155 and FSC237 except for three substitutions upstream of the deletion in non-coding sequences, and eight substitutions in a downstream pseudogene. The PCR fragments were cloned into the suicide vector pDM4 and the resulting plasmids pAL12 (pilC), pAL16 (pilQ), and pAL18 (pilT) (Table 2) were introduced into strain FSC237 by conjugal mating as previously described [7]. The in vitro growth rate of the different mutant strains were compared with the wild type strain by measuring OD at different time points, 0 h, 6 h and ON after dilution in Chamberlain medium.
RNA isolation and RT-PCR
Bacteria were grown for 18 h on plates, harvested and suspended in TRIzol reagent (Life Technologies). Total RNA was extracted and treated with RNase-free DNase I (Roche), phenol extracted, and precipitated by ethanol. An aliquot of the RNA (3 μg) was used to synthesize cDNA using random hexamers (final concentration 25 ng/μl) and Superscript III reverse transcriptase as described by the manufacturer (Life Technologies). In control experiments samples processed without addition of RT enzyme were used.
Animal infections
F. tularensis strains were grown for 16 h on BCGA before the bacteria were suspended in phosphate buffered saline (PBS) pH 7.4 to an OD540 = 1, which normally corresponds to approximately 2 × 109 bacteria/ml. The bacterial suspension was then diluted in PBS into two doses used for challenge, around 10 and 100 bacteria in a total volume of 100 μl. All bacterial infections were initiated by subcutaneous injections of 6-8 week old C57Black/6 female mice. The study was approved by the Local Ethical Committee on Laboratory Animals in Umeå, Sweden. For competitive index (CI) infections, the mice were infected with a 50:50 mixture of mutant and wild-type strains with around 50 bacteria of each strain. Mice were culled five days post-infection, and the spleens were homogenized in 1 ml of PBS and spread on BCGA. Individual colonies were analysed by PCR with primers specific for each mutation in order to examine the distribution of each strain. Spleens from at least three animals were collected for each pair of strains, and at least 200 colonies were analysed by PCR. The CI was calculated for each strain by dividing the ratio of mutant/wt after infection (determined with PCR) with the ratio of mutant/wt before infection (determined by viable count). Statistical analysis was performed with a GraphPad Prism computer software program using a paired Student's t-test (one-tailed) where P < 0.05 was regarded as significant.
Gel electrophoresis and Western blotting
Samples were boiled in the presence of SDS and Β-mercaptoethanol for 5 min and then separated on a 12% acrylamide gel by electrophoresis as described by Laemmli [31]. Proteins were transferred to Immobilon-P Transfer Membranes using a Trans-Blot Semi-Dry transfer cell (BioRad). Membranes were blocked overnight in Tris-buffered saline (TBS) with 5% nonfat dry milk. Membranes were probed with rabbit polyclonal anti-PilA sera [22] and a horseradish peroxidase-conjugated anti rabbit antibody (Amersham Pharmacia Biotech) was used as secondary antibody and the filters were developed by using the ECL Kit (Amersham Pharmacia Biotech) according to the instructions from the manufacturer.
Authors' contributions
ALF carried out major parts the molecular genetic studies, participated in analysing samples from the animal assay and drafted the manuscript. ENS carried out parts of the molecular genetic studies, participated in analysing samples from the animal assay and drafted the manuscript. IG carried out parts the molecular genetic studies. KK analysed samples from the animal assay and performed the transcriptional analysis. SM carried out parts of the molecular genetics studies. RT was supervising and coordinating parts of the molecular genetics studies. PO supervised and also carried out key parts of the animal work and was involved in supervising the molecular genetics work. LN was involved in analysing bacterial ratios from animal samples and editing of the manuscript. AS supervised the molecular genetics work for parts of the mutagenesis work. ÅF conceived of the study, participated in its design, coordination and helped to draft and edit the manuscript. All authors read and approved the final manuscript.
Contributor Information
Anna-Lena Forslund, Email: annalena.forslund@foi.se.
Emelie Näslund Salomonsson, Email: emelie.salomonsson@foi.se.
Igor Golovliov, Email: igor.golovlev@climi.umu.se.
Kerstin Kuoppa, Email: kerstin.kuoppa@foi.se.
Stephen Michell, Email: S.L.Michell@exeter.ac.uk.
Richard Titball, Email: R.W.Titball@exeter.ac.uk.
Petra Oyston, Email: PCOYSTON@mail.dstl.gov.uk.
Laila Noppa, Email: laila.noppa@foi.se.
Anders Sjöstedt, Email: anders.sjostedt@climi.umu.se.
Åke Forsberg, Email: ake.forsberg@molbiol.umu.se.
References
- Mörner T. The ecology of tularaemia. Rev Sci Tech. 1992;11:1123–1130. [PubMed] [Google Scholar]
- Tärnvik A. Nature of protective immunity to Francisella tularensis. Rev Infect Dis. 1989;11:440–451. [PubMed] [Google Scholar]
- Petersen J, Schriefer M. Tularemia: emergence/re-emergence. Vet Res. 2005;36:455–467. doi: 10.1051/vetres:2005006. [DOI] [PubMed] [Google Scholar]
- Whipp M, Davis J, Lum G, de Boer J, Zhou Y. et al. Characterization of a novicida-like subspecies of Francisella tularensis isolated in Australia. J Med Microbiol. 2003;52:839–842. doi: 10.1099/jmm.0.05245-0. [DOI] [PubMed] [Google Scholar]
- Larsson P, Oyston P, Chain P, Chu M, Duffield M. et al. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat Genet. 2005;37:153–159. doi: 10.1038/ng1499. [DOI] [PubMed] [Google Scholar]
- Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman T. et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I, Sjöstedt A, Mokrievich A, Pavlov V. A method for allelic replacement in Francisella tularensis. FEMS Microbiol Lett. 2003;222:273–280. doi: 10.1016/S0378-1097(03)00313-6. [DOI] [PubMed] [Google Scholar]
- Fullner K, Mekalanos J. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J, Whitchurch C, Alm R. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa--a review. Gene. 1996;179:147–155. doi: 10.1016/S0378-1119(96)00441-6. [DOI] [PubMed] [Google Scholar]
- Tønjum T, Koomey M. The pilus colonization factor of pathogenic neisserial species: organelle biogenesis and structure/function relationships--a review. Gene. 1997;192:155–163. doi: 10.1016/S0378-1119(97)00018-8. [DOI] [PubMed] [Google Scholar]
- Strom M, Nunn D, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S, Dyer D, Nassif X, Pelicic V, Forest K. 3 D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci USA. 2007;104:15888–15893. doi: 10.1073/pnas.0707581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther-Larsen H, Wolfgang M, Dunham S, van Putten J, Dorward D. et al. A conserved set of pilin-like molecules controls type IV pilus dynamics and organelle-associated functions in Neisseria gonorrhoeae. Mol Microbiol. 2005;56:903–917. doi: 10.1111/j.1365-2958.2005.04591.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, van Putten J, Hayes S, Dorward D, Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 2000;19:6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom M, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- Whitchurch C, Hobbs M, Livingston S, Krishnapillai V, Mattick J. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-V. [DOI] [PubMed] [Google Scholar]
- Skerker J, Berg H. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Monfett M, Maier T, Benach J, Frank D. et al. Type IV pili in Francisella tularensis: roles of pilF and pilT in fiber assembly, host cell adherence, and virulence. Infect Immun. 2008;76:2852–2861. doi: 10.1128/IAI.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X, Chakraborty S, Liu J, Thanassi D, Klose K. Characterization of the Francisella tularensis subsp. novicida type IV pilus. Microbiology. 2008;154:2139–2150. doi: 10.1099/mic.0.2008/018077-0. [DOI] [PubMed] [Google Scholar]
- Gil H, Benach J, Thanassi D. Presence of pili on the surface of Francisella tularensis. Infect Immun. 2004;72:3042–3047. doi: 10.1128/IAI.72.5.3042-3047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund A, Kuoppa K, Svensson K, Salomonsson E, Johansson A. et al. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol Microbiol. 2006;59:1818–1830. doi: 10.1111/j.1365-2958.2006.05061.x. [DOI] [PubMed] [Google Scholar]
- Svensson K, Larsson P, Johansson D, Byström M, Forsman M. et al. Evolution of subspecies of Francisella tularensis. J Bacteriol. 2005;187:3903–3908. doi: 10.1128/JB.187.11.3903-3908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson E, Kuoppa K, Forslund A, Zingmark C, Golovliov I. et al. Reintroduction of two deleted virulence loci restores full virulence to the live vaccine strain of Francisella tularensis. Infect Immun. 2009;77:3424–3431. doi: 10.1128/IAI.00196-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A, Bolton D, Pelletier M, Brittnacher M, Gallagher L. et al. Type IV pili-mediated secretion modulates Francisella virulence. Mol Microbiol. 2006;62:227–237. doi: 10.1111/j.1365-2958.2006.05365.x. [DOI] [PubMed] [Google Scholar]
- Rohmer L, Brittnacher M, Svensson K, Buckley D, Haugen E. et al. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect Immun. 2006;74:6895–6906. doi: 10.1128/IAI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonsson E, Forsberg A, Roos N, Holz C, Maier B. et al. Functional analyses of pilin-like proteins from Francisella tularensis: complementation of type IV pilus phenotypes in Neisseria gonorrhoeae. Microbiology. 2009;155:2546–2559. doi: 10.1099/mic.0.028183-0. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. 654 s. med var. pag. [Google Scholar]
- Thomas R, Titball R, Oyston P, Griffin K, Waters E. et al. The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect Immun. 2007;75:371–378. doi: 10.1128/IAI.01241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton D, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Bio/Technology. 1983. pp. 784–791. [DOI]