Abstract
Low oxygen tension–mediated transcription by hypoxia-inducible factors (HIF) has been reported to facilitate tumor progression, therapeutic resistance, and metastatic adaptation. One previously described target of hypoxia-mediated transcription is the cytokine/growth factor macrophage migration inhibitory factor (MIF). In studies designed to better understand hypoxia-stimulated MIF function, we have discovered that not only is MIF induced by hypoxia in pancreatic adenocarcinoma but MIF is also necessary for maximal hypoxia-induced HIF-1α expression. Cells lacking MIF are defective in hypoxia- and prolyl hydroxylase inhibitor–induced HIF-1α stabilization and subsequent transcription of glycolytic and angiogenic gene products. Moreover, COP9 signalosome subunit 5 (CSN5), a component of the COP9 signalosome previously reported to functionally interact with MIF, has recently been shown to interact with and stabilize HIF-1α. Our results indicate that MIF interacts with CSN5 in pancreatic cancer cells and that MIF-depleted cells display marked defects in hypoxia-induced CSN5/HIF-1α interactions. This functional interdependence between HIF-1α and MIF may represent an important and previously unrecognized protumorigenic axis.
Introduction
Hypoxia-inducible factor (HIF) transcription factors have been implicated in controlling the expression of a wide variety of genes implicated in apoptosis, angiogenesis, invasion, and metastasis (1, 2). Subsequent studies on the von Hippel-Lindau tumor suppressor protein (pVHL) revealed that wild-type pVHL is the recognition component of an E3-ubiquitin ligase that controls the ubiquitylation of HIF-1α subunits (3). Multiple studies have reported significant correlations between HIF-1α expression and disease progression, therapeutic resistance, and fatality in a number of common human cancers (4, 5).
Recent studies have suggested that COP9 signalosome subunit 5 (CSN5) functionally binds to the CODD domain of both HIF-1α and the pVHL tumor suppressor (6). High CSN5 expression generates a pVHL-independent form of CSN5 that stabilizes HIF-1α aerobically by inhibiting HIF-1α prolyl-564 hydroxylation. Aerobic CSN5 association with HIF-1α occurs independently of the CSN holocomplex, leading to HIF-1α stabilization independent of Cullin 2 deneddylation. CSN5 also associates with HIF-1α under hypoxia and is required for optimal hypoxia-mediated HIF-1α stabilization (6).
Classically defined as an inflammatory cytokine, macrophage migration inhibitory factor (MIF) has also been suggested to play multiple roles in various tumorigenic processes (7). Recent studies from our laboratory have shown that MIF is important for, and plays a direct role in, normal cell division and oncogenic cell transformation (8, 9). Further studies showed that both cells and mice lacking the functional MIF gene product are resistant to malignant transformation and carcinogenesis (9, 10). Additionally, several groups have reported that MIF promotes tumor growth and viability by supporting tumor angiogenesis. Specifically, MIF intratumoral expression is shown to have a strong correlation with angiogenic growth factor expression, tumor vessel density, and risk of recurrence after resection in a variety of different tumors (11-15). Thus, the ability of MIF to modulate replicative, migratory, and angiogenic processes suggests that MIF provides several levels of support to a developing neoplasm.
Several proteins identified by yeast two-hybrid screening to interact with MIF reportedly do so through a C-X-X-C thiol reductase catalytic domain within MIF (16-19). One such protein is a component of the COP9 signalosome, which makes up a part of the 26S proteasome (19). MIF was found to physically interact with CSN5 and to inhibit CSN5-mediated p27 degradation, c-jun NH2-terminal kinase activation, and activator protein-1 transcription (19). This observation, coupled with reports that MIF regulates other CSN-regulated ubiquitin E3 ligase targets (i.e., p53 and E2F), suggests that MIF could modulate multiple processes tied to Cullin-dependent E3 activity (10, 20, 21). Because MIF has been described as a hypoxia-inducible target (22-24) and HIF-1α functionally interacts with CSN5 (6), we sought to investigate whether there was any influence by MIF on hypoxia-induced cellular processes.
Materials and Methods
Cell culture
MIA-PaCa-2 and Panc-1 cells were cultured in DMEM (Gibco Life Technologies, Grand Island, NY) containing 10% heatinactivated fetal bovine serum (Gibco Life Technologies), 1% gentamicin, and 1% l-glutamine in a humidified incubator of 5% CO2 at 37°C. MIF−/− mice and their wild-type littermates were maintained on a mixed 129Sv × C57Bl/6 background (F3). Mouse embryonic fibroblasts (MEF) were generated from embryos at day 14.5 and grown in DMEM with 10% FCS and antibiotics as described (8, 25).
RNA interference
The targeted base sequence for human MIF was 5′-CCTTCTGGTGGGGAGAAAT-3′ (Dharmacon, Lafayette, CO). As a negative control, a commercially available small interfering RNA (siRNA) referred to as nonspecific control (Dharmacon) was used. Cells were incubated at 37°C for 48 or 72 h and subjected to further analysis. In all cases, cells were transfected with 50 nmol/L annealed siRNA oligos using Oligofectamine reagent (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol. SiRNA-transfected cells were exposed to normoxia (21% O2), hypoxia (1% O2), or anoxia (<0.2% O2), or 150 μmol/L cobalt(II) chloride hexahydrate (Sigma) was added to the medium for the indicated amount of time. For reconstitution experiments, conditioned medium from confluent MIA-PaCa-2 cells was mixed 1:1 with normal medium and pretreated with an isotype control monoclonal antibody (mAb; R&D Systems, Minneapolis, MN), anti-MIF mAb (R&D Systems), or the small-molecule antagonist of MIF, ISO-1 (a generous gift of Dr. Yousef Al-Abed, North Shore-Long Island Jewish Research Institute, Manhasset, NY; refs. 26-28) before replacing medium from nonsense siRNA– or MIF siRNA–transfected cells.
Western blotting and immunoprecipitations
Whole-cell lysates were collected with 1× radioimmunoprecipitation assay lysis buffer. Supernatant concentration was carried out with Microcon 10 centrifugal filter devices (Millipore, Bedford, MA). Nuclear and cytoplasmic extracts were prepared using the NE-PER kit (Pierce, Rockford, IL). All lysis buffers contained 1 mmol/L NaVO4, 2 mmol/L NaF, and a protease inhibitor cocktail (Roche Biochemical, Indianapolis, IN). For immunoprecipitations, cells were lysed in 1× immunoprecipitation lysis buffer [20 mmol/L Tris-HCl (pH 8.0), 137 mmol/L NaCl, 1 mmol/L EGTA, 1% Triton X-100, 10% glycerol, and 1.5 mmol/L MgCl2] and incubated for 3 h or overnight at 4°C with the antibody followed by the addition of 30 μL protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at 4°C. The beads were washed five times with 1× immunoprecipitation lysis buffer. Proteins were resolved using precast 10% to 20% SDS-polyacrylamide gels (Bio-Rad, Hercules, CA) and transferred to a polyvinylidene difluoride membrane (Millipore). Membranes were blocked and probed with a blocking solution of 5% nonfat dried milk, 1% goat serum in TBS containing 0.2% Tween 20 and were developed with enhanced chemiluminescence reagent (Pierce). Immunoblotting and immunoprecipitations were done with polyclonal and monoclonal antibodies directed against MIF (Santa Cruz Biotechnology and R&D Systems, respectively), HIF-1α (BD Transduction Laboratories, San Jose, CA for human and Novus Biologicals, Littleton, CO for mouse HIF-1α), CSN5 (Bethyl Laboratories, Montgomery, TX), lactate dehydrogenase A (LDH-A; Abcam, Cambridge, MA), inducible nitric oxide synthase (Santa Cruz Biotechnology), and β-actin (Sigma, St. Louis, MO).
Northern blotting
Total RNA was isolated with TRIzol (Life Technologies) from HIF-1α+/+ and HIF-1α−/− MEFs. Total RNA (10 μg/lane) was denatured with glyoxal and size fractionated by electrophoresis on 1.4% agarose/sodium phosphate gels. RNA was transferred to nylon membranes and UV cross-linked. Radiolabeled probes were generated by random priming (Rediprime, Amersham, Piscataway, NJ) of cDNAs representing the complete coding sequence of MIF. Blots were prehybridized and hybridized in ExpressHyb solution (Clontech, Mountain View, CA) at 65°C, washed several times in 2× SSC/0.05% SDS and 0.2× SSC/0.1% SDS at 65°C, exposed to a PhosphorImager plate overnight, and visualized on a Storm 860 PhosphorImager (Molecular Dynamics, Piscataway, NJ).
Human MIF plasma analysis
Plasma was collected from patients with locally advanced pancreatic cancer according to an Institutional Review Board–approved protocol. All patients had pathologically confirmed adenocarcinoma of the pancreas and specimens were collected before any therapy. Control specimens were collected from age-matched volunteers without a prior history of cancer. All specimens were aliquoted and immediately frozen at −80°C until MIF ELISAs were done. Premade human MIF ELISAs were used for plasma MIF analyses (R&D Systems).
Real-time PCR
Total RNA was extracted from the cells with RNeasy Mini Kit (Qiagen, Valencia, CA). The RNA was transcribed into the first strand cDNA with the Omniscript RT kit (Qiagen) according to the manufacturer’s description. The reactions were done using DNA Engine Opticon (Bio-Rad) and amplifications were carried out at a final volume of 25 μL containing 1.5 μL of cDNA, 5 μL of 5× Takara PCR mix (Takara Bio, Inc., Otsu, Shiga, Japan), forward and reverse primers at 0.3 μmol/L final concentration, and SYBR Green (Molecular Probes, Eugene, OR) at a final dilution ratio of 25,000. The specific primers were forward 5′-CAACATCACCATGCAGATTATGC-3′ and reverse 5′-GCTTTCGTTTTTGCCCCTTTC-3′ for vascular endothelial growth factor (VEGF), forward 5′-AGAACCGCTCCTACAGCAAG-3′ and reverse 5′-TAGGCGAAGGTGGAGTTGTT-3′ for MIF, forward 5′-CAAGGCCAACCGCGAGAAGA-3′ and reverse 5′-GGATAGCACAGCCTGGATAG-3′ for β-actin, and forward 5′-CGTTCCTTCGATCAGTTGTC-3′ and reverse 5′-TCAGTGGTGGCAGTGGTAGT-3′ for HIF-1α. Relative expression was determined using the ΔCt method.
Results
Hypoxia induces MIF expression and secretion from pancreatic adenocarcinoma cells in a HIF-1α-dependent manner
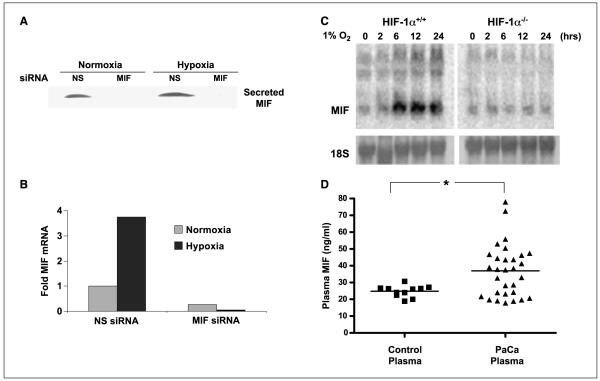
Prior studies have shown that MIF expression is increased in response to low oxygen tension in cancer cell lines (22, 23). Because pancreatic cancers are reportedly very hypoxic (29), we initiated a study to investigate MIF regulation by hypoxia in human pancreatic cancer cell lines. Our results indicate that two human pancreatic adenocarcinoma cell lines, MIA-PaCa-2 (Fig. 1) and PANC-1 (data not shown), are sensitive to hypoxia-induced transcriptional regulation of MIF. Specifically, we find that MIF secretion (Fig. 1A) and transcription (Fig. 1B) are markedly increased when subjected to a low-oxygen environment. Accompanying Fig. 1 are data from cells that had been transfected with short interfering RNA (siRNA) oligos designed against human MIF. Note the nearly complete loss of MIF in secreted protein (Fig. 1A) and message (Fig. 1B) when MIF siRNA is present.
Figure 1.
MIF transcription and secretion are enhanced by hypoxia, and patients with pancreatic adenocarcinoma have elevated levels of plasma MIF. A, MIA-PaCa-2 cells were transfected with nonsense (NS) or MIF siRNA. After 48 h, cells were placed in 0.5% FCS–containing media and placed under either normoxic or hypoxic conditions. After 24 h, cell supernatants were collected and concentrated. MIF protein levels were analyzed by Western blot. B, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA. After 48 h, cells were incubated for 16 h in either hypoxia or normoxia. RNA was isolated and cDNA was synthesized. Primers for MIF and β-actin were used in real-time PCR to determine levels of MIF transcript in the cells. Columns, ΔCt for each condition between MIF and β-actin; representative of three independent experiments. C, HIF-1α+/+ and HIF-1α−/− MEFs were challenged with 1% hypoxia for the indicated times and MIF mRNA levels were assessed by Northern blotting. D, plasma from healthy donors and pancreatic adenocarcinoma patients collected before chemotherapeutic treatment were assessed for MIF by ELISA. *, P < 0.05, Student’s t test (two tailed).
Although these and earlier studies have identified MIF as a hypoxia-inducible gene, there have been no reports on the relative requirements for HIF-1α in hypoxia-induced MIF transcription. To explore this possibility, MEFs from HIF-1α–deficient mice were compared with MEFs from littermate controls (30, 31). HIF-1α+/+ and HIF-1α−/− MEFs were exposed to hypoxia for differing times and then analyzed for relative MIF mRNA expression. As shown in Fig. 1C, Northern blot analysis reveals that whereas hypoxia rapidly and strongly stimulated MIF transcription in wild-type MEFs, HIF-1α–deficient MEFs had no increase in MIF message in response to hypoxia. These data provide evidence that MIF is an important and previously undescribed HIF-1α target gene.
Tumor hypoxia has been described as a strong prognostic indicator for neoplastic disease in general (32-34) and pancreatic cancer specifically (29). In an attempt to correlate our in vitro findings (Fig. 1A and B) to a clinical setting, we next measured MIF plasma levels in patients previously diagnosed with pancreatic adenocarcinoma. A pilot study was established to assess MIF levels in plasma samples from normal control donors and patients with pancreatic cancer who had yet to undergo any cancer therapy. As shown in Fig. 1D, the median serum MIF level for control sera was 24.8 ng/mL whereas pancreatic carcinoma patient’s mean MIF level was 36.8 ng/mL (P = 0.0382, two-tailed Student’s t test). These results are the first to show a positive correlation between MIF levels in the serum and pancreatic adenocarcinoma. Whereas these studies may be indicative of a potential use for MIF as a serum biomarker for pancreatic cancer, the possibility that MIF could represent a marker for intratumoral hypoxia or a novel therapeutic target is even more intriguing.
MIF is necessary for hypoxia-induced gene expression
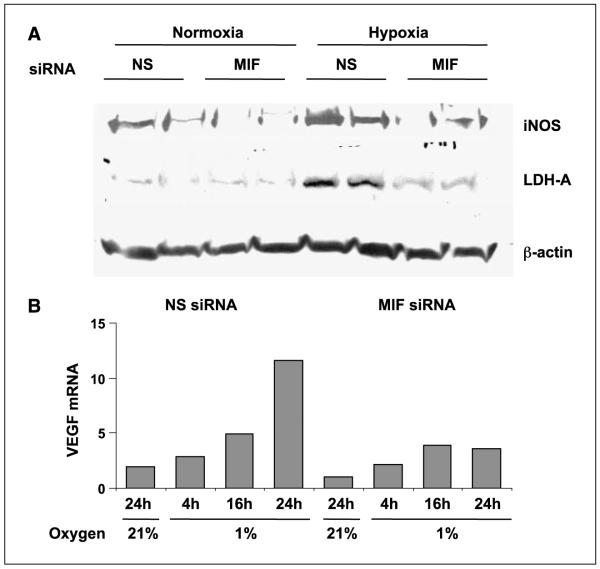
Several studies have described a unique contribution by MIF to the transcriptional activities of several important transcription factors (8, 9, 21, 35). To investigate whether HIF-1α-mediated MIF provides any functional contribution to hypoxia-induced transcriptional changes, short-interfering RNAs (siRNAs) directed against MIF were designed, optimized for use (see Fig. 1A), and transfected into human pancreatic adenocarcinoma cell lines. Control siRNA– and MIF siRNA–transfected cells were subjected to either ambient pO2 (normoxia) or 1% O2 (hypoxia) for 24 h and then examined for hypoxia-inducible gene product expression. Because low oxygen and/or nutrients trigger transcriptional increases in metabolic enzymes and angiogenic growth factors (36), we first looked at the hypoxic induction of two metabolic enzymes (i.e., inducible nitric oxide synthase and LDH-A). As shown in Fig. 2A, control siRNA–treated cells (nonsense) cultured under 1% O2 for 24 h had significant increases in both enzymes compared with cells cultured under ambient lab conditions, confirming previous studies (37, 38). Interestingly, pancreatic adenocarcinoma cells lacking MIF (siRNA to MIF) were refractory to hypoxia-induced increases in the expression of both inducible nitric oxide synthase and LDH-A (Fig. 2A).
Figure 2.
A functional requirement for MIF in hypoxia-induced gene expression. A, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA. Whole-cell lysates were prepared from cells after a 24-h exposure to either normoxia or hypoxia and immunoblotted for LDH-A and inducible nitric oxide synthase (iNOS). B, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA for 48 h, at which time cells were incubated for the indicated amount of time under hypoxia. RNA was harvested, cDNA synthesis was done, and real-time PCR for β-actin and VEGF was done as described in Materials and Methods. Columns, ΔCt of the average of duplicate reactions for each condition between VEGF and β-actin; representative of two independent experiments.
Hypoxia-induced expression of VEGF is a critical step in the malignant progression of tumors and is the target of multiple anticancer modalities (39). To determine whether hypoxia-induced VEGF transcription was similarly attenuated by MIF depletion in pancreatic carcinoma cells, we did real-time PCR analysis on cells with or without MIF that had been exposed to hypoxia for differing times. Our data indicate that maximal hypoxia-induced VEGF transcription is heavily reliant on MIF (Fig. 2B) and further suggest that MIF is an important contributor to hypoxia-mediated gene regulation. In a broader sense, these studies indicate that pancreatic tumorigenic potential, which is suggested to be influenced by hypoxia-initiated signaling (29), may be dependent on the presence of MIF.
MIF is required for maximal hypoxia-induced HIF-1α stabilization
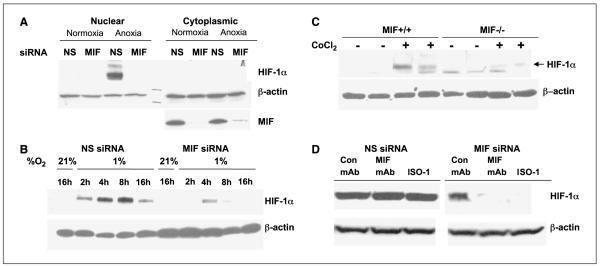
Work from several laboratories has suggested that some transcriptional targets of HIF-1α also regulate the expression/stabilization of HIF-α (40, 41). From the results above, we know that MIF is a target of HIF-1α gene transcription and is also indispensable for the efficient transcription of hypoxia-regulated gene products. Because of the well-documented modulation of hypoxia-induced gene expression by HIF-1α, we investigated whether HIF-1α stabilization by hypoxia was sensitive to loss of MIF. MIF-specific siRNA was transfected into MIA-PaCa-2 cells before being incubated in 0.2% O2 (anoxia) for 16 h. As shown in Fig. 3A, depletion of MIF by siRNA resulted in a loss of cytoplasmic MIF expression by >90% while not affecting β-actin expression. Whereas HIF-1α was efficiently stabilized in the nuclei by anoxia treatment in mock transfected (data not shown) or control siRNA–transfected cells, cells lacking MIF were completely deficient in HIF-α stabilization (Fig. 3A). To more thoroughly investigate the requirements for MIF in hypoxia-mediated HIF-1α stabilization, we did a time course study evaluating MIF-containing versus MIF-deficient pancreatic adenocarcinoma cells exposed to 1% O2 (hypoxia). Our findings reveal that there is a strong requirement for MIF in hypoxia-mediated stabilization at all time points evaluated (Fig. 3B).
Figure 3.
A requirement for MIF in hypoxia-induced HIF-1α expression. MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA for 48 h. Cells were then incubated for 16 h in anoxia (A) or the indicated amount of time in hypoxia (B). Cytoplasmic (A) and nuclear (A and B) extracts were prepared and assessed for HIF-1α followed by stripping and reprobing for β-actin and/or MIF. C, MIF+/+ and MIF−/− murine embryonic fibroblasts were cultured in duplicate under nearly confluent conditions with or without 150 μmol/L CoCl2 for 6 h. Nuclear extracts were immunoblotted for HIF-1α (top band, arrow) and then stripped and reprobed for β-actin. D, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA for 48 h. Conditioned medium from confluent MIA-PaCa-2 cells was mixed 1:1 with fresh medium, pretreated with isotype control mAb, anti-MIF mAb, or 100 μmol/L final concentration of the small-molecule inhibitor ISO-1, and used to replace media from nonsense- and MIF-transfected cells. After challenge for 4 h with 150 μmol/L CoCl2, nuclear extracts were assessed for HIF-1α and β-actin expression. A to D, representative of at least two independent experiments.
To determine whether the requirement for HIF-1α extends beyond human cancer cell lines and to verify that the requirement for MIF is not limited to cells treated with siRNAs, MIF+/+ and MIF−/− MEFs were evaluated (8, 25). As shown in Fig. 3C, homozygous deletion of MIF in mesenchymal cells results in impaired HIF-1α stabilization induced by an inhibitor of prolyl hydroxylases, CoCl2.
Finally, in an effort to “rescue” the loss of HIF-1α by exogenously added MIF, MIA-PaCa-2 conditioned supernatants containing MIF were incubated with control siRNA– and MIF siRNA–transfected cells. To evaluate the MIF specificity of the add-back, we pretreated the conditioned supernatants with neutralizing mAb to MIF or a well-characterized MIF small-molecule antagonist. As shown in Fig. 3D, control conditioned supernatants were almost fully able to rescue the HIF-1α stabilization induced by CoCl2 in MIF-depleted cells whereas conditioned supernatants previously inhibited or depleted of MIF were unable to do so. Combined, these results strongly suggest that the destabilizing effect observed on HIF-1α expression by siRNA-mediated MIF depletion is directly related to loss of MIF and further suggest that extracellular MIF is responsible for this effect.
MIF contributes to HIF stabilization by preventing proteasome-mediated degradation
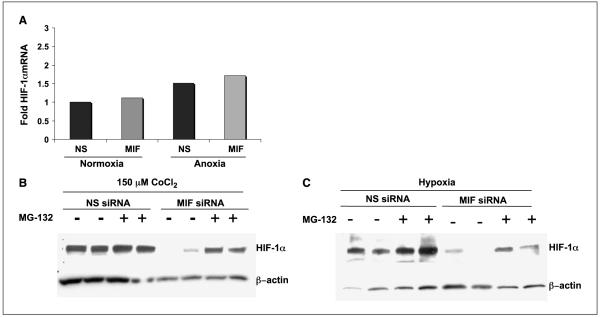
We next sought to investigate the point of control of MIF-dependent hypoxia- and prolyl hydroxylase inhibitorinduced HIF-1α expression. We first determined whether the defect in HIF-1α expression in MIF-depleted cells was due to faulty HIF-1α transcription. mRNA isolated from cells with or without MIF and exposed to either normoxic or hypoxic conditions was assessed for HIF-1α mRNA levels by real-time PCR. From the data shown in Fig. 4A, it is evident that MIF expression has no discernible effect on HIF-1α mRNA levels in either normoxic or hypoxic environment.
Figure 4.
HIF-1α destabilization by loss of MIF is 26S proteasome dependent. A, for RNA analysis of HIF-1α, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA oligos for 48 h. Cells were placed in either 21% or 1% O2 for 16 h and RNA was prepared, reverse transcribed, and analyzed by real-time PCR for HIF-1α and β-actin. Columns, ΔCt of the average of duplicate reactions for each condition between HIF-1α and β-actin; representative of two independent experiments. For proteasome inhibitor studies, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA. After 48 h, 10 μmol/L MG-132, a 26S proteasome inhibitor, was added to the indicated cells for 30 min before challenging for 6 h with either 150 μmol/L CoCl2 (B) or hypoxia (C). Equal amounts of nuclear fractions were analyzed by Western blot analysis. HIF-1α and β-actin antibodies were used for immunoblotting.
We next determined whether protein degradation might be involved in the MIF regulation of HIF expression. Proteasomal inhibitors were added to MIF-competent and MIF-depleted cells during either CoCl2-mediated HIF stabilization or hypoxia-induced HIF-1α stabilization. As shown in Fig. 4B, we observed a nearly complete rescue of HIF-1α expression in proteasome inhibitor–treated MIF-knockdown cells exposed to CoCl2 and a partial rescue of HIF-1α expression was observed in MIF-depleted cells exposed to hypoxia (Fig. 4C). Whereas these findings do not fully rule out a contributing effect by MIF on HIF-α translation, our results, combined with data described below, are more suggestive of a dominant role for MIF in modulating HIF-1α proteasomal degradation.
MIF binds to CSN5 in pancreatic adenocarcinoma cells and MIF depletion results in a loss of CSN5 binding and stabilization of HIF-1α
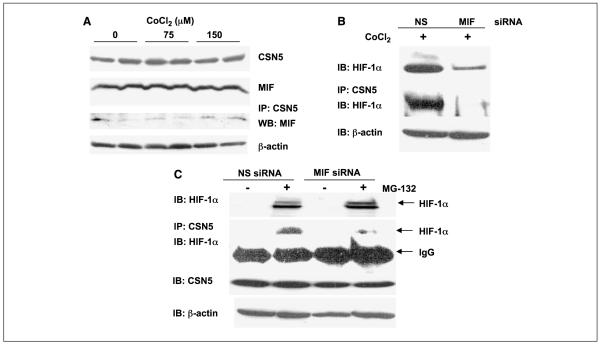
Because MIF is reported to modulate the function of CSN5 (19) and CSN5 was recently shown to be necessary for hypoxia-induced HIF-1α stabilization (6), we next sought to establish if there was any functional connection between MIF and CSN5 in HIF-1α stabilization. To determine if MIF interacts with CSN5 in pancreatic adenocarcinoma cells, coimmunoprecipitations were done from lysates of cells exposed to varying concentrations of CoCl2. As shown in Fig. 5A, MIF was efficiently coimmunoprecipitated with CSN5 under all conditions tested with a very moderate increase observed in the presence of CoCl2. Although not a dramatic effect, this apparent increase in MIF/CSN5 interaction was very reproducible (data not shown) and suggests that there may be some level of regulation that influences MIF/CSN5 intracellular binding.
Figure 5.
MIF interacts with CSN5 and promotes CSN5/HIF-1α interaction. A, MIA-PaCa-2 cells were incubated with indicated amounts of CoCl2 for 4 h. Cells were lysed and whole-cell extracts were analyzed by Western blotting for MIF CSN5 and β-actin or immunoprecipitated with a CSN5 antibody. CSN5 interaction with MIF was evaluated by Western blotting of coimmunoprecipitates with anti-MIF antibody. B, MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA. Forty-eight hours later, cells were incubated with or without 150 μmol/L CoCl2 for 4 h. Whole-cell extracts and nuclear extracts were obtained in parallel. Whole-cell extracts were immunoprecipitated with a CSN5 antibody. Nuclear extracts and coimmunoprecipitates were analyzed for HIF-1α by immunoblotting. C, forty-eight hours after MIA-PaCa-2 cells were transfected with nonsense or MIF siRNA, cells were incubated with or without 10 μmol/L MG-132 for 6 h. Both whole-cell extracts and nuclear extracts were obtained. Whole-cell extracts were immunoprecipitated with a CSN5 antibody. Nuclear extracts and coimmunoprecipitates were analyzed by Western blotting for HIF-1α whereas a small fraction of the whole-cell lysate was assessed for total CSN5. A to C, representative of at least three independent experiments.
Because binding of CSN5 acts to functionally stabilize HIF-1α (6), we next determined whether loss of MIF disrupts binding of HIF-1α to CSN5. Coimmunoprecipitation of the CSN5/HIF-1α complex from untreated and CoCl2-treated MIF-competent and MIF-depleted cells revealed that loss of MIF significantly decreased CSN5 binding to HIF-1α induced by prolyl hydroxylase inhibition (Fig. 5B). Although there seemed to be a significant difference between the HIF-1α expression in cell lysates versus the amount of HIF-1α being pulled down with CSN5, we could not rule out the possibility that the observed loss of CSN5/HIF-1α interaction in MIF-depleted cells was simply due to overall loss of HIF-1α stabilization.
To address this issue, we used a proteasome inhibitor to artificially stabilize HIF-1α in MIF-containing and MIF-depleted cells. Importantly, MIF-depleted pancreatic carcinoma cells treated with proteasome inhibitor contained an equivalent amount of stabilized HIF-1α as the cells containing MIF (Fig. 5C). This finding is consistent with our data suggesting a role for MIF in modulating HIF-1α protein degradation and not transcription or translation (Fig. 4). Cell lysates were then assessed for CSN5/HIF-1α interactions in MG-132-mediated HIF-1α-stabilized cells. Intriguingly, MG-132-treated, MIF-containing cells contained a significant amount of CSN5-bound HIF-1α whereas MIF-deficient cells contained only a small fraction of this CSN5/HIF-1α complex (Fig. 5C). It is important to note that CSN5 levels were unchanged by MIF status (Fig. 5C). Whereas more studies are needed to conclusively state that disrupted MIF binding to CSN5 accounts for the defect in HIF-1α/CSN5 interaction, these results are suggestive of a direct functional role for MIF in CSN5-dependent HIF-1α stabilization.
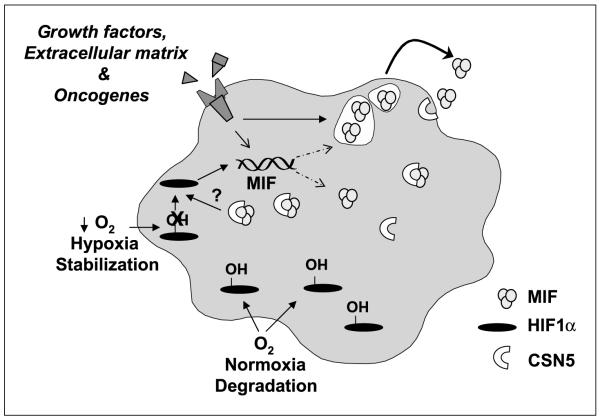
Combined, these findings reveal a novel coregulatory axis between MIF and HIF-1α in pancreatic adenocarcinoma cells (Fig. 6). Our data indicate that MIF expression and secretion is enhanced by hypoxia in a HIF-1α-dependent manner and, more importantly, that MIF is necessary for hypoxia-induced HIF-1α stabilization. Our results further suggest that MIF contributes to HIF-1α stabilization by facilitating CSN5 binding to HIF-1α and are consistent with prior reports describing MIF as a functional regulator of CSN5 bioactions (19).
Figure 6.
Schematic of proposed mechanism of the coregulatory axis between MIF and HIF-1α in pancreatic adenocarcinoma. MIF expression is coordinately regulated by several intracellular and extracellular stimuli. Consistent with prior observations, at least some intracellular MIF exists in a bound state with the COP9 signalosome subunit CSN5 (19). During hypoxia or prolyl hydroxylase inhibitor challenge, MIF is necessary for the previously described binding and stabilization of HIF-1α by CSN5 (6). Once stabilized, HIF-1α directs the transcription of many protumorigenic enzymes and growth factors, among these MIF (23). We propose that the net result of this coregulation between MIF and HIF-1α is an amplification of hypoxia-initiated responses and subsequent hypoxia-independent autocrine and paracrine effects of MIF (7).
Discussion
All solid tumors require microenvironmental adaptation throughout tumorigenesis. One of the hallmarks of this adaptive response is the development of intratumoral hypoxia that stimulates HIF-directed expression of protumorigenic/metastatic genes. This study has established that one of these gene products, MIF, is elevated in the serum of a subset of pancreatic cancer patients. We further describe a unique functional interrelationship between the extracellular cytokine/growth factor MIF and the transcription factor HIF-1α. This point is dramatically shown by our findings that cells lacking MIF are refractory to hypoxia- and prolyl hydroxylase inhibitor–induced HIF-1α stabilization and subsequent transcription of metabolic and angiogenic gene products. Combined, these studies are the first to show that (a) HIF-1α is necessary for hypoxia-induced MIF transcription; (b) MIF is necessary for maximal hypoxia-mediated HIF-1α stabilization; and (c) serum MIF is elevated in a subset of pancreatic cancers. These findings are particularly intriguing because there are only a small number of gene products that have been shown to be necessary for hypoxia-induced stabilization of HIF-1α and, of these, none were previously known to be secreted proteins.
Our data indicate that the requirement for MIF in hypoxia- or prolyl hydroxylase inhibitor-mediated HIF stabilization is at the level of proteasome-mediated degradation and MIF may prevent this by facilitating the binding of the COP 9 signalosome component CSN5 to HIF-1α. Although a number of CSN5-dependent cellular functions are associated with proteasomal regulation (42-44), the precise requirement for CSN5 in hypoxia-induced HIF-1α stabilization is presently unclear (6, 45). Whereas CSN5 overexpression in aerobic cells results in HIF-1α stabilization by inhibiting prolyl-564 hydroxylation and subsequent pVHL-dependent ubiquitylation (6, 45), cells cultured under low oxygen conditions would not be sensitive to this inhibition. Regardless of the mechanism of CSN5-dependent HIF-1α stabilization, our findings are highly suggestive of a role for MIF in regulating this process. Although beyond the scope of this study, future investigation into the mechanisms regulating both CSN5-dependent HIF-α stability and MIF-dependent regulation of CSN5 will yield critical insight into hypoxia-initiated HIF-α activity in tumors.
Several years ago, during a screen of squamous cell carcinoma lines, MIF was identified as one of the 10 candidate gene products of which the transcription was induced by hypoxia (23). Subsequent studies have shown that MIF is also a target gene of hypoglycemia and hypoxia-induced transcription in glioblastoma cell lines (22). More recently, Baugh et al. (24) showed that hypoxiainduced transcription from a MIF promoter–driven luciferase construct requires a HIF-response element found in the 5′ untranslated region of the human MIF gene. Combined with our findings that HIF-1α-deficient fibroblasts are resistant to hypoxia-induced MIF expression, these studies strongly support the conclusion that MIF is a target of hypoxia-induced HIF-1α.
From these results, we propose that a positive feedback loop exists between MIF and HIF-1α, which ultimately serves to amplify hypoxia-dependent transcription in human cancers. Combined with the numerous other proposed functions of MIF in tumorigenesis (10, 20, 21, 25, 46, 47), this cytokine becomes a very attractive target for cancer therapeutic modalities and potentially as a novel serum biomarker of tumor hypoxia.
Acknowledgments
Grant support: National Cancer Institute grant R01-CA102285 (R.A. Mitchell), Damon Runyon Clinical Investigator Award (A.C. Koong), and National Cancer Institute grant R01-CA102301 (W. Zundel).
References
- 1.Maxwell PH. The HIF pathway in cancer. Semin Cell Dev Biol. 2005;16:523–30. doi: 10.1016/j.semcdb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell PH, Ratcliffe PJ. Oxygen sensors and angiogenesis. Semin Cell Dev Biol. 2002;13:29–37. doi: 10.1006/scdb.2001.0287. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell PH, Wiesener MS, Chang GW, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–5. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 4.Brown JM. Tumor microenvironment and the response to anticancer therapy. Cancer Biol Ther. 2002;1:453–8. doi: 10.4161/cbt.1.5.157. [DOI] [PubMed] [Google Scholar]
- 5.Moeller BJ, Cao Y, Li CY, Dewhirst MW. Radiation activates HIF-1 to regulate vascular radiosensitivity in tumors: role of reoxygenation, free radicals, and stress granules. Cancer Cell. 2004;5:429–41. doi: 10.1016/s1535-6108(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 6.Bemis L, Chan DA, Finkielstein CV, et al. Distinct aerobic and hypoxic mechanisms of HIF-α regulation by CSN5. Genes Dev. 2004;18:739–44. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell RA. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell Signal. 2004;16:13–9. doi: 10.1016/j.cellsig.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Liao H, Bucala R, Mitchell RA. Adhesion-dependent signaling by macrophage migration inhibitory factor (MIF) J Biol Chem. 2003;278:76–81. doi: 10.1074/jbc.M208820200. [DOI] [PubMed] [Google Scholar]
- 9.Petrenko O, Fingerle-Rowson G, Peng T, Mitchell RA, Metz CN. Macrophage migration inhibitory factor deficiency is associated with altered cell growth and reduced susceptibility to Ras-mediated transformation. J Biol Chem. 2003;278:11078–85. doi: 10.1074/jbc.M211985200. [DOI] [PubMed] [Google Scholar]
- 10.Fingerle-Rowson G, Petrenko O, Metz CN, et al. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100:9354–9. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chesney J, Metz C, Bacher M, Peng T, Meinhardt A, Bucala R. An essential role for macrophage migration inhibitory factor (MIF) in angiogenesis and the growth of a murine lymphoma. Mol Med. 1999;5:181–91. [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Y, Law S, Huang X, et al. Macrophage migration inhibitory factor stimulates angiogenic factor expression and correlates with differentiation and lymph node status in patients with esophageal squamous cell carcinoma. Ann Surg. 2005;242:55–63. doi: 10.1097/01.sla.0000168555.97710.bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson JM, Coletta PL, Cuthbert RJ, et al. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 2005;129:1485–503. doi: 10.1053/j.gastro.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 14.Shun CT, Lin JT, Huang SP, Lin MT, Wu MS. Expression of macrophage migration inhibitory factor is associated with enhanced angiogenesis and advanced stage in gastric carcinomas. World J Gastroenterol. 2005;11:3767–71. doi: 10.3748/wjg.v11.i24.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hira E, Ono T, Dhar DK, et al. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 2005;103:588–98. doi: 10.1002/cncr.20818. [DOI] [PubMed] [Google Scholar]
- 16.Jung H, Kim T, Chae HZ, Kim KT, Ha H. Regulation of macrophage migration inhibitory factor (MIF) and thiol-specific antioxidant protein PAG by direct interaction. J Biol Chem. 2001;276:15504–10. doi: 10.1074/jbc.M009620200. [DOI] [PubMed] [Google Scholar]
- 17.Wadgaonkar R, Dudek SM, Zaiman AL, et al. Intracellular interaction of myosin light chain kinase with macrophage migration inhibition factor (MIF) in endothelium. J Cell Biochem. 2005;95:849–58. doi: 10.1002/jcb.20472. [DOI] [PubMed] [Google Scholar]
- 18.Potolicchio I, Santambrogio L, Strominger JL. Molecular interaction and enzymatic activity of macrophage migration inhibitory factor with immunorelevant peptides. J Biol Chem. 2003;278:30889–95. doi: 10.1074/jbc.M302854200. [DOI] [PubMed] [Google Scholar]
- 19.Kleemann R, Hausser A, Geiger G, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408:211–6. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell RA, Liao H, Chesney J, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci U S A. 2002;99:345–50. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrenko O, Moll UM. Macrophage migration inhibitory factor MIF interferes with the Rb-E2F pathway. Mol Cell. 2005;17:225–36. doi: 10.1016/j.molcel.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 22.Bacher M, Schrader J, Thompson N, et al. Up-regulation of macrophage migration inhibitory factor gene and protein expression in glial tumor cells during hypoxic and hypoglycemic stress indicates a critical role for angiogenesis in glioblastoma multiforme. Am J Pathol. 2003;162:11–7. doi: 10.1016/S0002-9440(10)63793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koong AC, Denko NC, Hudson KM, et al. Candidate genes for the hypoxic tumor phenotype. Cancer Res. 2000;60:883–7. [PubMed] [Google Scholar]
- 24.Baugh JA, Gantier M, Li L, Byrne A, Buckley A, Donnelly SC. Dual regulation of macrophage migration inhibitory factor (MIF) expression in hypoxia by CREB and HIF-1. Biochem Biophys Res Commun. 2006;347:895–903. doi: 10.1016/j.bbrc.2006.06.148. [DOI] [PubMed] [Google Scholar]
- 25.Swant JD, Rendon BE, Symons M, Mitchell RA. Rho GTPase-dependent signaling is required for macrophage migration inhibitory factor-mediated expression of cyclin D1. J Biol Chem. 2005;280:23066–72. doi: 10.1074/jbc.M500636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lubetsky JB, Dios A, Han J, et al. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J Biol Chem. 2002;277:24976–82. doi: 10.1074/jbc.M203220200. [DOI] [PubMed] [Google Scholar]
- 27.Al Abed Y, Dabideen D, Aljabari B, et al. ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem. 2005;280:36541–4. doi: 10.1074/jbc.C500243200. [DOI] [PubMed] [Google Scholar]
- 28.Nicoletti F, Creange A, Orlikowski D, et al. Macrophage migration inhibitory factor (MIF) seems crucially involved in Guillain-Barré syndrome and experimental allergic neuritis. J Neuroimmunol. 2005;168:168–74. doi: 10.1016/j.jneuroim.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 29.Koong AC, Mehta VK, Le QT, et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48:919–22. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 30.Ryan HE, Lo J, Johnson RS. HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–15. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zundel W, Schindler C, Haas-Kogan D, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14:391–6. [PMC free article] [PubMed] [Google Scholar]
- 32.Janssen HL, Haustermans KM, Balm AJ, Begg AC. Hypoxia in head and neck cancer: how much, how important? Head Neck. 2005;27:622–38. doi: 10.1002/hed.20223. [DOI] [PubMed] [Google Scholar]
- 33.Menon C, Fraker DL. Tumor oxygenation status as a prognostic marker. Cancer Lett. 2005;221:225–35. doi: 10.1016/j.canlet.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 34.Shannon AM, Bouchier-Hayes DJ, Condron CM, Toomey D. Tumour hypoxia, chemotherapeutic resistance and hypoxia-related therapies. Cancer Treat Rev. 2003;29:297–307. doi: 10.1016/s0305-7372(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 35.Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–82. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schumacker PT. Hypoxia-inducible factor-1 (HIF-1) Crit Care Med. 2005;33:S423–5. doi: 10.1097/01.ccm.0000191716.38566.e0. [DOI] [PubMed] [Google Scholar]
- 37.Melillo G, Musso T, Sica A, Taylor LS, Cox GW, Varesio L. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J Exp Med. 1995;182:1683–93. doi: 10.1084/jem.182.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blancher C, Moore JW, Talks KL, Houlbrook S, Harris AL. Relationship of hypoxia-inducible factor (HIF)-1α and HIF-2α expression to vascular endothelial growth factor induction and hypoxia survival in human breast cancer cell lines. Cancer Res. 2000;60:7106–13. [PubMed] [Google Scholar]
- 39.Zhong H, Bowen JP. Antiangiogenesis drug design: multiple pathways targeting tumor vasculature. Curr Med Chem. 2006;13:849–62. doi: 10.2174/092986706776361085. [DOI] [PubMed] [Google Scholar]
- 40.Epstein AC, Gleadle JM, McNeill LA, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 41.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1α. Science. 2003;302:1975–8. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 42.Wan M, Cao X, Wu Y, et al. Jab1 antagonizes TGF-β signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–6. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bech-Otschir D, Kraft R, Huang X, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–9. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1991;398:160–5. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 45.Richardson KS, Zundel W. The emerging role of the COP9 signalosome in cancer. Mol Cancer Res. 2005;3:645–53. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abe R, Peng T, Sailors J, Bucala R, Metz CN. Regulation of the CTL response by macrophage migration inhibitory factor. J Immunol. 2001;166:747–53. doi: 10.4049/jimmunol.166.2.747. [DOI] [PubMed] [Google Scholar]
- 47.Sun B, Nishihira J, Yoshiki T, et al. Macrophage migration inhibitory factor promotes tumor invasion and metastasis via the Rho-dependent pathway. Clin. Cancer Res. 2005;11:1050–8. [PubMed] [Google Scholar]