Abstract
Objectives
To investigate protein citrullination by the periodontal pathogen Porphyromonas gingivalis (P. gingivalis) as a potential mechanism for breaking tolerance to citrullinated proteins in rheumatoid arthritis (RA).
Methods
Expression of endogenous citrullinated proteins was analysed by immunoblotting of cell extracts from P. gingivalis and ten other oral bacteria. P. gingivalis knockout strains lacking the bacterial PAD or gingipains were created to assess the role of these enzymes in citrullination. Citrullination of human fibrinogen and α-enolase by P. gingivalis was studied by incubating live wild-type and knockouts with the proteins and analysing the products by immunoblotting and mass spectrometry.
Results
Endogenous protein citrullination was abundant in P. gingivalis but lacking in the other oral bacteria. Deletion of the bacterial PAD gene resulted in complete abrogation of protein citrullination. Inactivation of arginine-gingipains, but not lysine-gingipains, led to decreased citrullination. Incubation of wild-type P. gingivalis with fibrinogen or α-enolase caused degradation of the proteins and citrullination of the resulting peptides at carboxy-terminal arginine residues, which were identified by mass spectrometry.
Conclusion
We demonstrate that P. gingivalis is unique amongst the tested oral bacterial pathogens in its ability to citrullinate proteins. We further show that P. gingivalis rapidly generates citrullinated host peptides by proteolytic cleavage at arginine-X peptide bonds by arginine-gingipains followed by citrullination of carboxy-terminal arginines by bacterial PAD. Our results suggest a novel model where P. gingivalis-mediated citrullination of bacterial and host proteins provides a molecular mechanism for generating antigens driving the autoimmune response in RA.
Keywords: rheumatoid, gingivalis, citrullination, fibrinogen, autoimmunity
Introduction
Rheumatoid arthritis (RA) is characterized by disease-specific autoimmunity to citrullinated proteins. Citrullination is a post-translational modification of arginine residues, mediated by the family of peptidylarginine deiminases (PAD). Citrullinated fibrin(ogen) and α-enolase are two of the physiological proteins that are targeted by anti-citrullinated protein antibodies in RA (1–5). Fibrinogen is the precursor of fibrin and autoantibodies to citrullinated fibrin(ogen) are found in up to 66 % of patients with RA (6). Alpha-enolase is an evolutionary conserved, multifunctional protein (7), best known for its role in glucose metabolism, and more recently as a plasminogen-binding protein on the surface of various mammalian and prokaryotic cell types (8, 9). Autoantibodies to citrullinated α-enolase can be found in 40–60% of patients with RA (4–6). The pathogenicity of these autoantibodies may be mediated by immune-complex formation with citrullinated host proteins in the joint and activation of downstream inflammatory pathways via complement fixation and Fcγ receptor activation (10–13).
It is not yet known which factors trigger the breakdown of tolerance to citrullinated proteins. Protein citrullination is part of healthy physiology, such as citrullinated filaggrin in the skin (14), and part of the inflammatory response in general (15), whereas the autoantibody formation to citrullinated proteins is largely restricted to RA (16). For example, deposits of citrullinated fibrin can be found in a variety of inflammatory joint conditions without an accompanying autoantibody response (17, 18). Therefore, additional environmental and genetic risk factors are likely to be required. To date, tobacco exposure and the presence of certain alleles in the HLA-DRB1 locus with a common peptide-binding motif, collectively termed the ‘shared epitope’, have been identified as susceptibility factors for developing autoantibodies to citrullinated proteins (19, 20), in particular α-enolase and vimentin (21), but do not explain the total risk. Additional etiological pathways require consideration, with the periodontal pathogen Porphyromonas gingivalis (P. gingivalis) being a prime candidate for investigation.
Periodontitis, in which P. gingivalis is a major causative agent, is a chronic inflammatory disease of the supporting tissues of the teeth, with an estimated prevalence of 4.2% in the US population (22). P. gingivalis can be detected in 80–90% of periodontitis patients, and in 10–30% of healthy subjects (23, 24). The bacterium has recently attracted interest based on epidemiological links between RA and periodontitis (25) and the description of a novel bacterial PAD (26) (hereafter called PPAD), suggesting a potential etiological role for P. gingivalis in RA through the generation of citrullinated antigens. Periodontitis has similar pathophysiological mechanisms to RA, characterized by the resorption of the supporting bony structure around the teeth and mediated by a variety of pro-inflammatory molecules, including TNF-α, IL-1β, prostaglandin E2 and matrix metalloproteinases (27). A number of studies have indicated a positive association between the prevalence of periodontitis and RA (25, 28), even when adjusted for smoking, which is a major risk factor for both diseases. We have shown that RA-specific autoantibodies to CEP-1, the immunodominant B cell epitope of human α-enolase, cross-react with in vitro citrullinated enolase from P. gingivalis (5), raising the possibility of molecular mimicry between epitopes from citrullinated bacterial and human enolase. P. gingivalis is the only prokaryote described to date that expresses a functional bacterial PAD, though its physiological substrates are unknown, as are the molecular mechanisms of citrullination. PPAD displays no amino acid sequence similarity to the human PAD enzymes, and a previous study indicated that it might preferentially target carboxy-terminal arginine residues (26), in contrast to the human enzymes, which efficiently deiminate internal arginine residues (29). Citrullination of bacterial and host proteins and peptides by P. gingivalis PAD could therefore create new epitopes and, given the infectious context providing endogenous and exogenous danger signals, trigger a latent antibody response to citrullinated bacterial and host proteins in susceptible individuals.
Here, we aimed to elucidate the molecular requirements for bacterial and human protein citrullination by P. gingivalis PAD and thus advance our understanding of potential underlying mechanisms for the generation of citrullinated antigens and induction of autoimmunity in RA.
Materials and Methods
Bacterial strains and growth conditions
Porphyromonas gingivalis wild-type strain (W83), P. gingivalis clinical isolates (MaRL, D243, JH16, J430), obtained from patients with severe periodontitis, and P. gingivalis mutants (Δppad, ppad+, Δrgp, Δkgp, Δrgp+kgp) were grown in Schaedler anaerobe broth (Oxoid, UK), supplemented with 5% sheep blood, at 37°C in an anaerobic chamber (90% N2, 5% CO2, 5% H2). Erythromycin or tetracycline was used at 5 µg/mL or 1 µg/mL, respectively, on solid media. The concentrations were doubled for selective growth in liquid culture. Other anaerobic oral bacteria (Prevotella intermedia H13 (clinical isolate), Prevotella oralis ATCC 33269, Capnocytophaga gingivalis ATCC 33624, Capnocytophaga ochracea ATCC 27872) were grown in Schaedler anaerobe broth, supplemented with 2.5 µg/L vitamin K, at 37°C in an anaerobic chamber (90% N2, 5% CO2, 5% H2). Fusobacterium nucleatum ATCC 10953 was grown in Schaedler anaerobe broth in an anaerobic chamber with 80% N2, 10% CO2 and 10% H2 at 37°C. Aggregatibacter actinomycetemcomitans ATCC 43718 was grown in Tryptic soy broth (Sigma, UK), supplemented with 6% yeast extract and 8% glucose, in 5% CO2 at 37°C. Aerobic bacteria (Streptococcus constellatus ATCC 27823, Streptococcus gordonii ATCC 10558, Streptococcus sanguinis ATCC 10556, Streptococcus salivarius ATCC 7073) were grown on Columbia agar plates, supplemented with 8% defibrinated sheep blood or brain heart infusion broth.
Construction of P. gingivalis mutant strains
ppad mutant
A 1-kb region 3′ to the P. gingivalis ppad gene (Genbank accession number 2552184; locus tag PG1424) was amplified by PCR (primers: 5’-GCTCTAGATGGAATCCGTGAGACAATG and 5’-TAAGCATGCGATATTTGTCGGAAGGACTC) for insertion into XbaI and SphI sites of the pUC19 plasmid (New England Biolabs Inc., USA). An erythromycin resistance cassette ermF/ermAM from plasmid pVA2198 was amplified and inserted it into SmaI and XbaI sites of the modified pUC19 plasmid. The resultant plasmid was modified further by incorporating (i) an amplified 1-kb region 5′ to the ppad gene (primers: 5’-AAGAGCTCAAGCACGTAATAAGGACAATGA and 5’-TTATCCCGGGTGTTCCTGAACATATGATAAGATCT) into SacI and SmaI sites to create the deletional inactivation plasmid construct (pΔppad) or (ii) the entire ppad gene and a 1-kb region 5′ to the gene (primers: 5’-AAGAGCTCAAGCACGTAATAAGGACAATGA and 5’-TTATCCCGGGTGTCTACCTGAGGAGTATTCT) into SacI and SmaI sites to create the control mutant construct (pppad+) to control for possible polar effects. The correct placement and orientation of the DNA segments were confirmed by sequencing. The modified plasmid constructs were integrated into the P. gingivalis W83 genome by a double crossover recombination event by electroporation using standard protocols (30). Erythromycin-resistant clones were subcultured on selective plates and genomic integration confirmed by PCR, using primers from outside of the cloned regions surrounding the ppad gene.
rgp and kgp mutants
The general procedure for construction of the Δrgp and Δkgp mutants is described elsewhere (30). Homologous recombination of the Δkgp plasmid into the P. gingivalis Δrgp mutant genome resulted in a kgp-rgp-deficient mutant (Δrgp+kgp). The respective phenotypes were confirmed by enzymatic assays and western blot analysis.
Preparation of bacterial whole cell extracts
Bacterial cultures were grown in liquid media until early stationary phase. Twenty mL of culture was centrifuged at 10,000 × g for 15 min at 4°C. The resulting bacterial pellet was resuspended in phosphate buffered saline (PBS) and the optical density (at 600 nm) was measured and adjusted to 1.0 with PBS and sonicated on ice. Sodium azide (final concentration 0.02% v/v) was added to all samples as preservative.
SDS-PAGE
Protein samples were mixed with reducing 4× lithium dodecyl sulphate (LDS) sample buffer (Invitrogen, Paisley, UK), heated for 10 min at 70°C and resolved on 12% NuPAGE Bis-Tris gels (Invitrogen) using MOPS running buffer. After electrophoresis, proteins were stained with the Coomassie-based stain InstantBlue (Triple Red, Bucks, UK), or transferred to nitrocellulose membranes for immunoblotting. For analysis of fibrinogen/enolase-derived peptides, 10–20% Tricine gels, 2× Tricine sample buffer and Tricine SDS running buffer (all from Invitrogen) were used, and protein bands were visualised using a standard silver staining protocol.
Detection of citrullinated proteins by immunoblot and dot blot
Citrullinated proteins were detected using the anti-citrulline (modified) detection kit (Upstate, Millipore, Billerica, MA) in accordance with the manufacturer's instructions. For dot blotting, 10 µL of sample was spotted onto an equilibrated nitrocellulose membrane (0.1 µm, PROTRAN®) and allowed to dry before proceeding with the standard western blotting protocol. Controls were performed in which the modification step or the primary antibody were omitted, to control for non-specific binding by the primary antibody to structures other than modified citrulline side chains, or binding of the secondary antibody to proteins other than the primary antibody, respectively.
Analysis of fibrinogen and α-enolase citrullination by live P. gingivalis
P. gingivalis was cultured as described above and the OD600 measured. Bacterial cells were pelleted, washed in ice-cold PBS, and resuspended in assay buffer (10 mM HEPES, 150 mM NaCl, 1 mM CaCl2, pH 7.5, 10 mM L-Cysteine) to a final OD600 of 1.0. Purified fibrinogen was purchased from Sigma-Aldrich, St. Louis, MO (F3879) and recombinant human α-enolase was expressed in E. coli as previously described (31). The proteins were diluted in HEPES buffer at a concentration of 0.5 mg/mL. Equal volumes of protein solution and bacterial cell suspension were mixed and an aliquot immediately withdrawn (corresponds to time point t = 1 min). The cultures were then incubated at 37°C on a shaking platform, and further aliquots withdrawn after 1.5 h, 3 h and 6 h. Bacterial cells were immediately removed from all aliquots by centrifugation. The resulting supernatant was used for analysis by SDS-PAGE gels and immunoblotting as described and for protein precipitation with 15% meta-phosphoric acid, leaving small peptides in solution, and subsequent analysis by HPLC. Collected HPLC peak fractions were subjected to peptide analysis by mass spectrometry.
HPLC analysis
HPLC was performed with a Shimadzu VP Series chromatograph equipped with a Supelcosil LC-318 reversed-phase column (25 cm × 4.6 mm, Supelco, Sigma-Aldrich, St. Louis, MO). Samples (100 µL) were injected and eluted with a gradient of H2O/0.1% trifluoroacetic acid (TFA) (solution A) and 80% acetonitrile/0.08% TFA (solution B), and monitored at 215 nm with a Shimadzu SPD-10A UV-Vis detector. Peaks were collected manually and freeze-dried prior to analysis by mass spectrometry.
Mass spectrometry
In-gel digestion of proteins
Protein bands were excised with a scalpel and in-gel digestion was performed using a robotic system (Investigator ProGest, Genomic Solutions, Huntingdon, UK). The bands were washed in 100 mM ammonium bicarbonate buffer and dehydrated in 100% acetonitrile. Cysteine residues were reduced with 10 mM DTT, then carboamidomethylated with 55 mM iodoacetamide. Digestion was performed for six hours at 37 °C by addition of modified porcine trypsin (10 µL at 6.5 ng/µL in 25 mM ammonium bicarbonate) and peptides were recovered by sequential extraction with 25 mM ammonium bicarbonate buffer, 5% formic acid, and acetonitrile. Extracts were pooled, lyophilised and re-dissolved in 0.1% formic acid prior to mass spectrometry.
Mass spectrometry analysis
Tandem electrospray mass spectra were recorded with a Q-Tof hybrid quadrupole/orthogonal acceleration time-of-flight spectrometer (Waters, Manchester, UK) interfaced to a CapLC chromatograph. Freeze-dried peptide samples were redissolved in 0.1% formic acid, and 6 µL injected onto a Pepmap C18 column (300 µm × 0.5 cm; LC Packings, Amsterdam, The Netherlands), and eluted with an acetonitrile/0.1% formic acid gradient at 1 µl per minute. The capillary voltage was set to 3,500 V, and data-dependent product ion scans were performed on precursor ions with charge states of 2, 3 or 4 over a survey mass range of 400 to 1,400. The raw spectra were smoothed, deisotoped, transformed onto a singly charged m/z axis using a maximum entropy method as implemented in the peptide auto module of MassLynx (Waters UK) and saved in the peaklist (pkl) format prior to database searching. Proteins were identified by correlation of uninterpreted spectra to entries in SwissProt / TrEMBL using ProteinLynx Global Server (version 1.1, Waters, Manchester, UK) and a local installation of Mascot (version 2.2: www.matrixscience.com). The database used was a FASTA format composite constructed in house by merging SwissProt, TrEMBL and associated splice variants (releases of 26/05/09; 8,413,758 sequences). Searches were run in error tolerant mode and no mass or taxomic constraints were applied. The initial enzyme specificity was set to trypsin, but subsequent searches of the P. gingivalis digestions of fibrinogen and enolase were repeated with no enzyme specificity in order to match peptides resulting from the combination of gingipain activity with other enzymes such as amino- and carboxy-peptidases. All spectra matching citrullinated peptides were reviewed manually by interpretation of sequence-specific fragment ions to confirm presence and location of the citrulline residue and to exclude other modifications such as deamidation of aspartic acid which also result in a mass increase of 1 Da.
Results
Expression of endogenous citrullinated proteins is unique for P. gingivalis
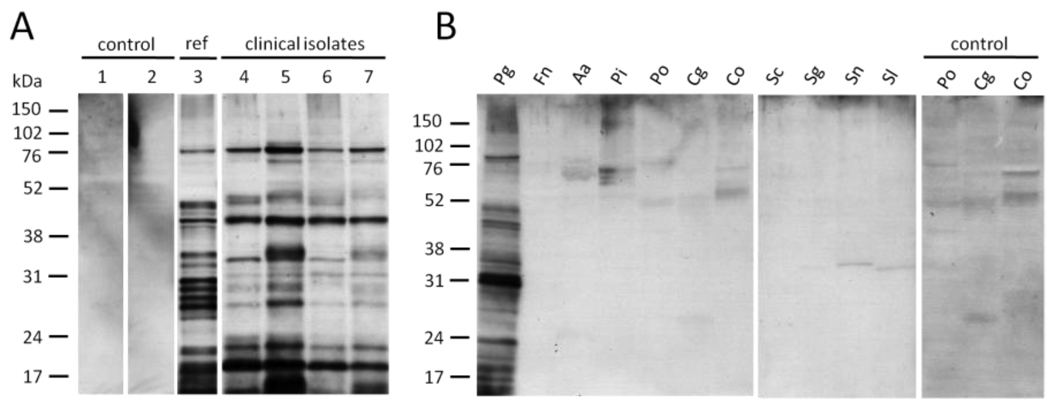
In order to test whether P. gingivalis citrullinates its own endogenous proteins, cultures of P. gingivalis, comprising a reference strain (W83) and four clinical isolates from patients with periodontal disease, were grown to stationary phase and whole cell lysates were analysed by immunoblotting using the anti-modified citrulline (AMC) antibody. We observed strong, distinct bands, with a similar pattern of citrullinated proteins in all strains tested (Fig. 1A). The gene encoding PPAD was detected in all tested P. gingivalis strains, including the clinical isolates, by PCR (data not shown). Subcellular fractionation of the wild-type further showed that the majority of citrullinated proteins are associated with the periplasm and the outer and inner membrane fractions (data not shown), which is a typical feature of bacterial virulence factors and antigens. To examine whether endogenous citrullination is a unique ability of P. gingivalis within the community of oral pathogens, we tested whole cell lysates of ten other oral organisms for the presence of citrullinated proteins. None were detected except in P. gingivalis (Fig. 1B), suggesting that functional PAD enzymes are absent from the other strains tested. Weak bands were noticeable in a number of strains, but were the result of nonspecific antibody binding (Fig. 1B ‘control’).
Figure 1.
Expression of endogenous citrullinated proteins is ubiquitous in P. gingivalis but absent in ten other oral bacteria. A, Protein citrullination in total cell extracts of the P. gingivalis wild-type reference strain (1–3) and four clinical isolates (4–7, corresponding to strains MaRL, D243, JH16, J430) was analysed by immunoblotting with the anti-modified citrulline (AMC) antibody. Lanes 1 and 2 are controls in which the modification step (1) or secondary antibody (2) has been omitted. The positions of the protein molecular mass markers in kilodalton (kDa) are indicated on the left. B, Total cell extracts of ten other prominent oral bacteria were tested for endogenous protein citrullination using the AMC antibody. Background signals were confirmed to stem from non-specific binding of the primary antibody (control). Pg: Porphyromonas gingivalis; Fn: Fusobacterium nucleatum; Aa: Aggregatibacter actinomycetemcomitans; Pi: Prevotella intermedia; Po: Prevotella oralis; Cg: Capnocytophaga gingivalis; Co: Capnocytophaga ochracea; Sc: Streptococcus constellatus; Sg: Streptococcus gordonii; Sn: Streptococcus sanguinis; Sl: Streptococcus salivarius.
Endogenous protein citrullination is dependent on the bacterial PAD enzyme in cooperation with protein cleavage by arginine-gingipains
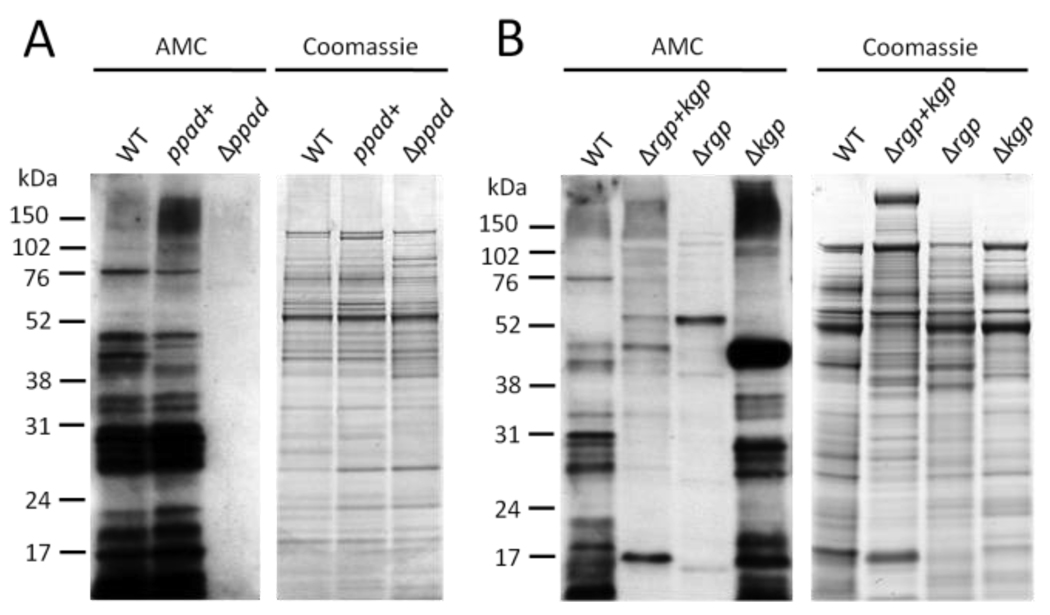
To confirm that the observed endogenous protein citrullination in P. gingivalis is due to the enzymatic activity of PPAD, and to rule out the possibility of a second, uncharacterized, bacterial peptidylarginine deiminase enzyme, we created a P. gingivalis W83 knockout strain (Δppad) by replacement of the entire ppad-encoding DNA sequence with an antibiotic cassette. A strain in which the antibiotic cassette was inserted behind the ppad gene was used as a control against polar effects from genetic manipulations (ppad+). Immunoblotting for citrullinated proteins showed that Δppad entirely lacks endogenous citrullinated proteins, while the wild-type strain (WT) and the control strain (ppad+) showed a similar pattern and intensity of citrullinated proteins (Fig. 2A). These data demonstrate that PPAD is essential for citrullination of endogenous proteins in P. gingivalis and that it possesses only one peptidylarginine deiminase. We then examined how citrullination depends on the activity of the major virulence factors in P. gingivalis, termed gingipains (32, 33). Gingipains are potent proteases and cleave various proteins/peptides after either arginine- (arginine-gingipains, Rgp) or lysine-residues (lysine-gingipains, Kgp), resulting in peptides with carboxy-terminal arginine- or lysine-residues. The reported preference of native PPAD for carboxy-terminal arginine residues in vitro (26) could be mediated through the activity of arginine-gingipains, and as such would have important consequences for the type of citrullinated peptides that can be generated by P. gingivalis. Hence, we studied P. gingivalis mutants lacking functional arginine-gingipains (Δrgp), lysine-gingipains (Δkgp), or both types of gingipains (Δrgp+kgp) for endogenous citrullination. Immunoblotting of whole cell lysates showed that there is a significantly decreased level, but not complete abrogation, of citrullinated proteins in the Δrgp and Δrgp+kgp strains, but not in the Δkgp strain (Fig. 2B), confirming that arginine-gingipains play a role in protein citrullination, probably by generating proteins with carboxy-terminal arginine residues that are subsequently citrullinated by PPAD. The residual citrullinated proteins seen in the Δrgp and Δrgp+kgp strains might be due to proteins naturally containing a carboxy-terminal arginine residue, which have not been proteolytically processed, and therefore appear at different molecular weights compared to the wildtype.
Figure 2.
Citrullination in P. gingivalis depends on the bacterial peptidylarginine deiminase and is influenced by arginine-gingipain mediated proteolytic cleavage of substrate proteins. A, A P. gingivalis mutant strain lacking the bacterial peptidylarginine deiminase gene (Δppad) was constructed and total cell extracts were analysed for the presence of citrullinated proteins (AMC). A control mutant containing the entire ppad gene and the antibiotic cassette (ppad+) was created to control for possible polar effects. The wild-type strain W83 was used as positive control (WT). B, P. gingivalis mutant strains lacking all proteolytic gingipain activity (Δrgp+kgp) or either arginine-gingipain (Δrgp) or lysine-gingipain (Δkgp) were analysed for the presence of citrullinated proteins by immunoblotting.
P. gingivalis rapidly generates citrullinated fibrinogen and α-enolase peptides by proteolytic cleavage at arginine-X peptide bonds followed by citrullination of carboxy-terminal arginines
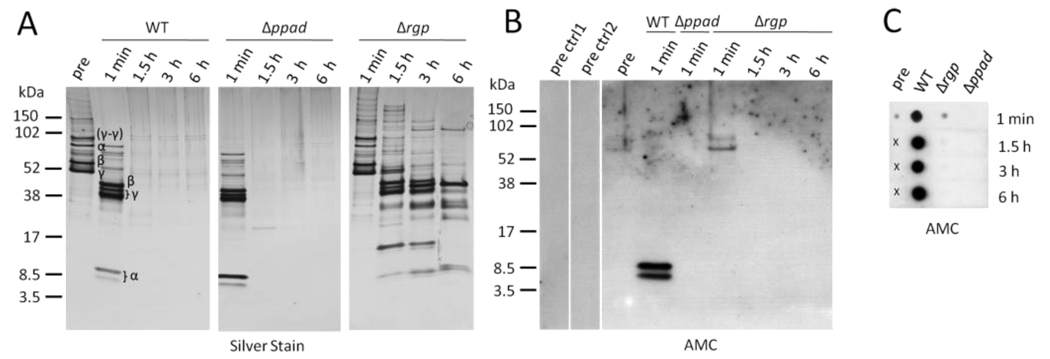
Having shown that citrullination of endogenous P. gingivalis proteins depends on the presence of PPAD and is influenced by arginine-gingipains, the question arose whether the same principles apply to human proteins. We initially chose human fibrinogen for this study as it is a major RA autoantigen in its citrullinated form (2, 3, 34) and a major part of the inflammatory response in general, due to its function in the coagulation and platelet aggregation cascade. Fibrinogen is also involved in the pathogenesis of periodontitis, where it is abundantly found in the periodontal lesion, being an established target protein of gingipains (35). We investigated the potential of P. gingivalis to citrullinate human fibrinogen using intact, live wild-type, Δppad and Δrgp knockout strains. Fibrinogen was rapidly cleaved by wild-type P. gingivalis (Fig. 3A), which is consistent with previous publications (36). A similar degradation pattern was observed with the Δppad strain, indicating that citrullination of substrate proteins, including gingipains themselves, is not essential for the proteolytic potency of gingipains. As expected, fibrinogen samples incubated with the Δrgp strain showed a considerably decreased proteolytic cleavage and a different pattern of cleaved peptides, with the residual proteolytic activity mainly due to cleavage by lysine-gingipains and other proteinases and peptidases from P. gingivalis (37–40). Our analysis of the identity of the protein bands by mass spectrometry confirmed that the majority were derived from any of the three fibrinogen chains (Fig. 3A). To exclude cleavage of protein by plasma-derived proteases which may contaminate fibrinogen, we further performed control reactions in which protein alone was incubated in assay buffer, and no such cleavage was detected. Next, we aimed to examine whether the cleaved fibrinogen fragments had been citrullinated by P. gingivalis. Immunoblotting of all samples that contained protein bands (see Fig. 3A) detected two citrullinated peptide bands around 8.5 kDa, mapping to the amino-terminal region of the fibrinogen α-chain, in samples incubated with P. gingivalis wild-type but not Δppad or Δrgp (Fig. 3B), confirming that Arg-X proteolytic cleavage of fibrinogen is a prerequisite for subsequent citrullination by PPAD. We further observed a weak positive signal in fibrinogen samples taken before incubation with P. gingivalis (‘pre’) and in the Δrgp ‘1 min’ sample, while the corresponding controls (‘pre ctrl1’: modification control; ‘pre ctrl2’: conjugate control) were negative (Fig. 3B), suggesting that purified human fibrinogen, as used in these experiments, is already endogenously citrullinated by human PADs. The fact that the majority of fibrinogen had been degraded within minutes, and that only two citrullinated proteins/peptides could be detected by immunoblotting, suggested that the majority of generated peptides was smaller than the size limit of the peptide gels (around 3 kDa) used for this analysis. Thus, we applied a dot blot technique that confirmed that wild-type P. gingivalis cells rapidly degrade and citrullinate fibrinogen into small citrullinated peptides (Fig. 3C), and that both arginine-gingipains and PPAD are required, as no positive signals were observed with the Δppad and Δrgp strains.
Figure 3.
P. gingivalis rapidly cleaves human fibrinogen through arginine-gingipain activity and the resulting peptides are citrullinated at the carboxy-terminus by bacterial peptidylarginine deiminase. A, Fibrinogen fragments after incubation with P. gingivalis wild-type (WT), mutants lacking peptidylarginine deiminase (Δppad), or arginine-gingipains (Δrgp), for 1 min, 1.5, 3 and 6 hours, were resolved by SDS-PAGE and visualised using silver staining. A fibrinogen sample prior to incubation with P. gingivalis served as control (pre). Protein bands were analysed by mass spectrometry (α, β, γ for fibrinogen α-, β-, and γ-chains). B, All samples with visible protein bands from A were tested for citrullination (AMC) by immunoblotting. Controls for the ‘pre’ sample were performed in which the modification step (pre ctrl1) or secondary antibody (pre ctrl2) were omitted. C, All samples from A were analysed for the presence of citrullinated peptides by dot blot. Crosses indicate areas on the membrane where samples were not applied.
We then aimed to identify the amino acid sequence of the citrullinated fibrinogen peptides and determine the position of the citrulline residue. To this end, we fractionated the peptides derived from the wild-type and Δppad strains by high-performance liquid chromatography (HPLC) and analysed the eluted peak fractions by liquid chromatography-tandem mass spectrometry (LC-MS/MS). We identified a total of 30 peptides derived from fibrinogen (not shown). The majority of identified peptides were the product of proteolytic cleavage after either an arginine or lysine residue, which is consistent with the results described above, but further proteolytic processing by other peptidases, in particular at the amino-terminus after glycine-, alanine- and serine-residues, was also evident. In samples incubated with P. gingivalis wild-type but not in Δppad, we found four peptides that contained a carboxy-terminal citrulline residue (Fig. 4): (1) 1ADSGEGDFLAEGGGVCit16, (2) 31PAPPPISGGGYCit42, (3) 253GGSTSYGTGSETESPCit268, (4) 540ESSSHHPGIAEFPSCit554. Carboxy-terminal arginine-containing peptides were detected only in the Δppad, but not in the wild-type (Table 1), suggesting that citrullination of fibrinogen is tightly linked to cleavage by arginine-gingipains. The combined data support the concept that target proteins such as human fibrinogen are cleaved by arginine-gingipains, generating suitable peptide substrates for subsequent citrullination at the exposed carboxy-terminal arginine residue by P. gingivalis PAD.
Figure 4.
Sequences of citrullinated peptides from human fibrinogen generated after incubation with P. gingivalis. Amino acid sequences of human fibrinogen α-chain (Fib A, SwissProt entry P02671) and β-chain (Fib B, SwissProt entry P02675) are shown. Peptides were detected using tandem mass spectrometry. Citrullinated peptides detected after incubation of fibrinogen with P. gingivalis wild-type are underlined. Fibrinopeptide A and B and the thrombin cleavage sites are indicated. Cit = citrulline.
Table 1.
Mass spectrometry analysis of citrullinated peptides generated after incubation of human fibrinogen or α-enolase with P. gingivalis wild-type (WT) and their respective arginated peptides generated with the Δppad strain.
| Sequence1 | Protein | Location (amino acids) |
Strain | m/z ratio2 | Mascot Score |
|---|---|---|---|---|---|
| -.ADSGEGDFLAEGGGVCit.G | Fibrinogen α- chain |
1–16 | WT | 769.29 (+2) | 56 |
| -.ADSGEGDFLAEGGGVR.G | Δppad | 768.77 (+2) | 99 | ||
| R.GGSTSYGTGSETESPCit.N | Fibrinogen α- chain |
253–268 | WT | 546.91 (+3) | 51 |
| R.GGSTSYGTGSETESPR.N | Δppad | 786.82 (+2) | 78 | ||
| K.ESSSHHPGIAEFPSCit.G | Fibrinogen α- chain |
540–554 | WT | 819.83 (+2) | 98 |
| S.SHHPGIAEFPSR.G | 543–554 | Δppad | 445.54 (+3) | 39 | |
| R.PAPPPISGGGYCit.A | Fibrinogen β- chain |
31–42 | WT | 585.30 (+2) | 26 |
| R.PAPPPISGGGYR.A | Δppad | 584.77 (+2) | 65 | ||
| S.TGIYEALELCit.D | α-enolase | 41–50 | WT | 583.30 (+2) | 24 |
| S.TGIYEALELR.D | Δppad | 582.79 (+2) | 56 | ||
Amino acids in the uncleaved proteins located carboxy- and amino-terminal to the identified peptides are indicated to demonstrate sites of proteolytic cleavage. Identified citrulline residues (Cit) are underlined.
m/z: mass-to-charge; numbers in brackets: peptide ion charge state
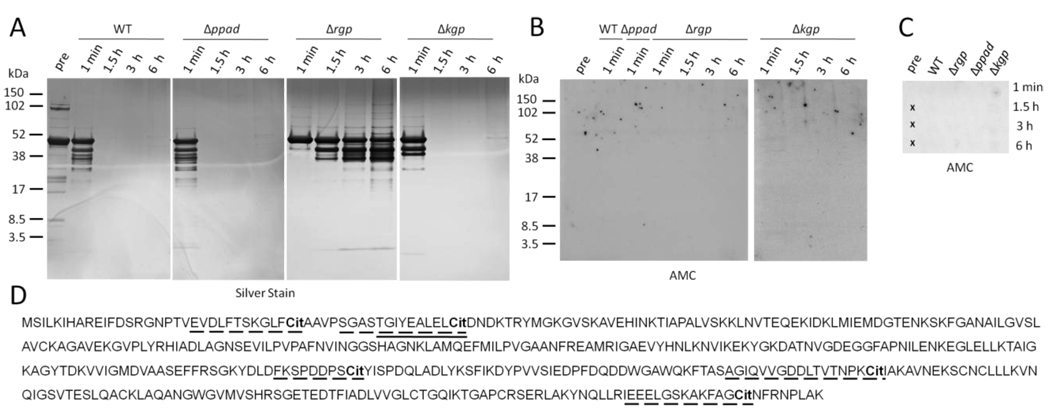
To further test this concept, we performed analogous experiments using recombinant human α-enolase. Similar to fibrinogen, enolase was rapidly degraded by the WT, Δppad and less so by the Δrgp strain (Fig. 5A). Using immunoblotting on peptide SDS-PAGE gels (Fig. 5B) and dot blot (Fig. 5C), no citrullination could be detected. Analysis of the WT and Δppad derived samples by mass spectrometry revealed only one citrullinated peptide in the WT (41TGIYEALELCit50, Fig. 5D and Table 1), amongst a total of 17 detected peptides. The arginine-containing counterpart of this citrullinated peptide was detected in the samples incubated with Δppad. Analogous to fibrinogen, no peptides with carboxy-terminal arginine were detected in the WT samples. The proportion of peptides cleaved at residues other than arginine and lysine was higher than that found in fibrinogen, suggesting extensive cleavage by non-argine/lysine specific peptidases. Combined with the higher relative number of lysine residues in α-enolase (8.8 % versus 6.9 % in fibrinogen), this might result in the generation of short peptides, some of which might be citrullinated but too short to be detectable using the methods employed. We therefore incubated P. gingivalis Δkgp with α-enolase (Fig. 5A–C), and could detect five citrullinated peptides by mass spectrometry (Fig. 5D), confirming that PPAD is able to citrullinate α-enolase peptides. Using the AMC dot blot, which relies on long peptides that are hydrophobic enough to bind to the membrane, we observed weak positive signals with the Δkgp strain, which decreased with time (Fig. 5C), again suggesting extensive proteolytic degradation by other proteinases.
Figure 5.
Human α-enolase is rapidly cleaved by P. gingivalis gingipains and citrullinated peptides are detectable by mass spectrometry. A, Experiments were performed analogous to fibrinogen (Figure 3). Fragments of human α-enolase after incubation with P. gingivalis wild-type (WT), mutants lacking peptidylarginine deiminase (Δppad), arginine-gingipains (Δrgp), or lysine-gingipains (Δkgp), for 1 min, 1.5, 3 and 6 hours, were resolved by SDS-PAGE and visualised using silver staining. An enolase sample prior to incubation with P. gingivalis served as control (pre). B, All samples with visible protein bands from A were tested for citrullination (AMC) by immunoblotting. C, All samples from A were analysed for citrullinated peptides by dot blot. Crosses indicate areas on the membrane where samples were not applied. D, Citrullinated peptides detected by mass spectrometry after incubation of enolase with P. gingivalis wild-type and Δkgp are underlined with a continuous and dashed line, respectively. Amino acid sequence of human α-enolase: SwissProt entry P06733. Cit = citrulline.
Discussion
In the present study we provide evidence that the periodontal pathogen P. gingivalis is an alternative source in the human host for generating citrullinated proteins and peptides. The underlying mechanism - proteolytic cleavage and subsequent citrullination at carboxy-terminal arginine residues - differs from that of the human PAD enzymes, which citrullinate internal arginine residues in whole proteins most efficiently. This finding suggests that protein citrullination by PPAD has the potential to generate epitopes to which immunological tolerance does not exist, not only due to the presence of foreign citrullinated proteins from the bacterium, but also through a foreign mode of proteolytic processing and post-translational modification of host antigens. It also indicates that citrullination of bacterial proteins at internal arginines, as a potential mechanism for triggering autoantibodies via molecular mimicry (5), is more likely to be due to the action of human PAD enzymes present at the site of inflammation.
Endogenous citrullinated proteins were detected exclusively in P. gingivalis, out of eleven oral bacterial species tested, indicating that a bacterial PAD gene is expressed or active only in this bacterium amongst those tested. To substantiate this finding, we performed similarity searches using BLAST and Psi Blast (41). This revealed numerous orthologues distantly related to PPAD among prokaryotes, including several of the oral organisms we tested. Most share the predicted conserved catalytic residues of PPAD and other members of the guanidino-group modifying enzymes superfamily, although they most likely possess agmatine iminohydrolase or arginine deiminase rather than peptidylarginine deiminase activity (42).
Using fibrinogen as a model antigen, we show that P. gingivalis rapidly generates small fibrinogen peptides with carboxy-terminal citrulline residues. Fibrinopeptide A, which normally results from thrombin cleavage of the fibrinogen α-chain after arginine-16, was also detected in its citrullinated form (1ADSGEGDFLAEGGGVcit16) in samples incubated with P. gingivalis wild-type, but only in the native, arginine-containing form in the Δppad samples. It is known that P. gingivalis gingipain-mediated degradation of human fibrinogen inhibits fibrinogen polymerization and results in the localised bleeding tendency which is typical for chronic periodontitis (35). A recent study has shown that, in intact fibrinogen, internal citrullination at arginine-16 by mammalian PAD impairs thrombin-catalyzed cleavage and fibrin polymerization (43), indicating at least two possible pathogenic roles for citrullinated fibrinogen in RA, serving as an autoantigen and disturbing the coagulation cascade and linked pathways. The pathophysiological role of fibrinogen peptides with carboxy-terminal citrulline residues, generated by the concerted action of gingipain and PPAD, is yet unknown, opening up a novel area for future investigations. Similarly, it is known that P. gingivalis arginine-gingipains cleave a number of other human proteins, releasing biologically active peptides with important roles in immunity and inflammation, such as C5a (44) and bradykinin (45, 46), and simultaneous citrullination of these peptides by P. gingivalis PAD might have a previously unappreciated role in human disease.
The lower levels of detectable citrullination of α-enolase peptides with the P. gingivalis wild-type are likely to be the result of a combination of physiological and technical factors. Enolase has a lower percentage of arginine residues (3.9%) compared to fibrinogen (5.2%), and a higher percentage of lysine residues (enolase 8.8% versus fibrinogen 6.9%). Further, it appears to be more extensively cleaved by non-arg/lys peptidases. Combined, this would result in fewer suitable PPAD substrates, and overall very small peptides, which would not be detected using the methods employed. Thus, lower levels of detectable citrullination may simply be due to a relative paucity of the substrate, and technical shortcomings with the detection of short peptides.
Here, we have demonstrated that P. gingivalis efficiently citrullinates its own proteins and peptides from host fibrinogen, and, to a lesser extent, α-enolase. The two major findings of this study, that proteolytic processing is required for citrullination by P. gingivalis and that host peptides with exclusively carboxy-terminal citrulline residues are generated, provides a strong basis for future in vivo studies, aimed at identifying citrullinated peptides at the site of gingival inflammation and exploring their potency for triggering a T- and/or B -cell response. Citrullinated host peptides, generated by P. gingivalis, are likely to expose epitopes previously hidden from immune surveillance, which, in the context of bacterial infection in a genetically susceptible host, may trigger an immune response. The slightly increased prevalence of anti-citrullinated protein antibodies reported in patients with periodontitis compared to healthy controls (47, 48) supports this concept, but the lower frequency and titre than that found in RA suggest that P. gingivalis infection is not sufficient on its own for the mature autoimmune response. However, once tolerance is breached, we predict that exposure to host proteins in the inflamed joint, citrullinated by human PADs (31), leads to intra- and inter-molecular epitope spreading to additional peptides from the initiating porteins and other autoantigens. Thus we propose a “two-hit” model of RA, based on (i) tolerance breakdown to specific citrullinated peptides generated by P. gingivalis at the site of gingival inflammation, followed by (ii) epitope-spreading to other host citrullinated proteins in the inflamed joint. This self-sustaining immune response would then result in the chronic and destructive inflammation that typifies RA. The unique nature of the bacterial deiminase, along with its location on the cell-surface of the bacterium (26), provides a target for treatment designed to prevent this otherwise incurable disease.
Acknowledgments
We are thankful to Drs Andrzej Kozik, Maria Rapala–Kozik, and Anna Golda from the Jagiellonian University Krakow, Poland, for their help with HPLC analysis and to Mr Anto Jose, Glasgow Dental School, Glasgow, UK, for preparation of bacterial strains. N.W., R.W., K.L. and P.V. were supported by a grant from the Arthritis Research Campaign, UK (DKCR F33018), S.C. was supported by Medical Research Scotland 168RFG, and J.P. by National Institutes of Health, USA, (DE 09761) and Department of Scientific Research, Polish Ministry of Science and Education (1642/B/P01/2008/35). The Faculty of Biochemistry, Biophysics and Biotechnology of the Jagiellonian University Krakow is a beneficent of the structural funds from the European Union (grant No: POIG.02.01.00-12-064/08 – “Molecular biotechnology for health”). We also acknowledge the European Union funded “AutoCure” (Curing Autoimmune Rheumatic Disease) Consortium for providing some of the funding to KL and PV and fostering international collaborations.
Footnotes
Author contributions
N.W. designed and performed experiments, analysed data, and wrote the paper; R.W. designed experiments, performed mass spectrometry and analysed data; A.S., S.E. and S.C. cultivated bacterial strains and prepared cell extracts; K.-A.N. built the knockout constructs; K.L. performed initial experiments on endogenous citrullination; A.K. built the α-enolase expression construct; J.P. co-supervised the study, designed experiments and analysed data. P.V. co-supervised the study, designed experiments, analysed data, and supervised writing of the paper. A.S., S.E., S.C., R.W., K.-A.N., K.L., A.K. and J.P. also contributed to the preparation of the manuscript.
References
- 1.Wegner N, Lundberg K, Kinloch A, Fisher B, Malmström V, Feldmann M, et al. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunological Reviews. 2010;233(1):34–54. doi: 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- 2.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, et al. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J. Immunol. 2001;166(6):4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 3.Sebbag M, Moinard N, Auger I, Clavel C, Arnaud J, Nogueira L, et al. Epitopes of human fibrin recognized by the rheumatoid arthritis-specific autoantibodies to citrullinated proteins. Eur. J. Immunol. 2006;36(8):2250–2263. doi: 10.1002/eji.200535790. [DOI] [PubMed] [Google Scholar]
- 4.Kinloch A, Tatzer V, Wait R, Peston D, Lundberg K, Donatien P, et al. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther. 2005;7(6):R1421–R1429. doi: 10.1186/ar1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundberg K, Kinloch A, Fisher BA, Wegner N, Wait R, Charles P. et al. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58(10):3009–3019. doi: 10.1002/art.23936. [DOI] [PubMed] [Google Scholar]
- 6.Snir O, Widhe M, von Spee C, Lindberg J, Padyukov L, Lundberg K, et al. Multiple antibody reactivities to citrullinated antigens in sera from patients with rheumatoid arthritis: association with HLA-DRB1 alleles. Ann. Rheum. Dis. 2009;68(5):736–743. doi: 10.1136/ard.2008.091355. [DOI] [PubMed] [Google Scholar]
- 7.Wegner N, Wait R, Venables PJ. Evolutionarily conserved antigens in autoimmune disease: Implications for an infective aetiology. Int J Biochem Cell Biol. 2009;41(2):390–397. doi: 10.1016/j.biocel.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Wygrecka M, Marsh LM, Morty RE, Henneke I, Guenther A, Lohmeyer J, et al. Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood. 2009;113(22):5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 9.Pancholi V, Fischetti VA. alpha-enolase a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273(23):14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 10.Clavel C, Nogueira L, Laurent L, Iobagiu C, Vincent C, Sebbag M, et al. Induction of macrophage secretion of tumor necrosis factor alpha through Fcgamma receptor IIa engagement by rheumatoid arthritis-specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum. 2008;58(3):678–688. doi: 10.1002/art.23284. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, Okeke NL, Sharpe O, Batliwalla FM, Lee AT, Ho PP, et al. Circulating immune complexes contain citrullinated fibrinogen in rheumatoid arthritis. Arthritis Res. Ther. 2008;10(4):R94. doi: 10.1186/ar2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trouw LA, Haisma EM, Levarht EWN, Woude Dvd, Ioan-Facsinay A, Daha MR, et al. Anti-cyclic citrullinated peptide antibodies from rheumatoid arthritis patients activate complement via both the classical and alternative pathways. Arthritis Rheum. 2009;60(7):1923–1931. doi: 10.1002/art.24622. [DOI] [PubMed] [Google Scholar]
- 13.Hill JA, Bell DA, Brintnell W, Yue D, Wehrli B, Jevnikar AM. et al. Arthritis induced by posttranslationally modified (citrullinated) fibrinogen in DR4-IE transgenic mice. J. Exp. Med. 2008;205(4):967–979. doi: 10.1084/jem.20072051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI. Citrullination: a posttranslational modification in health and disease. Int J Biochem Cell Biol. 2006;38(10):1662–1677. doi: 10.1016/j.biocel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 15.Makrygiannakis D, af Klint E, Lundberg IE, Lofberg R, Ulfgren AK, Klareskog L, et al. Citrullination is an inflammation-dependent process. Ann. Rheum. Dis. 2006;65(9):1219–1222. doi: 10.1136/ard.2005.049403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Venrooij WJ, Zendman AJ. Anti-CCP2 Antibodies: An Overview and Perspective of the Diagnostic Abilities of this Serological Marker for Early Rheumatoid Arthritis. Clin Rev Allergy Immunol. 2008;34(1):36–39. doi: 10.1007/s12016-007-8029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vossenaar ER, Smeets TJ, Kraan MC, Raats JM, van Venrooij WJ. Tak PP. The presence of citrullinated proteins is not specific for rheumatoid synovial tissue. Arthritis Rheum. 2004;50(11):3485–3494. doi: 10.1002/art.20584. [DOI] [PubMed] [Google Scholar]
- 18.Chapuy-Regaud S, Sebbag M, Baeten D, Clavel C, Foulquier C, De Keyser F. et al. Fibrin deimination in synovial tissue is not specific for rheumatoid arthritis but commonly occurs during synovitides. J. Immunol. 2005;174(8):5057–5064. doi: 10.4049/jimmunol.174.8.5057. [DOI] [PubMed] [Google Scholar]
- 19.van der Helm-van Mil AH, Verpoort KN, Breedveld FC, Huizinga TW, Toes RE. de Vries RR. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anti-cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54(4):1117–1121. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 20.Klareskog L, Stolt P, Lundberg K, Kallberg H, Bengtsson C, Grunewald J, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54(1):38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 21.Mahdi H, Fisher BA, Kallberg H, Plant D, Malmstrom V, Ronnelid J, et al. Specific interaction between genotype, smoking and autoimmunity to citrullinated alpha-enolase in the etiology of rheumatoid arthritis. Nat Genet. 2009;41(12):1319–1324. doi: 10.1038/ng.480. [DOI] [PubMed] [Google Scholar]
- 22.Borrell LN, Burt BA, Taylor GW. Prevalence and Trends in Periodontitis in the USA: from the NHANES III to the NHANES, 1988 to 2000. Journal of Dental Research. 2005;84(10):924–930. doi: 10.1177/154405910508401010. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis are associated with severity of periodontal tissue destruction. J Periodontol. 2001;72(10):1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 24.Griffen AL, Lyons SR, Becker MR, Moeschberger ML, Leys EJ. Porphyromonas gingivalis strain variability and periodontitis. J Clin Microbiol. 1999;37(12):4028–4033. doi: 10.1128/jcm.37.12.4028-4033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Pablo P, Chapple ILC, Buckley CD, Dietrich T. Periodontitis in systemic rheumatic diseases. Nat. Rev. Rheumatol. 2009;5(4):218–224. doi: 10.1038/nrrheum.2009.28. [DOI] [PubMed] [Google Scholar]
- 26.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 1999;67(7):3248–3256. doi: 10.1128/iai.67.7.3248-3256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graves D. Cytokines that promote periodontal tissue destruction. J. Periodontol. 2008;79(8 Suppl):1585–1591. doi: 10.1902/jop.2008.080183. [DOI] [PubMed] [Google Scholar]
- 28.de Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. J. Rheumatol. 2008;35(1):70–76. [PubMed] [Google Scholar]
- 29.Sugawara K, Oikawa Y, Ouchi T. Identification and properties of peptidylarginine deiminase from rabbit skeletal muscle. J. Biochem. 1982;91(3):1065–1071. doi: 10.1093/oxfordjournals.jbchem.a133755. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen K-A, Travis J. Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J. Bacteriol. 2007;189(3):833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinloch A, Lundberg K, Wait R, Wegner N, Lim NH, Zendman AJW, et al. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58(8):2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 32.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalis proteinases as virulence factors in the development of periodontitis. J. Periodontal Res. 1997;32(1 Pt 2):120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 33.Potempa J, Sroka A, Imamura T, Travis J. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 2003;4(6):397–407. doi: 10.2174/1389203033487036. [DOI] [PubMed] [Google Scholar]
- 34.Klareskog L, Ronnelid J, Lundberg K, Padyukov L, Alfredsson L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu. Rev. Immunol. 2008;26:651–675. doi: 10.1146/annurev.immunol.26.021607.090244. [DOI] [PubMed] [Google Scholar]
- 35.Imamura T, Potempa J, Pike RN, Moore JN, Barton MH, Travis J. Effect of free and vesicle-bound cysteine proteinases of Porphyromonas gingivalis on plasma clot formation: implications for bleeding tendency at periodontitis sites. Infect Immun. 1995;63(12):4877–4882. doi: 10.1128/iai.63.12.4877-4882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ally N, Whisstock JC, Sieprawska-Lupa M, Potempa J, Le Bonniec BF, Travis J, et al. Characterization of the specificity of arginine-specific gingipains from Porphyromonas gingivalis reveals active site differences between different forms of the enzymes. Biochemistry. 2003;42(40):11693–11700. doi: 10.1021/bi0349726. [DOI] [PubMed] [Google Scholar]
- 37.Nelson D, Potempa J, Kordula T, Travis J. Purification and characterization of a novel cysteine proteinase (periodontain) from Porphyromonas gingivalis. Evidence for a role in the inactivation of human alpha1-proteinase inhibitor. J Biol Chem. 1999;274(18):12245–12251. doi: 10.1074/jbc.274.18.12245. [DOI] [PubMed] [Google Scholar]
- 38.Jagels MA, Travis J, Potempa J, Pike R, Hugli TE. Proteolytic inactivation of the leukocyte C5a receptor by proteinases derived from Porphyromonas gingivalis. Infect Immun. 1996;64(6):1984–1991. doi: 10.1128/iai.64.6.1984-1991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masuda K, Yoshioka M, Hinode D, Nakamura R. Purification and Characterization of Arginine Carboxypeptidase Produced by Porphyromonas gingivalis. Infect. Immun. 2002;70(4):1807–1815. doi: 10.1128/IAI.70.4.1807-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banbula A, Bugno M, Goldstein J, Yen J, Nelson D, Travis J, et al. Emerging Family of Proline-Specific Peptidases of Porphyromonas gingivalis: Purification and Characterization of Serine Dipeptidyl Peptidase, a Structural and Functional Homologue of Mammalian Prolyl Dipeptidyl Peptidase IV. Infect. Immun. 2000;68(3):1176–1182. doi: 10.1128/iai.68.3.1176-1182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Shirai H, Mokrab Y, Mizuguchi K. The guanidino-group modifying enzymes: structural basis for their diversity and commonality. Proteins. 2006;64(4):1010–1023. doi: 10.1002/prot.20863. [DOI] [PubMed] [Google Scholar]
- 43.Nakayama-Hamada M, Suzuki A, Furukawa H, Yamada R, Yamamoto K. Citrullinated Fibrinogen Inhibits Thrombin-catalysed Fibrin Polymerization. J Biochem. 2008;144(3):393–398. doi: 10.1093/jb/mvn079. [DOI] [PubMed] [Google Scholar]
- 44.Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J Immunol. 2007;178(11):7242–7250. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- 45.Monteiro AC, Scovino A, Raposo S, Gaze VM, Cruz C, Svensjo E, et al. Kinin danger signals proteolytically released by gingipain induce Fimbriae-specific IFN-gamma- and IL-17-producing T cells in mice infected intramucosally with Porphyromonas gingivalis. J Immunol. 2009;183(6):3700–3711. doi: 10.4049/jimmunol.0900895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imamura T, Pike RN, Potempa J, Travis J. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalis induces vascular permeability enhancement through activation of the kallikrein/kinin pathway. J Clin Invest. 1994;94(1):361–367. doi: 10.1172/JCI117330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC, et al. Antibody responses to Porphyromonas gingivalis (P. gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int. Immunopharmacol. 2009;9(1):38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Havemose-Poulsen A, Westergaard J, Stoltze K, Skjodt H, Danneskiold-Samsoe B, Locht H, et al. Periodontal and hematological characteristics associated with aggressive periodontitis, juvenile idiopathic arthritis, and rheumatoid arthritis. J. Periodontol. 2006;77(2):280–288. doi: 10.1902/jop.2006.050051. [DOI] [PubMed] [Google Scholar]