Summary
Single-gene mutants with extended lifespan have been described in several model organisms. We performed a genome-wide screen for long-lived mutants in Escherichia coli which revealed strains lacking TCA cycle-related genes that exhibit longer stationary phase survival and increased resistance to heat stress compared to wild-type. Extended lifespan in the sdhA mutant, lacking subunit A of succinate dehydrogenase, is associated with reduced production of superoxide and increased stress resistance. On the other hand, the longer lifespan of the lipoic acid synthase mutant (lipA) is associated with reduced oxygen consumption and requires the acetate-producing enzyme pyruvate oxidase, as well as acetyl-CoA synthetase, the enzyme that converts extracellular acetate to acetyl-CoA. The hypoxia-inducible transcription factor ArcA, acting independently of acetate metabolism, is also required for maximum lifespan extension in the lipA and lpdA mutants, indicating that these mutations promote entry into a mode normally associated with a low-oxygen environment. Since analogous changes from respiration to fermentation have been observed in long-lived Saccharomyces cerevisiae and Caenorhabditis elegans strains, such metabolic alterations may represent an evolutionarily conserved strategy to extend lifespan.
Keywords: lifespan, acetate, Escherichia coli, hypoxia, superoxide, stress resistance
Introduction
The existence of a germ line that is distinct from somatic tissue has been proposed as a prerequisite for the evolution of senescence (Partridge & Barton 1993). A key prediction of this theory was disproved by the observation that one of the two daughter cells that result from the morphologically symmetrical division of an individual Escherichia coli cell displays reduced growth rate with successive generations, which is the hallmark of reproductive senescence (Stewart et al. 2005). At the population level, the growth rate of an Escherichia coli batch culture (maintained in the same medium without addition or removal of any material) gradually declines and proliferation eventually ceases, despite the presence of extracellular nutrients that could support a further production of biomass (our unpublished observation), marking the onset of stationary phase. As stationary phase progresses, an increasing fraction of the E. coli population becomes unable to resume growth upon transfer to fresh nutrient medium, subsequently loses membrane integrity assessed using fluorescent dyes and is therefore considered dead (Ericsson et al. 2000; Finkel 2006).
Using the fluorescent nucleic acid stain propidium iodide, a good correlation was found between the loss of proliferating potential and the loss of membrane integrity (Ericsson et al. 2000); thus, there does not seem to be a substantial fraction of a stationary phase E. coli population in LB medium that loses culturability but maintains viability. We therefore decided to use the formation of colonies from cells sampled from a stationary phase population (colony forming units, CFU) as a measure of the viability of that population. The progressive loss of culturability / viability observed in stationary phase results in the loss of 90-99% of the initial population and is reminiscent of the stationary phase survival of the budding yeast Saccharomyces cerevisiae, which has been introduced by our lab as a model system for the study of aging and lifespan in higher organisms (Fabrizio & Longo 2003). For consistency with our work in yeast and to distinguish it from reproductive lifespan we call survival in stationary phase “chronological lifespan”.
Most previous work on stationary phase E. coli has focused on the characterization of the organism's physiology as compared to log phase cells. Particular focus has been given to the σs subunit of the RNA polymerase, encoded by rpoS, which is the master regulator of several stationary phase-inducible genes and phenotypes, such as resistance to heat and oxidative stress (Hengge-Aronis 2002). Numerous strains with a more rapid loss of stationary phase viability than wild-type (wt) have been described (Groat et al. 1986; Visick et al. 1998), but there have been few reports of mutants with extended survival. Loss of the toxin-antitoxin cell death system encoded by hipBA causes resistance to hydrogen peroxide and extended stationary phase survival (Kawano et al. 2009). A microscopy-based screen of a transposon-mutagenized collection of E. coli mutants revealed that a strain lacking the response regulator RssB has a reduced proportion of dead cells during stationary phase and is also resistant to heat, oxidative and osmotic stress (Fontaine et al. 2008). The authors attributed these results to the stabilization of RpoS in the rssB strain. Finally, addition of ethanol has been shown to delay the viability loss of a stationary phase culture, also in an RpoS-dependent manner (Vulic & Kolter 2002).
We used the KEIO collection, which consists of single-gene deletion strains of every non-essential protein-coding gene in E. coli (Baba et al. 2006), to comprehensively screen for long-lived mutants by a spectrophotometric method. The screen revealed three mutants with extended chronological lifespan that are also stress–resistant. Our results provide evidence for the role of novel pathways in the regulation of prokaryotic survival and suggest that some fundamental metabolic processes lie at the center of survival regulation in organisms on either side of the line separating prokaryotes from eukaryotes.
Results
Genome-wide screen reveals three long-lived, heat shock-resistant strains
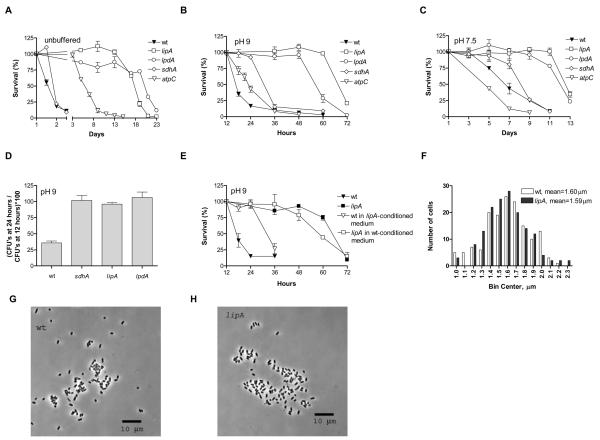
We employed a screening strategy similar to the one used in S. cerevisiae (Powers et al. 2006) to identify E. coli strains with increased stationary phase survival (Figure S1 and Text S1). Briefly, the mutants of the KEIO collection were maintained in batch culture in 96-well plates and were used to inoculate fresh cultures at two different time-points when more than 90% of a wild-type population is no longer viable (data not shown). The values of the optical density at 600 nm obtained after outgrowth of these fresh cultures normalized by each strain's growth rate were used as a proxy measure of the number of cells that were still alive at these time-points. All strains were thus ranked for both sampled time-points and top-ranking strains were then individually tested for stationary phase survival (Table S1). Most of the top-ranking strains displayed delayed entry into stationary phase (gradual increase in biomass several hours after the wt has ceased proliferation) and not extended stationary phase survival; these strains were not further tested. The four strains shown in figure 1A reach stationary phase at approximately the same time as wt, but maintain 100% survival for longer periods than wt.
Figure 1. Survival of E. coli mutants recovered from a genome-wide screen for extended lifespan.
Stationary phase survival in unbuffered LB medium (A), LB medium buffered to pH 9 (B) and LB medium buffered to pH 7.5 (C). Cell density was also equalized to approximately 1.5 × 109 cells per milliliter for the buffered survival experiments. (D) Ratio of colony-forming units per milliliter at 24 over 12 hours in stationary phase for the indicated strains at pH 9 in the MG1655 genetic background. (E) Survival of wt switched to lipA-conditioned medium and of the lipA strain switched to wt-conditioned medium. (F) Length distribution of 155 cells of wt and the lipA strain (cells outside the shown fields of view were also measured). (G) Phase contrast image of early stationary phase cells of wt. (H) Corresponding image for the lipA strain. See also figure S2.
We performed all experiments in LB, a peptide-rich, complex nutrient medium which contains traces of glucose (present in the yeast extract found in LB) that are consumed by a wt strain within the first 90 minutes of incubation (Baev et al. 2006). The metabolism of amino acids in this medium is accompanied by the excretion of ammonia that results in an extracellular stationary phase pH of 8.5-9 (Pruss et al. 1994; Farrell & Finkel 2003). The majority of the long-lived strains have a slower growth rate (Figure S2D) and lower saturation density than wt and also exhibit a slightly acidic pH at stationary phase (Table S2). We compared the survival of these strains to wt in several different ways, shown in Figure 1. All strains survive longer than wt when no adjustments are made (Figure 1A). To dissect the effect of pH and lower cell density on the observed lifespan extension, we equalized the cell density of all strains to approximately 1.5 × 109 cells per milliliter of culture and adjusted the pH upon stationary phase entry to either 9 or 7.5 using the biological buffers AMPSO or HEPES respectively. Only strains that lived longer in both alkaline and neutral conditions were investigated further (Fig. 1B and C). The CFU titers for the survival experiments shown in Figure 1A-C are shown in Figure S2A-C. The extended survival of the sdhA, lipA and lpdA mutants was confirmed in the commonly used wt strain MG1655 (Figure 1D).
To rule out the possibility that the hypoxic conditions generated by extended incubation in an orbitally shaking test-tube play a role in the observed lifespan extension, we compared the survival of wt and the longest-lived strain, lipA, in 10-ml cultures maintained in 125-ml flasks with loose-fitting caps, which provides a more thorough aeration of the cultures. The survival extension of the lipA strain is not diminished under these conditions (Figure S2E). We also found that the extended survival of the lipA mutant is largely unaffected by incubation in cell-free conditioned medium obtained from a stationary phase wt culture (Figure 1E). Therefore, the observed lifespan extension is to a large extent independent of the potentially retarded utilization of the carbon and energy available in LB by the lipA mutant, which grows slowly and saturates at a low cell density compared to wt..
It is important to note that despite slower growth rates and lower saturation densities, the lipA and lpdA strains appear to reach stationary phase at the same time as wt, since log-phase populations of the three strains reach a plateau at the same time, as shown by the respective growth curves (Figure S2D). Therefore, the observed lifespan extension is not due to delayed entry of the mutants in stationary phase. On the other hand, the survival of wt is extended by incubation in conditioned medium obtained from a stationary phase lipA culture (Figure 1E), pointing towards the existence of lifespan-enhancing substances generated by the lipA mutant during stationary phase, lifespan-shortening substances generated by wt, or both. It is unlikely that the observed lifespan extension of wt in lipA-conditioned medium is solely due to the increased availability of carbon and energy sources in the mutant's medium, since the wt survives even longer when incubated in 0.5% NaCl, without any extracellular carbon or energy source (compare wt in lipA-conditioned medium in figure 1E to wt 0.5% NaCl in figure 6E).
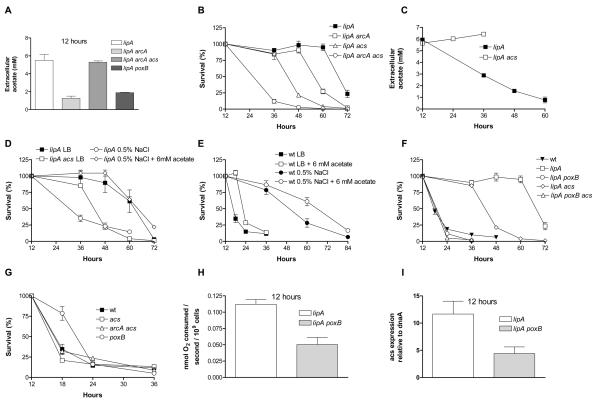
Figure 6. Acetate production and uptake are required for the extended lifespan of the lipA mutant.
(A) Extracellular acetate concentration of the lipA, lipA arcA, lipA arcA acs and lipA poxB strains at late stationary phase. (B) Effect of deletion of arcA, acs or both on the survival of the lipA mutant. (C) Extracellular acetate concentration of the lipA and lipA acs strains over time. No data were collected after 36 hours for the latter because of loss of viability. (D) Survival of the lipA and lipA acs strains in LB and of the former strain in 0.5% NaCl with or without the addition of 6 mM acetate. (E) Survival of wt in LB and in 0.5% NaCl with or without the addition of 6 mM acetate. (F) Effect of the deletion of poxB, acs or both on the survival of the lipA strain. (G) Stationary phase survival of the acs, arcA acs and poxB strains. (H) Effect of poxB deletion on the rate of oxygen consumption of the lipA strain at late stationary phase. (I) Expression of acs in the lipA and lipA poxB strains at late stationary phase. See also figures S4 and S5.
All subsequent experiments were performed in the following way, unless otherwise stated: cultures were incubated for 12 hours in test tubes in unbuffered LB, at which point cell density and pH were adjusted to approximately 1.5 × 109 cells per milliliter and pH 9 respectively, simulating the stationary phase conditions generated by wt in this medium. We define this as the zero-time-point in all experiments and all time periods denoted in the figures refer to the time elapsed after this point. Therefore, in the text the 1-hour time-point is referred to as ‘early stationary phase’ and the 12-hour time-point is referred to as ‘late stationary phase’. No loss of viability is observed in wt during this 12-hour period.
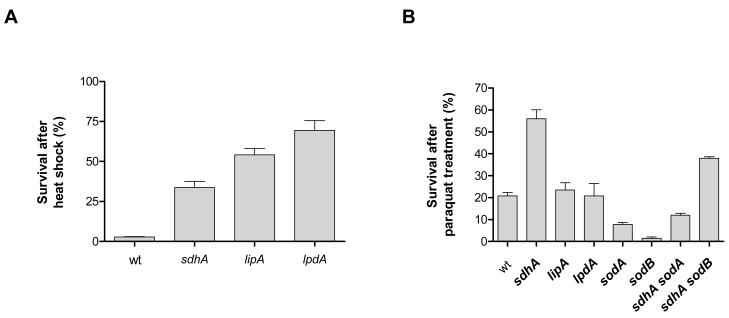
Since long-lived mutants in other model organisms are typically resistant to heat and oxidative stress (Miller 2009), we tested the survival of the three mutants after treatment with the superoxide-generating agent paraquat or after a high-temperature incubation. All long-lived strains were more resistant than wt to heat shock (Figure 2A) and the sdhA strain is more resistant than wt to the viability loss caused by treatment with paraquat (Figure 2B). Fumarate reductase, the protein that functionally replaces succinate dehydrogenase under anaerobic conditions (Maklashina et al. 1998), has been shown to react with paraquat (Jones & Garland 1977). It is therefore possible that SdhA is also an electron donor in the redox-cycling catalyzed by paraquat, which would explain the resistance of the strain lacking this protein to the lethal effects of paraquat. However, SdhA may also affect paraquat redox-cycling indirectly.
Figure 2. Stress resistance of long-lived mutants.
Survival of stationary phase cultures after 4 minutes of incubation at 55°C (A) or 12 hours of incubation with 500 μM paraquat at 37 °C (B).
To further investigate the mechanistic basis of the resistance of the sdhA mutant to paraquat, we tested the dependence of the phenotype on the proteins that catalyze the conversion of superoxide to oxygen and hydrogen peroxide, the manganese-containing superoxide dismutase SodA and its iron counterpart, SodB. We found that lack of SodA increases the sensitivity to paraquat in both a wt or sdhA background. The sodB mutant is more sensitive to paraquat than the sodA strain; this pronounced sensitivity is reversed by the deletion of sdhA (Figure 2B), consistent with the hypothesis that redox cycling between SdhA and paraquat is responsible for the enhanced sensitivity of the sodB mutant to the paraquat-induced viability loss. Long-lived organisms in other model systems are often smaller than their wt counterparts (Longo & Finch 2003). We measured the length of several stationary phase cells of both wt and the lipA mutant using phase contrast microscopy and found no difference in cell length between the two strains (Figure 1F-H). In conclusion, the extended lifespan of the three mutants we describe is associated with resistance to heat stress and is independent of differences in growth rate, saturation density, external pH and cell size.
Differences in metabolic physiology among long-lived mutants
lpdA encodes lipoamide dehydrogenase, a common component of the 2-ketoglutarate dehydrogenase (2-KGDH) complex of the TCA cycle and of the pyruvate dehydrogenase (PDH) complex (Smith & Neidhardt 1983a). Its function is to oxidize the protein-bound lipoic acid used during the oxidative decarboxylation of 2-ketoglutarate to succinyl-CoA and of pyruvate to acetyl-CoA. lipA encodes lipoic acid synthase, a protein that catalyses the formation of the carbon-sulfur bonds required to produce the lipoic acid used in the aforementioned reactions (Miller et al. 2000). Lack of either LipA or LpdA therefore results in the inactivation of both the 2-ketoglutarate dehydrogenase and the pyruvate dehydrogenase complexes. Lipoic acid is also a cofactor for the glycine cleavage system (Vanden Boom et al. 1991). However, inactivation of the T component protein of the glycine cleavage system (gcvT) has no effect on survival (Figure S2F). Inactivation of the E1 component of the PDH (aceE) or the 2-KGDH (sucA) complex individually or in combination also has no positive effect on survival (Figure S2F). Therefore, although LipA and LpdA are not known to function independently of the PDH, 2-KGDH and glycine cleavage complexes, our data indicate that lack of either LipA or LpdA extends lifespan by causing metabolic changes that are distinct from those caused by inactivation of the protein complexes in which they participate (see also Discussion).
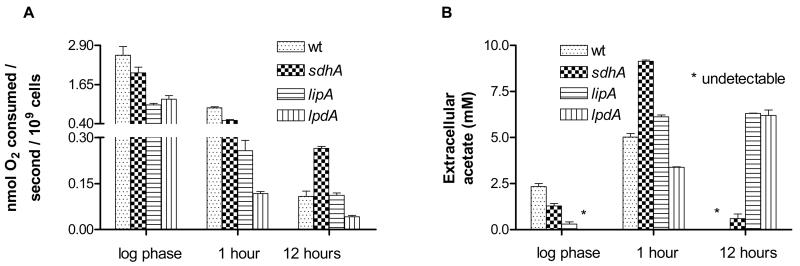
We first attempted to gain a mechanistic insight into the lifespan extension of the lipA and lpdA mutants by measuring the oxygen consumption of these strains over time. As shown in figure 3A, both strains consume less oxygen than wt at log phase and early stationary phase and the lpdA mutant also respires less at late stationary phase. On the other hand, the sdhA mutant displays a profile similar to wt, the only difference being a higher rate of oxygen consumption at late stationary phase. E. coli is a facultative anaerobe, able to grow and survive both in the presence of oxygen and also in the absence of any electron acceptors (Clark 1989). When oxygen respiration is not possible either due to the lack of oxygen or due to a genetic block in the respiratory chain, the pyruvate formed by the decarboxylation of amino acids in LB medium is converted to one of four fermentation products, namely lactate, succinate, acetate or ethanol. The relative amount of these by-products is mostly dictated by the need to maintain a physiological NADH/NAD+ ratio (Clark 1989).
Figure 3. Characterization of the metabolic physiology of long-lived mutants.
Rate of oxygen consumption (A) and extracellular acetate concentration (B) of long-lived mutants and wt over time. ‘1 hour’ and ‘12 hours’ refer to time-points in stationary phase, the onset of which is defined as 12 hours after the inoculation of each culture; ‘12 hours’ or ‘late stationary phase’ is therefore equivalent to 24 hours after the cultures' inoculation and ‘1 hour’ or ‘early stationary phase’ is equivalent to 13 hours after the cultures' inoculation.
Since the lipA and lpdA strains are genetically unable to perform respiratory metabolism at the level of wt, we measured the concentration of extracellular acetate over time. As reported in the literature, we found that the wild-type strain consumes the acetate it initially produced resulting in no detectable acetate at late stationary phase (Kumari et al. 1995), the sdhA mutant showing a similar behavior. On the other hand, the lipA and lpdA strains maintain a high extracellular acetate concentration at late stationary phase (Figure 3B). It is worth noting that the acetate produced by wt in the presence of oxygen is thought to be the consequence of the inability of the otherwise functional TCA cycle and the respiratory chain to utilize all the available acetyl-CoA produced by pyruvate dehydrogenase (Wolfe 2005). Therefore, the results presented in figure 3 demonstrate that at least two different mechanisms can lead to extended survival in E. coli; one, exemplified by the sdhA mutant, that involves a wt-like pattern of oxygen consumption and acetate metabolism and another, exemplified by the lipA and lpdA strains, that is characterized by reduced oxygen consumption and sustained presence of acetate in the extracellular environment.
Decreased superoxide production is associated with extended survival in the sdhA mutant
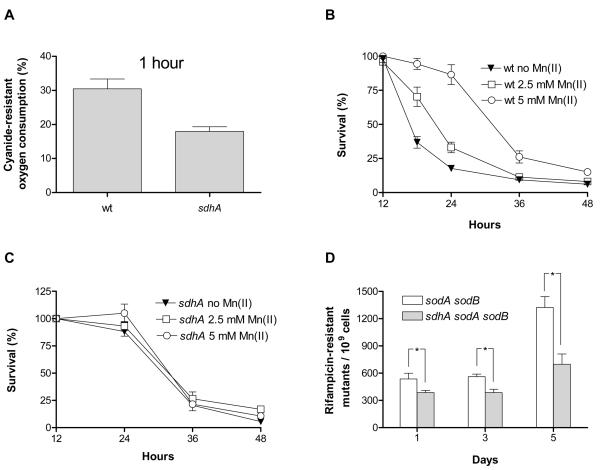
Since the mutant lacking subunit A of succinate dehydrogenase is the only one that is resistant to the superoxide generator paraquat (Figure 2B) and since this enzyme has also been shown to be a source of superoxide in vitro, possibly through its bound flavin adenine dinucleotide cofactor (Messner & Imlay 2002), we measured superoxide production in wild-type and the sdhA mutant by monitoring cyanide-resistant oxygen consumption. Cyanide acts as an inhibitor of the oxygen-consuming cytochrome oxidase and the oxygen consumption observed in its presence can be used as an approximation for the intracellular production of superoxide (Hassan & Fridovich 1977). Confirming previous in vitro results, the sdhA strain produced less superoxide at stationary phase compared to wt (Figure 4A).
Figure 4. Extended lifespan is associated with reduced superoxide production in the sdhA mutant.
Cyanide-resistant oxygen consumption of wt and the sdhA mutant at early stationary phase (A). Effect of manganese (II) chloride addition at early stationary phase on the lifespan of wt (B) and the sdhA mutant (C). (D) Time-dependent frequency of rifampicin-resistant mutants of the sodA sodB and sdhA sodA sodB strains during a stationary phase survival experiment at pH 7.5. Asterisks denote that the shown differences are significant at the p<0.05 level. See also figure S3.
Nystrom et al. reported no effect of the overproduction of superoxide dismutase A (sodA) on the survival of E. coli in minimal glucose medium (Nystrom et al. 1996). We attempted to perform the same experiment in LB medium, but observed a strong selection against retention of the SodA-overexpressing plasmid pDT1-5 (Touati 1983) through stationary phase despite the presence of ampicillin, to which the plasmid confers resistance (data not shown). On the other hand, the sodB-overexpressing plasmid pHS1-7 (Carlioz & Touati 1986) was retained throughout stationary phase, but produced no effect on stationary phase survival (Figure S3A). Although both SodA and SodB scavenge superoxide generated in the cytosol, it is possible that overexpression of both is required to have an effect on lifespan, as we have previously observed in yeast (Fabrizio et al. 2003). In fact, the SodA- and SodB-deficient mutants display distinct phenotypes (Carlioz & Touati 1986), consistent with the possibility that the contribution of these two enzymes on the physiology of E. coli is not identical. As an alternative method, we tested the effect of manganese, a known superoxide scavenger (Chang & Kosman 1989) on the survival of wt and the sdhA strain. Stationary phase addition of manganese (II) chloride produced a dose-dependent lifespan extension in wild-type, but did not further increase the lifespan of the sdhA mutant (Figures 4B and 4C).
Because superoxide has been shown to enhance DNA mutations (Benov & Fridovich 1996), we investigated the effect of succinate dehydrogenase on stationary phase mutation frequency, by measuring the occurrence of rifampicin-resistant mutants over time. Rifampicin is a drug that inhibits bacterial growth by binding to the β subunit of RNA polymerase, encoded by rpoB. Mutations in rpoB allow proliferation in the presence of this drug and the occurrence of such mutants has been used as a measure of mutation frequency in E. coli (Garibyan et al. 2003). No increase in mutation frequency was observed for both wt and the sdhA strain (Figure S3B). However, deletion of sdhA attenuated the time-dependent increase in the occurrence of rifampicin-resistant mutants in the strain devoid of both SodA and SodB (Figure 4D), consistent with the possibility that SdhA-produced superoxide contributes to the increased DNA damage observed over time in sodA sodB mutants. Note that the decreased mutation frequency in sdhA sodA sodB compared to sodA sodB is not growth rate-dependent since the two strains have similar growth curves (Figure S3C) and also rifampicin plates were checked for several days for the appearance of resistant colonies (see also Experimental procedures). Although these data are consistent with a role for SdhA-produced superoxide in promoting aging and death in E. coli, further studies are required to rule out the possibility that the lifespan extension caused by Mn (II) is superoxide-independent.
ArcA is required for the fully extended lifespan of the lipA and lpdA mutants
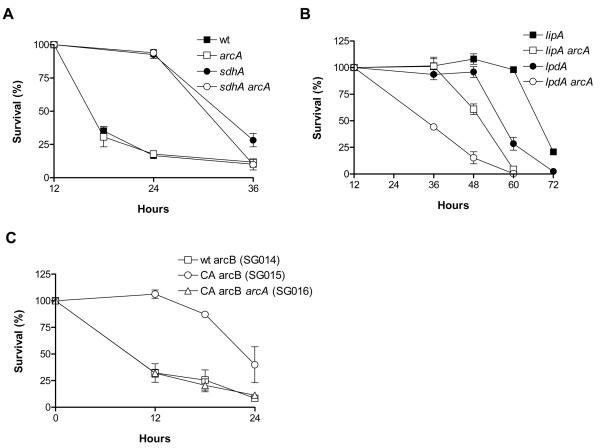
ArcA is a transcription factor that suppresses the expression of genes involved in respiration and activates genes involved in fermentative metabolism, thus contributing to the adaptation of E. coli to hypoxic conditions (Iuchi & Lin 1991). Lack of ArcA causes reduced stationary phase survival under glucose starvation conditions, which was attributed to the deregulation of cellular redox balance and the uncontrolled drainage of the cells' endogenous energy reserves (Nystrom et al. 1996). Because the PDH and 2-KGDH complexes, in which LipA and LpdA participate, are among the most drastically downregulated in response to oxygen shortage (Smith & Neidhardt 1983b), and both the lipA and lpdA strains consume less oxygen compared to wt (Figure 3A), we tested whether the hypoxia transcription factor ArcA becomes essential in these strains, even under relative abundance of environmental oxygen. We therefore tested the effect of ArcA deletion on the survival of these mutants and found that ArcA is required for maximum lifespan extension in both (Figure 5B). ArcA is also required for the extended survival of the mutants at an extracellular pH of 7.5 (Figure S4C). In contrast, the survival of both the wt and the sdhA strain was unaffected by the loss of ArcA (Figure 5A).
Figure 5. ArcA is necessary and sufficient for lifespan extension.
(A) Effect of arcA deletion on the survival of the lipA and lpdA strains. (B) Effect of arcA deletion on the survival of wt and the sdhA strain. (C) Effect of constitutive activation (CA) of arcB on the survival of wt and its dependence on arcA. The genotype of the shown strains are shown in table S2.
ArcA forms a typical two-component signal transduction module, along with the membrane protein ArcB, which is activated in response to hypoxia and then activates ArcA by phosphorylation (Georgellis et al. 2001). Replacement of the wt arcB gene with an allele that is constitutively active, due to its fusion with Tar (a methyl-accepting chemotaxis protein for sensing aspartate (Kwon et al. 2003)) results in an ArcA-dependent lifespan extension, of a smaller magnitude compared to the one observed in the lipA and lpdA mutants (Figure 5C). Thus, ArcA is necessary for the fully extended lifespan of the lipA and lpdA strains and its constitutive activation is sufficient to extend the lifespan of wt.
The fully extended lifespan of the lipA mutant is entirely dependent on pyruvate oxidase and partly dependent on acetyl-CoA synthetase
ArcA has been shown to contribute to acetate formation (Vemuri et al. 2006). We found that the extended survival of the lipA and lpdA strains is associated with a sustained presence of extracellular acetate during stationary phase (Figure 3B) and that the extended lifespan of these mutants is partially dependent on arcA (Figure 5B). Based on these results, we sought to determine the effect of arcA deletion on the extracellular acetate concentration of these mutants. Lack of ArcA results in a ~6-fold decrease in the concentration of acetate in the medium of the lipA mutant (Figure 6A), but has no effect on extracellular acetate in the lpdA strain (figure S4A).
Extracellular acetate can be converted to acetyl-CoA via two pathways, one catalyzed by the acetate kinase / phosphotransacetylase (AckA / Pta) enzyme pair and the other by acetyl-CoA synthetase (Acs) (Wolfe 2005). Measurement of extracellular acetate concentration over time reveals that the conversion of acetate to acetyl-CoA by Acs is responsible for the gradual disappearance of the metabolite from the extracellular environment of the lipA strain (Figure 6C), whereas the AckA / Pta enzyme pair is not (Figure S4F). This difference might be related to the lower Km of Acs for acetate, which renders it more suitable for acetate concentrations lower than 10 mM (Kumari et al. 1995). The observed trend of decreased expression of both ackA and pta in the lipA strain (Figures S4G-H) might also be a contributing factor for the non-participation of this enzyme pair in acetate uptake in the lipA strain. We ruled out the possibility that the disappearance of acetate from the medium of the lipA strain is due to degradation or conversion to another substance by recovering acetate added at 10 mM from spent medium of the lipA mutant after several days of incubation at 37°C (data not shown).
Prompted by the correlation between extracellular acetate concentration and lengthened survival revealed by the deletion of ArcA, we tested the effect of the lack of acs in the survival of the lipA strain. Lack of Acs shortens the survival of the lipA mutant (Figure 6B), whereas lack of Pta does not (Figure S4E). Acs mediates the gradual uptake of acetate from the medium in the lipA mutant also at pH 7.5 (data not shown), but it is not required for extended survival under these conditions (Figure S4D). Although the gradual uptake of acetate and its conversion to acetyl-CoA is required for extended survival at basic pH, the availability of extracellular acetate is not limiting for the lifespan of the lipA strain, as addition of acetate at a time-point when it has been almost depleted from the medium does not produce a further lifespan extension (Figure S4B). Deletion of acs from the lipA arcA mutant restores extracellular acetate to the level observed in the lipA strain (Figure 6A). However, the additive detrimental effect of arcA and acs deletion on the lifespan of the lipA mutant (Figure 6B) shows that ArcA and Acs independently contribute to the extended lifespan of this strain.
To confirm the dependence of the extended survival of the lipA strain on acetate by non-genetic means, a stationary phase population of the strain grown in LB was washed once with a solution of 86 mM NaCl (to remove all extracellular metabolites, including acetate) and subsequently maintained in 86 mM NaCl (the concentration used in LB). No acetate was detectable in a saline-resuspended culture of the lipA mutant both immediately after transfer to NaCl and 12 hours after the transfer (data not shown). The survival of the lipA mutant in NaCl is similar to that of the lipA acs strain in LB (Figure 6D). Adding back extracellular acetate at the concentration found in a late stationary phase culture of the lipA mutant maintained in LB (6 mM) right after transfer to NaCl restores the lifespan of the mutant to the level observed in LB (Figure 6D).
On the other hand, as reported previously (Vulic & Kolter 2002), the survival of wt is extended in 86 mM NaCl compared to incubation in LB (Figure 6E). Addition of 6 mM acetate to a wt culture maintained in NaCl causes a small survival increase, bringing the lifespan of wt close to the level of the lipA mutant under the same conditions. Hence, transfer of stationary phase populations of wt and the lipA strain from LB to 86 mM NaCl has opposite effects in terms of survival. The lifespan of wt increases, possibly due to the removal of death-accelerating substances present in spent LB medium, whereas the lifespan of the lipA strain is diminished due to the removal of the survival-extending effect of extracellular acetate present in spent LB medium of that strain.
Individual deletion of several genes encoding proteins known to utilize acetyl-CoA as a substrate had no effect on the survival of the lipA strain (Figure S4E). Lack of Acs causes a major reduction in the rate of oxygen consumption of wt, whereas the lipA acs strain does not consume less oxygen than the lipA strain (Figure S5A). These observations are consistent with the explanation that the acetyl-CoA formed by the uptake of extracellular acetate is used by the TCA cycle producing reducing equivalents that are subsequently fed in the electron transport chain in the wt, but not in the lipA strain. Thus, conversion of extracellular acetate to intracellular acetyl-CoA by Acs is required for the extended lifespan of the lipA mutant, but our genetic analysis could not identify the downstream effect of the produced acetyl-CoA.
Next, we tested the individual contribution of the known acetate-producing proteins to the lifespan of the lipA strain. Lack of phosphotransacetylase has no effect on the survival (Figure S4E) and the extracellular acetate concentration (Figure S4F) of the lipA mutant. We therefore turned our attention to pyruvate oxidase (PoxB), a lipid-activated enzyme that converts pyruvate to acetate and carbon dioxide and in the process supplies electrons to the electron transport chain (Koland et al. 1984). In the absence of PoxB, the acetate concentration of the lipA strain is halved and its lifespan is reduced to that of wt (Figures 6A, F), while the poxB strain does not live shorter than wt (Figure 6G). The presence of the biological buffer AMPSO in the survival experiments shown in figure 6F maintains the pH at 9 despite any differences in the concentration of extracellular acetate among certain strains (data not shown); therefore, the observed effects on survival are not due to differences in the extracellular pH. Note, however, that lack of PoxB has no effect on the survival of the lipA mutant at pH 7.5 (Figure S4D).
Finally, we sought to determine whether acetate production by PoxB and acetate uptake by Acs function in the same pathway to extend the lifespan of the lipA mutant. The epistasis results shown in figure 6F support this hypothesis, since the lipA poxB acs strain has a similar lifespan as the lipA poxB strain. Prompted by the finding that the transcription of acs is reduced in the absence of poxB (Kumari et al. 2000) we measured the level of acs expression by RT PCR in the lipA and lipA poxB strains. Similar to published findings (Kumari et al. 2000), the expression of acs shows a 2.5-fold reduction in the lipA poxB mutant compared to the lipA mutant (Figure 6I). Since pyruvate oxidase is part of the electron transport chain and the lipA poxB mutant consumes less oxygen than the lipA strain (Figure 6H), we tested the contribution of PoxB to the level of ATP found in the lipA mutant, using a luciferase-based assay, but found no difference between the lipA and lipA poxB strains (Figure S5D). The results presented in this section demonstrate that pyruvate oxidase and acetyl-CoA synthetase function in the same pathway to extend the lifespan of the lipA mutant via the metabolism of acetate.
Discussion
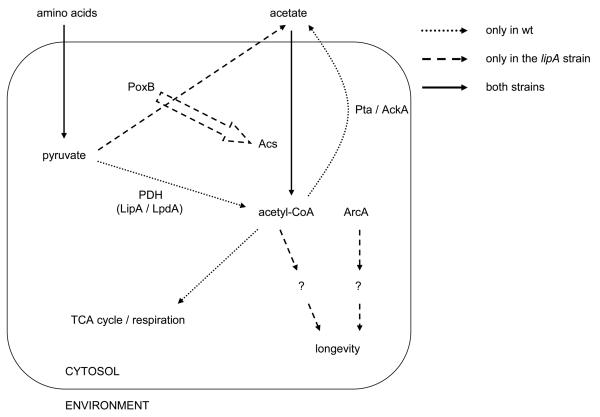
Our genome-wide screen for E. coli mutants with prolonged stationary phase survival revealed three strains, which live longer independently of the alkaline conditions that incubation in LB medium normally generates and also independently of reduced growth rates and saturation densities. Lack of subunit A of succinate dehydrogenase leads to increased stress resistance and extended lifespan, possibly linked to reduced superoxide generaiton. On the other hand, the extended lifespan of the longest-lived mutant, lipA, is entirely dependent on the conversion of pyruvate to acetyl-CoA via acetate by the pyruvate oxidase / acetyl-CoA synthetase (PoxB / Acs) enzyme pair. The hypoxia transcription factor ArcA contributes to the extended lifespan of the lipA strain, but it does so independently of acetate metabolism (Figure 7).
Figure 7.
Model for lifespan regulation by LipA in Escherichia coli. In the wt strain, pyruvate generated from the metabolism of amino acids present in LB medium is converted to acetyl-CoA by the PDH complex; acetyl-CoA is subsequently consumed by the TCA cycle or converted to acetate by Pta / AckA as the capacity of the TCA cycle to consume acetyl-CoA becomes limiting. In the lipA strain, the PDH complex is inactive and pyruvate is instead converted to acetate and carbon dioxide by PoxB. In both strains, extracellular acetate is taken up and converted to acetyl-CoA by Acs. The PoxB / Acs bypass of the PDH complex is required for lifespan extension in the lipA strain. The hypoxia-inducible transcription factor ArcA contributes to the extended lifespan of the lipA strain independently of the PoxB / Acs bypass. The block arrow denotes the activation of acs transcription by PoxB. PDH: Pyruvate dehydrogenase complex, Pta / AckA: phosphotransacetylase / acetate kinase, PoxB: pyruvate oxidase, Acs: acetyl-CoA synthetase, LipA: lipoic acid synthase, LpdA: lipoamide dehydrogenase.
Succinate dehydrogenase is a tetrameric protein complex catalyzing the conversion of succinate to fumarate in the TCA cycle coupled to electron transport to the ubiquinone pool (Yu & Yu 1980). The enzymatically active part of the complex, subunit A encoded by sdhA, is a well-established source of superoxide in the electron transport chain of E. coli through its covalently bound flavin cofactor, which is an efficient single-electron donor to molecular oxygen (Messner & Imlay 2002). We are showing that lack of this enzyme results in a reduced rate of superoxide production in early stationary phase which is accompanied by extended stationary phase survival (Figure 4). In fact, the accumulation of oxidatively damaged macromolecules in the form of protein carbonyls has been reported in stationary phase E. coli (Dukan & Nystrom 1998). More recently, these damaged proteins were shown to preferentially accumulate in cells that are about to lose viability and show lower expression of both cytosolic superoxide dismutases (Desnues et al. 2003). Mutants lacking proteins that provide defense against oxidative stress such as superoxide dismutases and catalases have reduced lifespan (Eisenstark et al. 1992) and incubation of stationary phase E. coli in the absence of oxygen results in a slower rate of viability loss (Conter et al. 2001). Observations of this kind implicate oxidative stress as a possible causative factor in the deterioration of stationary phase E. coli and are consistent with our finding of extended lifespan in the sdhA strain. We have previously shown that increased scavenging of superoxide extends the lifespan of S. cerevisiae (Fabrizio et al. 2003). However, the overexpression of both SOD1 and SOD2 caused a 30% life span extension versus the 3-fold extension observed in mutants lacking signal transduction genes (Fabrizio et al. 2003) in agreement with the longer lifespan of the lipA and lpdA mutants compared to that of the sdhA mutant reported in this study.
The adaptation of the metabolic physiology of E. coli to changes in oxygen availability mostly occurs at the level of gene expression through the action of the transcription factors Fnr and ArcA (Iuchi & Lin 1991). Experiments quantifying the transcriptional and functional changes elicited by varying oxygen tensions led to the current model that Fnr is activated under anaerobic conditions, whereas ArcA is active under microaerobic conditions (Levanon et al. 2005). ArcA suppresses the expression of TCA cycle genes such as citrate synthase (gltA), while activating the expression of genes required for energy generation under limited oxygen availability such as the cytochrome d terminal oxidase operon, cydAB (Lynch & Lin 1996). Furthermore, a microarray analysis of the response of E. coli to oxygen limitation also placed lpdA among the most down-regulated genes in response to the absence of oxygen (Salmon et al. 2005). We found that the lipA and lpdA mutants consume less oxygen than wt (Figure 3A) and that their fully extended lifespan is dependent on ArcA (Figure 5B). Taken together, these observations suggest that maximum lifespan extension in these mutants is dependent on physiological changes that are normally induced by hypoxic conditions in wt. Inactivation of the protein complexes LipA and LpdA participate in, individually or in combination, had no positive effect on survival (Figure S2F). It is therefore possible that lack of LipA or LpdA is specifically required to induce the ArcA-dependent physiological changes leading to extended survival and that inactivation of compnents of the PDH, 2-KGDH or glycine cleavage systems is not sufficient to induce these changes.
Nystrom et al. reported that the strain lacking ArcA survives poorly during glucose starvation-induced stationary phase (Nystrom et al. 1996). This transcription factor was shown to be required for the down-regulation of TCA cycle genes including sdhA and lpdA upon stationary phase entry (Nystrom et al. 1996). The arcA mutant also consumed more oxygen than its wt counterpart. The requirement of ArcA for survival under these conditions was attributed to the minimization of oxidative damage caused by unchecked respiration and possibly to the regulation of the utilization of endogenous carbon reserves, such as membrane lipids (Nystrom et al. 1996). We found ArcA to be required only for the extended survival of the lipA and lpdA mutants, but not for the survival of wt (Figures 5A, B). The difference between our results and those of Nystrom et al. is most likely due to the different medium used, minimal glucose in their study versus LB in ours. Similar to anaerobiosis, glucose, which is absent from LB medium, is well known to cause the suppression of enzymes of the TCA cycle (Halpern et al. 1964), since energy can be produced solely through the glycolytic Embden-Meyerhof-Parnas pathway. On the other hand, incubation of wt (and the sdhA mutant) in LB is expected to elicit the activation of the TCA cycle for the generation of energy using amino acids, whereas TCA cycle genes are down-regulated in the lipA and lpdA mutants due to the combined loss of the PDH and 2-KGDH complexes (Li et al. 2006). Hence, the results of that study are consistent with ours in showing that ArcA is required for survival under metabolic conditions that do not rely on the function of the TCA cycle for energy generation (wt in glucose minimal medium and lipA and lpdA strains in LB).
The transcriptional regulator hypoxia-inducible factor 1 (HIF-1) mediates changes in genes expression in response to hypoxia in organisms as diverse as humans and nematode worms (Semenza 2000; Shen et al. 2005) and it can therefore be considered as the functional homolog of ArcA, although the two proteins do not display significant sequence similarity. Three different groups recently reported the involvement of HIF-1 in the regulation of the lifespan of C. elegans. Mehta et al. reported that loss of VHL-1, the protein responsible for the degradation of HIF-1 under normoxic conditions, leads to extended lifespan (Mehta et al. 2009). Zhang et al. found that HIF-1 overexpression also leads to extended lifespan (Zhang et al. 2009). Lastly, Chen et al. showed that loss of HIF-1 causes lifespan extension under rich nutrient conditions, but failed to show lifespan extension under dietary restriction (Chen et al. 2009). These studies reveal complex nutrient-dependent interactions between HIF-1 and lifespan regulation in C. elegans and, along with our results, point towards the adaptive response to oxygen shortage as a novel, evolutionarily conserved mechanism of lifespan extension.
Oxygen shortage in E. coli results in the production of acetate (Phue et al. 2005). E. coli maintained in batch culture in LB undergo what has been described as the acetate switch, whereby the acetate initially produced by the culture is subsequently taken up and utilized by the TCA cycle for energy generation and biosynthesis (Wolfe 2005). The phosphotransacetylase / acetate kinase enzyme pair converts acetyl-CoA to acetate (Yang et al. 1999) and the excreted acetate is then taken up by acetyl-CoA synthetase (Kumari et al. 1995). Importantly, acetate metabolism has no effect on the survival of wt, since both the pta mutant, which makes no acetate (Hahm et al. 1994) and the acs mutant which cannot take up acetate (Kumari et al. 1995) survive as long as wt (Figures S5E and 6G respectively). The PDH complex, which converts pyruvate to acetyl-CoA in the wt strain is not functioning in the lipA mutant and as a consequence Pta is not involved in acetate production in this mutant (Figure S4F).
We are showing that this mutant bypasses the PDH complex through the function of the PoxB / Acs enzyme pair which converts pyruvate to acetyl-CoA via an acetate intermediate. Furthermore, this metabolic adaptation is fundamental for the extended lifespan of the lipA mutant (Figures 6B, F). Note that the PoxB / Acs bypass of the PDH complex has been previously described in growing cultures (Abdel-Hamid et al. 2001; Wolfe 2005). The much smaller magnitude of the survival-extending effect of extracellular acetate in the wt compared to the lipA strain (Figure 6D, E) might be due to the differential utilization of the molecule by the two strains; consumption by the TCA cycle, producing reducing equivalents used in the electron transport chain in the wt versus an unidentified, yet non-TCA cycle-dependent utilization in the lipA mutant.
Thus, although the metabolism of acetate in the lipA mutant is broadly similar to that of wt (initial production from pyruvate followed by assimilation by acetyl-CoA synthetase), the extended lifespan of the lipA mutant is dependent on the PoxB-Acs bypass of the PDH complex (Figure 6B, F), whereas neither the production of acetate by Pta-AckA nor its uptake by Acs affect the survival of wt (Figure 6G, S5C). The slower pace of acetate uptake in the lipA strain, as well as its non-utilization by the TCA cycle, are also likely factors favoring the lengthened survival of this mutant. Note that the elimination of extracellular acetate was recently shown to be partly responsible for dietary restriction-induced lifespan extension in wild-type S. cerevisiae (Burtner et al. 2009).
In conclusion, we found that mutants that genetically promote aspects of hypoxic metabolism extend the stationary phase survival of Escherichia coli, in an ArcA- and acetate-dependent manner. Our lab recently reported the metabolic switch from TCA cycle/respiration to glycolysis and glycerol production as a central component of the lifespan extension observed in the Saccharomyces cerevisiae Tor1Δ and Sch9Δ mutants (Wei et al. 2009). Such mechanisms of lifespan extension are expected to be dependent on a broad metabolic repertoire that confers the ability to grow and survive both in the presence and in the absence of oxygen. A metabolic model for lifespan extension in the nematode worm Caenorhabditis elegans, which cannot survive in the absence of oxygen, also invoked the reduced use of aerobic respiration in favor of fermentative malate dismutation, producing acetate and succinate, as a common metabolic adaptation of most long-lived mutants described for this species (Rea & Johnson 2003). Also, several studies recently implicated HIF-1, the functional homolog of ArcA found in metazoans in lifespan regulation in C. elegans (Chen et al. 2009; Mehta et al. 2009; Zhang et al. 2009). Thus, the metabolic alterations leading to extended stationary phase survival in the bacterium Escherichia coli might reveal an evolutionary conservation of lifespan-regulating mechanisms that is not purely phenomenological.
Experimental Procedures
Strains and genetic manipulations
The wild-type Escherichia coli strain BW25113 and its respective single-gene knock-outs were provided by the KEIO collection (Baba et al. 2006). Reference (Baba et al. 2006) also describes the pedigree of BW25113, a K-12 derivative. To create strains deleted for multiple genes, the kanamycin cassette was excised using FLP-mediated recombination, resulting in deletions carrying only a single FRT site (denoted, for example lipA::FRT in table S3), as described in (Datsenko & Wanner 2000), with the only difference that the non-selective incubation took place at 37°C and not at 43°C. Kanamycin alleles were transduced by bacteriophage P1 using standard techniques and the correct insertion was verified by PCR using primer K1 described in (Baba et al. 2006), along with a locus-specific primer annealing to a sequence upstream of the disrupted locus. Genomic sequence information was obtained from the “Profiling of the E. coli Chromosome” web site (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp). All strains used in the study are shown in table S3.
Survival experiments
LB medium consisted of 1% bacto tryptone, 0.5% yeast extract and 0.5% NaCl w/v. Cultures were inoculated 1:1000 using an overnight culture created by inoculating 2-3 colonies from an LB plate to 1 ml of LB. Cultures were grown in 3 ml's of LB in 16-mm diameter test tubes rotating orbitally at 220 RPM for 12 hours, at which point cell density was adjusted to ~1.5 × 109 CFU per ml by resuspending a pellet containing the desired number of cells in cell-free spent medium of the same strain for the long-lived strains with reduced saturation cell density. AMPSO or HEPES was added to 100 mM to achieve a stationary phase pH of 9 or 7.5 respectively. Due to the different stationary phase pH reached spontaneously by each strain, the pH of the buffers had to be adjusted accordingly for each strain. Spontaneous and adjusted pH was quantified using a pH electrode and pH test strips. All cultures were grown and maintained at 37°C and 70% relative humidity and colony-forming units (CFU) were enumerated over time by removing an aliquot, serially diluting in 0.5% NaCl, followed by colony enumeration after plating on LB plates that were incubated at 37°C. For the ‘high aeration’ experiment shown in figure S2E, cultures were grown and maintained in 10-ml volume in orbitally shaking 125-ml Erlenmeyer flasks. For experiments shown in figure S4D-E, medium collected after centrifugation of an early stationary phase (12 hours of incubation) wt culture was filter-sterilized by passing through a 0.22 μm filter and its pH adjusted to 9 by the addition of 100 mM AMPSO. Early stationary phase cultures of wt and the lipA mutant were centrifuged, washed once with 0.5% NaCl and the washed pellet was then resuspended in 30 μl of 0.5% NaCl and transferred to the conditioned medium prepared as above. For survival experiments in 0.5% NaCl, the same washing procedure was followed and cells were then resuspended in 2 ml 0.5% NaCl to which appropriately buffered AMPSO was added to 100 mM.
Phase contrast microscopy
Cultures of wt and the lipA strain were processed as described above with AMPSO. 1 hour after the AMPSO processing, cultures were washed once with 0.5% NaCl and 5 μl were spread onto a glass slide and air-dried. The dried spots were incubated for 5′ with 100% methanol to fix the cells and then washed 3 times with phosphate buffered saline, pH 7.4. Images were obtained under 100x magnification and cell length measured using the ImageJ software.
Stress resistance
For the heat shock experiment, cultures were processed as described above with AMPSO, returned to the incubator and 4 hours later (16 hours after inoculation) were subjected to either a 4-minute incubation in a 55°C waterbath without shaking and CFU enumerated before and after the treatment.
For the paraquat experiment, cultures were processed as described above with AMPSO and paraquat (methyl viologen dichloride hydrate obtained from Sigma-Aldrich) was added to 500 μM at the time of processing (12 hours after inoculation). CFU were enumerated before and after a 12-hour incubation at 37°C.
Oxygen consumption and acetate quantification
Oxygen consumption measurements were performed with 2 ml of culture stirred by a magnetic stir bar in a 37°C waterbath using a Clark-type electrode. Conversion to nanomoles of oxygen consumed was done by assuming that the liquid culture contains the same amount of oxygen as water equilibrated with 21% oxygen in 1 atmosphere pressure, which is 5.02 μl/ml (manufacturer's manual) and was further normalized by the number of CFU present. Cyanide-resistant respiration was measured after a 5-minute incubation in the 37°C water bath with 1 mM sodium cyanide. Data were recorded until a straight line trace was obtained indicating that a steady state of oxygen consumption had been reached. Extracellular acetate concentration was quantified on cell-free samples obtained by centrifugation using the R-Biopharm acetic acid kit (catalog number 10148261035) according to the manufacturer's instructions.
acs mRNA quantification
1-ml aliquots of late stationary phase cultures (24 hours after inoculation) were added to a 95μl ethanol + 5μl water-equilibrated acidic phenol mixture and rapidly centrifuged for 45 seconds at 4°C. RNA was subsequently extracted using the MasterPure complete DNA and RNA purification kit (Epicentre Biotechnologies, catalog number MC85200) according to the manufacturer's instructions. 3 μg of RNA were used per reverse transcription reaction using Superscript III reverse transcriptase (Invitrogen) and random hexamers as primers according to the manufacturer's instructions. 50 ng of reverse-trasncribed RNA were then used as substrate for real-time PCR. The expression level of three different housekeeping genes was measured (rpoA, frr and dnaA); dnaA was found to be highly expressed at similar levels in both the lipA and lipA poxB strains and was therefore used for normalization of the values obtained for acs. Standard curves were constructed for each assayed transcript and used for quantification.
Mutation frequency measurement
Cultures were processed as described above with HEPES (pH 7.5), with the only difference that cultures were maintained in a volume of 25 ml in 125-ml Erlenmeyer flasks. Rifampicin-resistant mutants were quantified by plating on LB plates containing the antibiotic at 120 μg/ml after washing once with 0.5% NaCl. The colonies grown in rifampicin were enumerated and normalized by the total number of CFU at each time-point. Note that rifampicin plates were checked for 3 days after plating and late-appearing colonies (the rifampicin resistance of which was confirmed) were counted.
Supplementary Material
Schematic representation of the absorbance-based, genome-wide screen for extended stationary phase survival in E. coli.
Figure S2, related to figure 1. (A) to (C) CFU titers of experiments shown in figures 1A to 1C. (D) Growth curves as CFU counts of wt and the long-lived strains. (E) Survival of 10-ml cultures of wt and the lipA strain in 125-ml flasks. (F) Survival of the shown strains.
Figure S3, related to figure 4. (A) Effect of the overexpression of SodB (pHS1-7) on the survival of wt (pBR322 empty vector). (B) Time-dependent frequency of rifampicin-resistant mutants in wt and the sdhA strain. (C) Growth curves as CFU counts of the shown strains. (D) Stationary phase survival of all strains included in the mutation frequency experiment.
Figure S4, related to figure 6. (A) Extracellular acetate concentration at late stationary phase of the shown mutants. (B) Effect of sodium chloride or sodium acetate added to 6 mM at the time-point designated by the arrow on the survival of the lipA mutant. (C,D) Stationary phase survival of the shown mutants at pH 7.5. (E) Stationary phase survival of the shown mutants at pH 9. (F) Extracellular acetate concentration of the shown mutants over time. (G, H) Expression of pta and ackA in wt and the lipA strain over time.
(A) Rate of oxygen consumption of the shown mutants at early stationary phase. (B) ATP levels of the shown strains, measured using firefly luciferase. (C) Survival of the wt and pta strains.
Acknowledgements
This work was funded by NIH grant to VDL. Stavros Gonidakis was partially supported by the Myronis fellowship from the Graduate School of the University of Southern California and the department of Integrative and Evolutionary Biology of the University of Southern California.
References
- Abdel-Hamid AM, Attwood MM, Guest JR. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology. 2001;147:1483–1498. doi: 10.1099/00221287-147-6-1483. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baev MV, Baev D, Radek AJ, Campbell JW. Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of sugars, alcohols, and organic acids with transcriptional microarrays. Appl Microbiol Biotechnol. 2006;71:310–316. doi: 10.1007/s00253-006-0317-6. [DOI] [PubMed] [Google Scholar]
- Benov L, Fridovich I. The rate of adaptive mutagenesis in Escherichia coli is enhanced by oxygen (superoxide) Mutat Res. 1996;357:231–236. doi: 10.1016/0027-5107(96)00128-5. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? Embo J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang EC, Kosman DJ. Intracellular Mn (II)-associated superoxide scavenging activity protects Cu,Zn superoxide dismutase-deficient Saccharomyces cerevisiae against dioxygen stress. J Biol Chem. 1989;264:12172–12178. [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DP. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;5:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Conter A, Gangneux C, Suzanne M, Gutierrez C. Survival of Escherichia coli during long-term starvation: effects of aeration, NaCl, and the rpoS and osmC gene products. Res Microbiol. 2001;152:17–26. doi: 10.1016/s0923-2508(00)01164-5. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues B, Cuny C, Gregori G, Dukan S, Aguilaniu H, Nystrom T. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 2003;4:400–404. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukan S, Nystrom T. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 1998;12:3431–3441. doi: 10.1101/gad.12.21.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstark A, Miller C, Jones J, Leven S. Escherichia coli genes involved in cell survival during dormancy: role of oxidative stress. Biochem Biophys Res Commun. 1992;188:1054–1059. doi: 10.1016/0006-291x(92)91338-q. [DOI] [PubMed] [Google Scholar]
- Ericsson M, Hanstorp D, Hagberg P, Enger J, Nystrom T. Sorting out bacterial viability with optical tweezers. J Bacteriol. 2000;182:5551–5555. doi: 10.1128/jb.182.19.5551-5555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Liou LL, Moy VN, Diaspro A, Valentine JS, Gralla EB, Longo VD. SOD2 functions downstream of Sch9 to extend longevity in yeast. Genetics. 2003;163:35–46. doi: 10.1093/genetics/163.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Farrell MJ, Finkel SE. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol. 2003;185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Fontaine F, Stewart EJ, Lindner AB, Taddei F. Mutations in two global regulators lower individual mortality in Escherichia coli. Mol Microbiol. 2008;67:2–14. doi: 10.1111/j.1365-2958.2007.05988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, Diep A, Hu K, Iverson A, Yang H, Miller JH. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair (Amst) 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin EC. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- Groat RG, Schultz JE, Zychlinsky E, Bockman A, Matin A. Starvation proteins in Escherichia coli: kinetics of synthesis and role in starvation survival. J Bacteriol. 1986;168:486–493. doi: 10.1128/jb.168.2.486-493.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm DH, Pan J, Rhee JS. Characterization and evaluation of a pta (phosphotransacetylase) negative mutant of Escherichia coli HB101 as production host of foreign lipase. Appl Microbiol Biotechnol. 1994;42:100–107. doi: 10.1007/BF00170231. [DOI] [PubMed] [Google Scholar]
- Halpern YS, Even-Shoshan A, Artman M. Effect of Glucose on the Utilization of Succinate and the Activity of Tricarboxylic Acid-Cycle Enzymes in Escherichia Coli. Biochim Biophys Acta. 1964;93:228–236. doi: 10.1016/0304-4165(64)90370-8. [DOI] [PubMed] [Google Scholar]
- Hassan HM, Fridovich I. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J Biol Chem. 1977;252:7667–7672. [PubMed] [Google Scholar]
- Hengge-Aronis R. Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol. 2002;4:341–346. [PubMed] [Google Scholar]
- Iuchi S, Lin EC. Adaptation of Escherichia coli to respiratory conditions: regulation of gene expression. Cell. 1991;66:5–7. doi: 10.1016/0092-8674(91)90130-q. [DOI] [PubMed] [Google Scholar]
- Jones RW, Garland PB. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1977;164:199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano H, Hirokawa Y, Mori H. Long-term survival of Escherichia coli lacking the HipBA toxin-antitoxin system during prolonged cultivation. Biosci Biotechnol Biochem. 2009;73:117–123. doi: 10.1271/bbb.80531. [DOI] [PubMed] [Google Scholar]
- Koland JG, Miller MJ, Gennis RB. Reconstitution of the membrane-bound, ubiquinone-dependent pyruvate oxidase respiratory chain of Escherichia coli with the cytochrome d terminal oxidase. Biochemistry. 1984;23:445–453. doi: 10.1021/bi00298a008. [DOI] [PubMed] [Google Scholar]
- Kumari S, Beatty CM, Browning DF, Busby SJ, Simel EJ, Hovel-Miner G, Wolfe AJ. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–4179. doi: 10.1128/jb.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon O, Georgellis D, Lin EC. Rotational on-off switching of a hybrid membrane sensor kinase Tar-ArcB in Escherichia coli. J Biol Chem. 2003;278:13192–13195. doi: 10.1074/jbc.M210647200. [DOI] [PubMed] [Google Scholar]
- Levanon SS, San KY, Bennett GN. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng. 2005;89:556–564. doi: 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- Li M, Ho PY, Yao S, Shimizu K. Effect of lpdA gene knockout on the metabolism in Escherichia coli based on enzyme activities, intracellular metabolite concentrations and metabolic flux analysis by 13C-labeling experiments. J Biotechnol. 2006;122:254–266. doi: 10.1016/j.jbiotec.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maklashina E, Berthold DA, Cecchini G. Anaerobic expression of Escherichia coli succinate dehydrogenase: functional replacement of fumarate reductase in the respiratory chain during anaerobic growth. J Bacteriol. 1998;180:5989–5996. doi: 10.1128/jb.180.22.5989-5996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M. Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science. 2009;324:1196–1198. doi: 10.1126/science.1173507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- Miller JR, Busby RW, Jordan SW, Cheek J, Henshaw TF, Ashley GW, Broderick JB, Cronan JE, Jr., Marletta MA. Escherichia coli LipA is a lipoyl synthase: in vitro biosynthesis of lipoylated pyruvate dehydrogenase complex from octanoyl-acyl carrier protein. Biochemistry. 2000;39:15166–15178. doi: 10.1021/bi002060n. [DOI] [PubMed] [Google Scholar]
- Miller RA. Cell stress and aging: new emphasis on multiplex resistance mechanisms. J Gerontol A Biol Sci Med Sci. 2009;64:179–182. doi: 10.1093/gerona/gln072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom T, Larsson C, Gustafsson L. Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. Embo J. 1996;15:3219–3228. [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Barton NH. Optimality, mutation and the evolution of ageing. Nature. 1993;362:305–311. doi: 10.1038/362305a0. [DOI] [PubMed] [Google Scholar]
- Phue JN, Noronha SB, Hattacharyya R, Wolfe AJ, Shiloach J. Glucose metabolism at high density growth of E. coli B and E. coli K: differences in metabolic pathways are responsible for efficient glucose utilization in E. coli B as determined by microarrays and Northern blot analyses. Biotechnol Bioeng. 2005;90:805–820. doi: 10.1002/bit.20478. [DOI] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss BM, Nelms JM, Park C, Wolfe AJ. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Johnson TE. A metabolic model for life span determination in Caenorhabditis elegans. Dev Cell. 2003;5:197–203. doi: 10.1016/s1534-5807(03)00242-9. [DOI] [PubMed] [Google Scholar]
- Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005;280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88:1474–1480. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the HIF-1 hypoxia-inducible factor during hypoxia response in Caenorhabditis elegans. J Biol Chem. 2005;280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- Smith MW, Neidhardt FC. 2-Oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J Bacteriol. 1983a;156:81–88. doi: 10.1128/jb.156.1.81-88.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Neidhardt FC. Proteins induced by aerobiosis in Escherichia coli. J Bacteriol. 1983b;154:344–350. doi: 10.1128/jb.154.1.344-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati D. Cloning and mapping of the manganese superoxide dismutase gene (sodA) of Escherichia coli K-12. J Bacteriol. 1983;155:1078–1087. doi: 10.1128/jb.155.3.1078-1087.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Boom TJ, Reed KE, Cronan JE., Jr. Lipoic acid metabolism in Escherichia coli: isolation of null mutants defective in lipoic acid biosynthesis, molecular cloning and characterization of the E. coli lip locus, and identification of the lipoylated protein of the glycine cleavage system. J Bacteriol. 1991;173:6411–6420. doi: 10.1128/jb.173.20.6411-6420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Eiteman MA, Altman E. Increased recombinant protein production in Escherichia coli strains with overexpressed water-forming NADH oxidase and a deleted ArcA regulatory protein. Biotechnol Bioeng. 2006;94:538–542. doi: 10.1002/bit.20853. [DOI] [PubMed] [Google Scholar]
- Visick JE, Cai H, Clarke S. The L-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol. 1998;180:2623–2629. doi: 10.1128/jb.180.10.2623-2629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulic M, Kolter R. Alcohol-induced delay of viability loss in stationary-phase cultures of Escherichia coli. J Bacteriol. 2002;184:2898–2905. doi: 10.1128/JB.184.11.2898-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Fabrizio P, Madia F, Hu J, Ge H, Li LM, Longo VD. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YT, Bennett GN, San KY. Effect of inactivation of nuo and ackA-pta on redistribution of metabolic fluxes in Escherichia coli. Biotechnol Bioeng. 1999;65:291–297. [PubMed] [Google Scholar]
- Yu L, Yu CA. Interaction between succinate dehydrogenase and ubiquinone-binding protein from succinate-ubiquinone reductase. Biochim Biophys Acta. 1980;593:24–38. doi: 10.1016/0005-2728(80)90005-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348. doi: 10.1371/journal.pone.0006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the absorbance-based, genome-wide screen for extended stationary phase survival in E. coli.
Figure S2, related to figure 1. (A) to (C) CFU titers of experiments shown in figures 1A to 1C. (D) Growth curves as CFU counts of wt and the long-lived strains. (E) Survival of 10-ml cultures of wt and the lipA strain in 125-ml flasks. (F) Survival of the shown strains.
Figure S3, related to figure 4. (A) Effect of the overexpression of SodB (pHS1-7) on the survival of wt (pBR322 empty vector). (B) Time-dependent frequency of rifampicin-resistant mutants in wt and the sdhA strain. (C) Growth curves as CFU counts of the shown strains. (D) Stationary phase survival of all strains included in the mutation frequency experiment.
Figure S4, related to figure 6. (A) Extracellular acetate concentration at late stationary phase of the shown mutants. (B) Effect of sodium chloride or sodium acetate added to 6 mM at the time-point designated by the arrow on the survival of the lipA mutant. (C,D) Stationary phase survival of the shown mutants at pH 7.5. (E) Stationary phase survival of the shown mutants at pH 9. (F) Extracellular acetate concentration of the shown mutants over time. (G, H) Expression of pta and ackA in wt and the lipA strain over time.
(A) Rate of oxygen consumption of the shown mutants at early stationary phase. (B) ATP levels of the shown strains, measured using firefly luciferase. (C) Survival of the wt and pta strains.