Abstract
The term ‘transcriptional interference’ (TI) is widely used but poorly defined in the literature. There are a variety of methods by which one can interfere with the process or the product of transcription but the term TI usually refers to the direct negative impact of one transcriptional activity on a second transcriptional activity in cis. Two recent studies, one examining Saccharomyces cerevisiae and the other Escherichia coli, clearly show TI at one promoter caused by the arrival of a transcribing complex initiating at a distant promoter. TI is potentially widespread throughout biology; therefore, it is timely to assess exactly its nature, significance and operative mechanisms. In this article, we will address the following questions: what is TI, how important and widespread is it, how does it work and where should we focus our future research efforts?
What is transcriptional interference?
In this article, we wish to define transcriptional interference (TI) specifically as the suppressive influence of one transcriptional process, directly and in cis on a second transcriptional process. Our definition of TI (see Glossary) excludes the kind of interference that results from the following: (i) the binding of a repressor to its operator overlapping a promoter [1]; (ii) promoter modification, such as methylation [2]; (iii) hindering the progress of an elongating RNA polymerase (RNAP) by DNA-bound obstacles (other than a second RNAP) [3]; (iv) the inactivation of RNAP by RNA regulators [4]; (v) the insulation of an enhancer site [5]; and (vi) RNA interference (RNAi) in which the product of one transcriptional unit interferes with the half-life of the product of a second transcriptional unit [6]. We exclude from our definition of TI examples whereby transcription interferes directly with a cellular activity rather than with transcription associated with cellular activity (e.g. the interference with chromosome replication in Saccharomyces cerevisiae as a result of transcription across its site of initiation [7]). We also exclude those cases of ‘negative interference’ whereby one transcriptional process, directly and in cis, enhances rather than suppresses a second transcriptional process, such as the fortuitous positioning of a gene within an active chromatin domain [8], chromatin remodelling that promotes intergenic transcription [9] and transcriptional coupling in which a promoter is activated by the activity of an upstream divergent promoter [10].
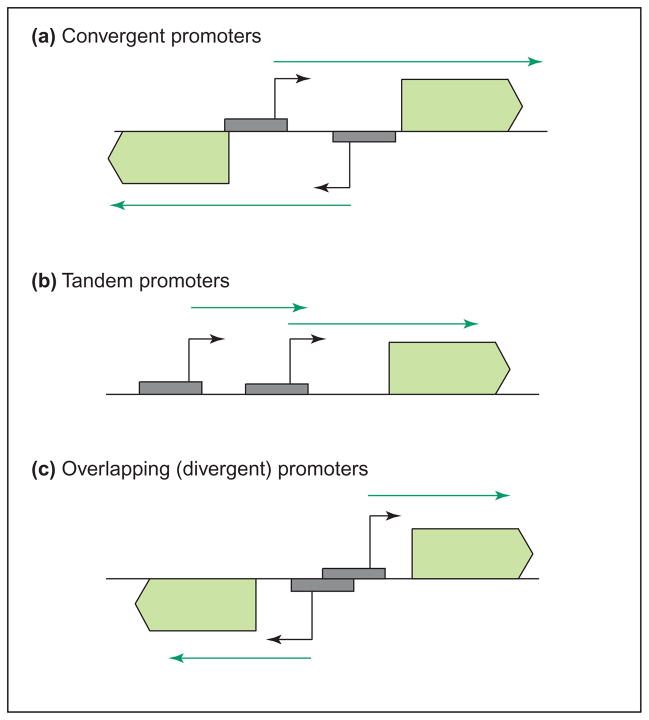
TI is often asymmetric and results from the existence of two promoters, the strong (aggressive) promoter reducing the expression of the weak (sensitive) promoter (Figure 1). These promoters can be either: (i) convergent promoters directing converging transcripts that overlap for at least part of their sequence (Figure 1a); (ii) tandem promoters, one upstream of the other but transcribing in the same direction, with their transcripts possibly but not necessarily overlapping (Figure 1b); or (iii) overlapping promoters, either divergent, convergent or tandem, in which the two RNAP-binding sites share at least a common DNA sequence (Figure 1c).
Figure 1.
Promoter arrangements that can lead to transcriptional interference (TI). TI can arise as a result of several different promoter arrangements: (a) convergent promoters, such as the coliphage 186 lytic and lysogenic promoters [20]; (b) tandem promoters, for example, the promoters of yeast SRG1 and SER3 [21]; (c) overlapping promoters, such as the E. coli aroP P1 and P3 promoters [22]. Although the example of overlapping promoters shown in (c) shows a divergent promoter pair, both tandem and convergent promoter pairs can also overlap. RNAP-binding sites are indicated by grey boxes, the starting points (+1) of transcription are shown as black arrows and transcripts as green arrows.
What is the importance of transcriptional interference?
In a genome, interfering promoters can exist naturally either as an integral part of a genetic network or as a reflection of the arrival of a transposable element. Alternatively, they can exist as a result of either an intended experimental manipulation (e.g. in the studies of Proudfoot [11–13], Eszterhas et al. [14], and Padidam and Cao [15]) or an unintended manipulation (e.g. the insertion of foreign cloned DNA [16–18]). We first turn our attention to the importance of the natural occurrence of TI.
In a genetic network, it appears that TI provides a platform for gene regulation. This can be illustrated by the following examples.
Convergent promoters: in the lysis–lysogeny switch of the temperate coliphage 186, the strong lytic promoter reduces the activity of the weaker convergent lysogenic promoter (which lies 62-bp downstream) by 5.6 times [19,20] (Box 1). The repression of the strong lytic promoter by the immunity repressor prevents the lytic promoter interfering with the convergent lysogenic promoter and so provides the positive autoregulation important in maintaining a stable lysogenic state [19].
Tandem promoters: during growth of S. cerevisiae in a rich medium, the upstream SRG1 promoter is active and its activity interferes with the tandem downstream promoter of the serine biosynthetic gene SER3, thereby blocking the unnecessary biosynthesis of serine [21]. The presumption is that in a nutritionally poor medium, repression of the upstream SRG1 promoter by an as yet uncharacterized mechanism relieves its interference of the promoter of SER3, enabling SER3 expression and synthesis of serine (Box 1).
Overlapping promoters: in Escherichia coli, the P1 promoter of aroP, the gene encoding the permease responsible for the general import of aromatic amino acids, is overlapped by the divergent promoter P3. RNAP binds to P3 in the presence of the aromatic amino acid bound form of the TyrR protein, thus reducing the activity of P1 [22]. This system provides a feedback mechanism whereby the intracellular accumulation of any (or all) of the aromatic amino acids blocks their transport into the cell from the extracellular medium by this permease.
Box 1. Recent advances in transcriptional interference.
Two recent papers underpin the timeliness of a review on TI.
Callen et al. [20], in studies of the lysis–lysogeny switch in the temperate coliphage 186, used a single copy LacZ-reporter system demonstrate a 5.6-times reduction in the activity of the weaker lysogenic promoter by the activity of the stronger convergent lytic promoter, which lies 62-bp downstream. The two promoters were variously rearranged to test different models of interference, and mechanisms involving RNA–RNA hybridisation and promoter competition were excluded. Terminating transcription from the strong promoter before it reached the sensitive promoter dramatically reduced interference, indicating a requirement for the passage of a converging RNAP across the sensitive promoter. Based on in vitro experiments showing a slow rate of escape for open complexes at the sensitive promoter, and their sensitivity to ‘head-on’ collisions with an elongating RNAP, a ‘sitting duck’ model of interference was proposed (see Figure 2b) and supported with in vivo permanganate footprinting. The authors concluded that interference was not caused by occlusion or collision between elongating complexes, and supported their model by the analysis of a different set of prokaryotic convergent promoters that showed a lower level of interference of a sensitive promoter known to have a reduced ability to form sitting ducks.
Martens et al. [21], in studies with Saccharomyces cerevisiae, used chromatin immuno-precipitation assays and transcription run-on experiments to determine the presence of transcription upstream of the SER3 promoter under repressive conditions. Using northern analysis and primer-extension analysis, they determined the existence of an in-tandem transcript, initiating 480-bp upstream of the SER3 TATA element and terminating in its vicinity. Mutational inactivation of the associated TATA element eliminated repression of the SER3 promoter and they named this non-coding sequence the SER3 regulatory gene 1 (SRG1). The inhibition was cis-limited and therefore not RNA-mediated. Premature termination of this transcript before the SER3 upstream activating sequence (UAS) de-repressed the SER3 promoter, eliminating promoter competition as the cause of inhibition. When they replaced the SER3 UAS with a known activator site they obtained activation of the SER3 promoter that remained sensitive to repression by SRG1. They concluded that transcription of SRG1 interfered with binding of the activator(s) for transcription from the SER3 promoter, but did not discriminate between preventing the activator from binding by occlusion or ejecting the bound activators by the sitting duck mechanism.
In higher eukaryotes, TI has been demonstrated with artificial promoter arrangements [14,18], but examples also exist of gene regulation by TI for genes within their natural chromosomal context. Thus, the N-ras gene, which belongs to a family of small GTPases, is down-regulated by transcription of the unrelated upstream unr gene [23]. In the mouse, deletion of the promoter of unr leads to elevated transcription of N-ras and embryonic lethality for the homozygous mutant. An analogous situation applies to the defect in expression of the human α-globin genes in α-thalassemia, as a result of mutational read-through from the upstream promoter of the α2-globin gene [24].
In addition to these examples, TI is exploited in the regulation of some cellular activities, such as the initiation of plasmid replication [25,26]. There is also the potential of TI to: (i) contribute to the overall interference accorded to RNAi, caused by the presence of in cis intergenic convergent promoters [13,27,28]; and (ii) provide a different gene product, by alternative gene splicing from the extended transcript, following the interference of a downstream promoter by the activity of an upstream in-tandem promoter [29].
The importance of TI associated with transposable elements results from the transcription of flanking DNA from a promoter within the transposable element, as illustrated in the pioneering work of McClintock [30], in which she determined that the basis of variation in kernel pigmentation in maize was due to the in cis transcriptional activity of ‘controlling (transposable) elements’. Based on this potential for disturbing the expression of neighbouring genes, Whitelaw et al. [31,32] proposed a role for retrotransposons as epigenetic mediators of phenotypic variation in mammals. This is consistent with the abundance of retrotransposons in mammalian genomes – Smit [33] estimates that retroelements form >40% of a mammalian genome. In a similar vein, Nigumann et al. [34] recognized the potential of long interspersed nuclear elements (LINES) for ‘epigenetic control of different cellular genes’ in the human genome, based on the frequent transcription of neighbouring DNA from the antisense promoter located in the 5′ untranslated region (UTR) of the human L1 retrotransposon.
How widespread is transcriptional interference?
Nasser et al. [35] have suggested that the use of promoter interactions as a mechanism of gene regulation co-evolved with factor-dependent regulation or that there was a primordial RNA-polymerase-dependent homeostatic regulation and an extra level of control by transcription factors has evolved during the course of evolution. Examples of TI as a means of gene regulation are common in the genomes of ‘extrachromosomal’ elements such as bacteriophages [19,36–38], transposable elements [39], insertion sequences [40] and plasmids [25,26]. Perhaps the highly compact nature of these genomes favours the evolution of convergent transcription to regulate gene expression. In addition, natural examples of TI have been described in bacteria [22,35,41,42] and yeast [21,43,44], but less frequently for higher eukaryotes [23,24,34,45].
Prediction of the transcription units and promoters of E. coli is relatively straightforward, enabling an estimate of the occurrence of promoter arrangements that are likely to give rise to TI. Analysis of the 4462 known or predicted E. coli promoters in the RegulonDB database (v.4) [46], reveals 166 non-overlapping tandem promoters pairs (with starts sites that are >70-bp apart) and 54 non-overlapping convergent promoter pairs with starts sites >40-bp and <200-bp apart [47]. In addition, there are 54 convergent, 89 divergent and 292 tandem promoters whose start sites are separated by <40 bp and so could be classified as overlapping promoters [25]. Thus, in E. coli there is much potential for interference because of the tandem, convergent or overlapping arrangement of promoters. In addition, transcriptome analysis using oligonucleotide tiling arrays [48] of E. coli identified a set of 1102 transcripts in the intergenic regions of the genome, 334 of which could not be classified as either part of operon elements, 5′UTRs or 3′UTRs. Many of these transcripts might be antisense non-coding RNAs produced as a result of convergent transcription.
The highly compact, gene-rich genome of S. cerevisiae contains several adjacent and closely spaced gene sets with a bias towards divergent transcription suggestive of evolutionary pressure against convergent transcription [13,49]. The difficulty of reliably predicting promoter sequences of eukaryotes in silico [50] has perhaps led to an underestimation of the frequency of promoter arrangements that are likely to give rise to TI. However, the recent wave of transcriptome analyses hints that TI might be a relatively widespread form of gene regulation, in addition to conventional factor-dependent regulation [51]. For example, measurement of transcript levels in yeast has enabled calculation of the degree of correlation (either positive or negative) of expression levels of adjacent gene pairs [52]. A subset of those with a negative correlation of transcription levels might represent examples of genes regulated by TI. In higher eukaryotes, large intron size allows for nested genes (i.e. genes that reside within the intronic regions of other genes [53]), providing the potential for TI as a result of either tandem or convergent transcription. Analysis of the sequence of human chromosomes 20, 21 and 22 [54] revealed the existence of overlapping sequences for 182 genes, of which 106 were arranged convergently (sharing either their 5′ or 3′ regions), 35 were arranged in tandem and 41 were examples of a gene within a gene (either tandem or convergent). Thus there is great potential for TI.
Transcriptome analysis of several organisms, including Arabidopsis [55], Drosophila [56], mouse [57] and human [58–60] all reveal elevated levels of non-coding RNAs resulting from intergenic transcription. For example, analysis of the Drosophila transcriptome [56] indicates that non-coding RNAs are abundant and developmentally regulated, with 41% of the non-coding regions of the genome being expressed. In addition, in each of these genomes there are numerous sense–antisense pairs of transcripts [27]. In the mouse, for example, 2431 sense–antisense pairs have been identified [57], suggesting the potential for TI as a significant mechanism of gene regulation.
How does transcriptional interference work?
Although reports of TI in both prokaryotes and eukaryotes have been published over many years, few studies have addressed the underlying mechanisms. The earlier publications that have influenced our understanding of the mechanisms involved are in vivo studies on convergent promoters by Ward and Murray [61], an in vitro study on promoters arranged both convergently and in-tandem by Horowitz and Platt [62] and, in particular, the in vivo study of a convergent promoter pair by Elledge and Davis [63]. In eukaryotes, pioneering in vivo work has been performed by the Proudfoot laboratory [11] on promoter pairs that are arranged tandemly.
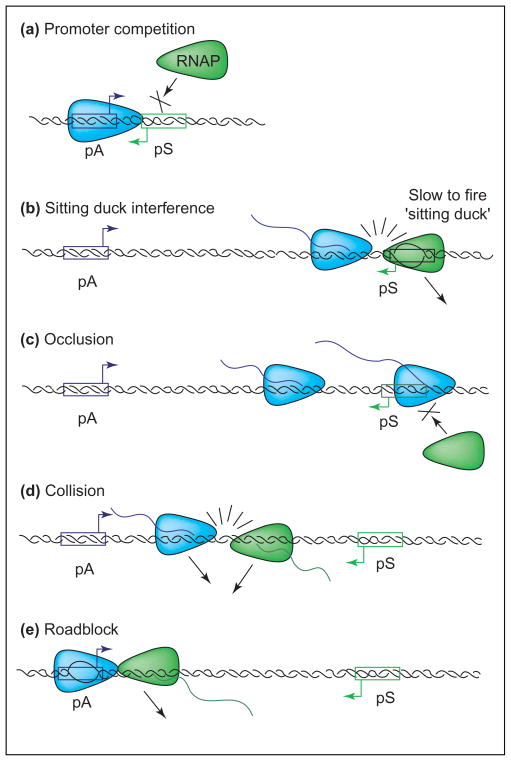
Of the mechanisms of TI that we will outline (Figure 2), three are implemented at the initiation phase of transcription and two at the elongation stage.
Figure 2.
Mechanisms of transcriptional interference (TI). Five possible mechanisms by which TI can occur are: (a) promoter competition; (b) sitting duck interference; (c) occlusion; (d) collision; and (e) roadblock. For the example shown here where a strong (aggressive) promoter pA is oriented convergently to a weak (sensitive) promoter pS, all five mechanisms are possible. For promoters arranged in tandem, all mechanisms except the collision mechanism [shown in (d)] can apply. When the promoters are arranged divergently, only the promoter competition mechanism shown in (a) can apply, but only when the promoters are also overlapping.
Promoter competition
TI arises because the occupation of one promoter by RNAP precludes its occupation of the second promoter. Thus, restricting RNAP-binding to one promoter enhances the activity of the second (Figure 2a). In the overlapping promoter example described earlier, Wang et al. [22], used diagnostic DNaseI-footprint analysis to show that RNAP occupation of the promoter P3 precluded RNAP occupation of P1, the promoter of aroP.
Promoter competition is also used to describe TI in eukaryotes in which two promoters share the same enhancer site: the transcription factors associated with the enhancer, while interacting with the first promoter, are precluded from interacting with the second promoter [43,64]. A duplication of that enhancer site eliminates the interference [65].
Sitting duck mechanism
If the RNAP complex at the sensitive promoter is slow to transit from the open complex to the elongation complex (i.e. it is slow to ‘fire’) it can be considered a ‘sitting duck’, available to be ‘hit’ and dislodged by the arrival of an elongation complex originating from a strong convergent (or tandem) promoter (Figure 2b). The sitting duck mechanism was proposed by Callen et al. [20] to explain the observation in coliphage 186 that interference of the weak lysogenic promoter by the strong lytic promoter disappeared if transcription from the aggressive promoter was terminated before it reached the sitting duck complex at the sensitive promoter. Although their studies involved prokaryotic promoters, the concept could equally well apply in eukaryotic systems, should the transcribing RNA polymerase remove from the DNA either the initiation complex bound at the sensitive promoter or transcription factors associated with it. This is discussed in the following section.
Mathematical modelling [47] of TI indicates that sitting duck interference increases as the ratio of the strength of the aggressive to the sensitive promoter increases. Interference as a result of the sitting duck mechanism is maximal when the rate of formation of the initiation complex at the sensitive promoter is equal to its rate of firing. Should the ratio of these rates be different from 1, then sitting duck interference is reduced; if it is <1, then the roost of the sitting duck is short and less likely to be hit, and if it is >1 then any sitting duck dislodged is rapidly replaced by another. Sitting duck interference is the dominant mechanism when the two promoters are close together (but not overlapping), because under these circumstances collisions between elongating polymerases make a minimal contribution to interference.
Occlusion
This term was first introduced by Adhya and Gottesman [41] to explain the TI caused by the limitation in time available for an RNAP to bind to a promoter as a result of its transient occupation by the passage of an elongating RNAP initiating at a tandem promoter (Figure 2c). However, unless the upstream promoter is very strong, calculations suggest that interference as a result of occlusion is minor. Thus, for E. coli RNAP the transit time across a standard promoter is 2–3 s, and therefore this type of interference, even by a strong promoter such as phage lambda pL, which fires once every 4.5 s [66], would only reduce the activity of the sensitive promoter by about half. The thirty-fold interference originally observed was probably due to sitting duck interference, unless the transit time across the sensitive promoter was extended as a result of pausing. The extent of interference caused by occlusion thus depends on the size of the sensitive promoter, the strength (rate of firing) of the aggressive promoter and the speed of transcription across the sensitive promoter. It applies to both convergent and tandem promoters.
In several eukaryotic studies [14,67], TI attributed to the loss of transcription-factor binding as a result of transcriptional activity from an interfering promoter has been inappropriately termed occlusion. However, as recognized by Bateman and Paule [68] and others [23], the same experimental result would be seen whether the absence of a bound transcription factor reflected the prevention of binding (occlusion) or dislodgement (sitting duck), and we suggest a distinction be maintained. Because transcription-factor-binding is part of the transcriptional process, interference with this binding is captured within our definition of TI, which we purposely avoided restricting to RNAP–RNAP interactions. Similar to prokaryotic systems, the likelihood of interference by occlusion in eukaryotic systems will depend on the frequency with which the interfering polymerase transcribes over the site, the rate of transcription and the size of the site. Again, similar to prokaryotic systems, we suspect that occlusion will make a significant contribution to TI only for a strong aggressive promoter.
Collisions
Collisions between converging elongation complexes lead to the premature termination of the transcriptional progress of one or both complexes (Figure 2d). This was clearly shown by Prescott and Proudfoot [13] with the gal7 and gal10 genes of S. cerevisiae when these genes and their associated promoters were arranged convergently and the termination signals in the intergenic space were then deleted. This observation could well underpin the evolutionary pressure against convergent transcription in the yeast genome [49]. It is not known whether both polymerases stall during collision and both ‘fall off’, whether one polymerase (randomly) is rescued from that stalled state by host factors and the other polymerase ‘falls off’ [69] or whether a cooperative ‘push’ from following polymerases can influence the result [70]. We found that interference with transcription from the weak pL promoter of coliphage 186 increases the further its distance from the convergent strong pR promoter [20]. Our modelling [47] confirms the expectation that TI caused by collisions should increase as the distance between two convergent promoters is increased and as their activity is increased. Natural examples of TI as a result of collision in genome networks are rare, with the convergent promoters pR and pRE of coliphage lambda being a possibility and thereby an inviting follow-up of the seminal study of Ward and Murray [61].
Roadblock
The DNA-bound Lac repressor has been shown to block the progress of RNAP initiating upstream of the ‘roadblock’ or obstacle [3] (Figure 2e). Theoretically, an open complex bound at a promoter could act as a roadblock to the progress of an RNAP from another promoter. The in vitro data from Callen et al. [20] indicate that the open complex at the coliphage 186 pL promoter does not hinder the progress of an elongating RNAP from pR. It is conceivable that some open complexes are so tightly bound that they could act as roadblocks rather than sitting ducks, but this possibility has not been confirmed.
Outlook
Eszterhas et al. [14] set out to obtain some systematic understanding of the mechanisms operating in TI in mouse leukemic cells. They studied the expression of two reporter genes each bounded by a CMV promoter and an SV40 large T antigen polyadenylation signal. They studied all arrangements (convergent, divergent and tandem) in both orientations and integrated them at two different chromosomal loci and concluded that ‘the two transcriptional units interfere with each other in ways that cannot be fully explained by simple models’, certainly not by the simple models we have presented here. Furthermore, genome context modulated the interference that was observed. Most notably, the transcription of the downstream gene influenced transcription of the upstream gene and was itself influenced by transcription of the upstream gene, despite the positioning of a polyA signal between these genes. Again, convergent transcription showed mutual interference despite the presence of a polyA signal. Indeed, recent work indicates that RNA polymerase II can continue past the polyA site [71,72]. Thus, although some confirmation of the efficiency and precise location of termination in this system is required, the complexity of both chromatin structure [73] and different transcription-initiation complexes [74] will present challenges in the design of mechanistic studies of TI in higher eukaryotes. However, as with the development of molecular biology itself, studies in simple systems might provide direction. Mathematical modelling [47] is one approach to provide focus for future studies on the important parameters for each TI mechanism. Furthermore, examining the consequences of RNAP meeting a DNA-bound obstacle, such as Lac repressor [3] will provide vital clues for future studies that will help us understand what happens when an elongating RNAP meets a variety of DNA-bound obstacles, including, for example, a RNAP-like obstacle.
Concluding remarks
TI occurs in most genomes and has probably persisted during evolution because of its potential use in regulating gene expression. A variety of mechanisms have been identified and successfully studied, at least in simple systems. A major aim for the future is to exploit mathematical modelling to characterize TI, and a major issue to explore further is the fate of an elongating RNAP when it meets a DNA-bound obstacle, either moving or stationary.
Acknowledgments
We wish to acknowledge the help of the other members of the Egan laboratory and of Kim Sneppen (NORDITA) in writing this article. Research in the Egan laboratory was funded by the Australian Research Council and the National Institutes of Health.
Glossary
- Transcriptional interference
the suppressive influence of one transcriptional process, directly and in cis, on a second transcriptional process
- Promoter competition
occupation by RNA polymerase (RNAP) of one promoter directly precludes RNA-polymerase binding at a second promoter. This can be extended to include competition between two promoters for the same enhancer
- Occlusion
transcription across a promoter from an external promoter transiently precludes its occupation by RNAP and/or associated transcription factors
- Sitting duck
a transcriptional-initiation complex that is slow to ‘fire’ is a sitting duck, and can be ‘hit’ by a RNA polymerase transcribing from an external promoter. This can be extended to include dislodgement of transcription factors by an RNA polymerase transcribing from an external promoter
- Collision
of two converging transcription elongation complexes
- Roadblock
an immobile block to the progress of a transcription-elongation complex
References
- 1.Rojo F. Mechanisms of transcriptional repression. Curr Opin Microbiol. 2001;4:145–151. doi: 10.1016/s1369-5274(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 2.Delaval K, Feil R. Epigenetic regulation of mammalian genomic imprinting. Curr Opin Genet Dev. 2004;14:188–195. doi: 10.1016/j.gde.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Epshtein V, et al. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 2003;22:4719–4727. doi: 10.1093/emboj/cdg452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wassarman KM. RNA regulators of transcription. Nat Struct Mol Biol. 2004;11:803–804. doi: 10.1038/nsmb0904-803. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr Opin Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 6.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka S, et al. Transcription through the yeast origin of replication ARS1 ends at the ABFI binding site and affects extrachromosomal maintenance of minichromosomes. Nucleic Acids Res. 1994;22:3904–3910. doi: 10.1093/nar/22.19.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cajiao I, et al. Bystander gene activation by a locus control region. EMBO J. 2004;23:3854–3863. doi: 10.1038/sj.emboj.7600365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gribnau J, et al. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol Cell. 2000;5:377–386. doi: 10.1016/s1097-2765(00)80432-3. [DOI] [PubMed] [Google Scholar]
- 10.Opel ML, et al. The effects of DNA supercoiling on the expression of operons of the ilv regulon of Escherichia coli suggest a physiological rationale for divergently transcribed operons. Mol Microbiol. 2001;39:1109–1115. doi: 10.1111/j.1365-2958.2001.02309.x. [DOI] [PubMed] [Google Scholar]
- 11.Proudfoot NJ. Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 12.Greger IH, et al. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 1998;26:1294–1301. doi: 10.1093/nar/26.5.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci U S A. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eszterhas SK, et al. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol Cell Biol. 2002;22:469–479. doi: 10.1128/MCB.22.2.469-479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padidam M, Cao Y. Elimination of transcriptional interference between tandem genes in plant cells. Biotechniques. 2001;31:328–334. doi: 10.2144/01312st04. [DOI] [PubMed] [Google Scholar]
- 16.Ingelbrecht I, et al. Transcriptional interference in transgenic plants. Gene. 1991;109:239–242. doi: 10.1016/0378-1119(91)90614-h. [DOI] [PubMed] [Google Scholar]
- 17.Olson EN, et al. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 18.Thompson AJ, Myatt SC. Tetracycline-dependent activation of an upstream promoter reveals transcriptional interference between tandem genes within T-DNA in tomato. Plant Mol Biol. 1997;34:687–692. doi: 10.1023/a:1005888616748. [DOI] [PubMed] [Google Scholar]
- 19.Dodd IB, Egan JB. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol Microbiol. 2002;45:697–710. doi: 10.1046/j.1365-2958.2002.03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callen BP, et al. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Martens JA, et al. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 22.Wang P, et al. Demonstration that the TyrR protein and RNA polymerase complex formed at the divergent P3 promoter inhibits binding of RNA polymerase to the major promoter, P1, of the aroP gene of Escherichia coli. J Bacteriol. 1998;180:5466–5472. doi: 10.1128/jb.180.20.5466-5472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boussadia O, et al. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 1997;420:20–24. doi: 10.1016/s0014-5793(97)01479-8. [DOI] [PubMed] [Google Scholar]
- 24.Whitelaw E, Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagura-Burdzy G, Thomas CM. Dissection of the switch between genes for replication and transfer of promiscuous plasmid RK2: basis of the dominance of trfAp over trbAp and specificity for KorA in controlling the switch. J Mol Biol. 1997;265:507–518. doi: 10.1006/jmbi.1996.0747. [DOI] [PubMed] [Google Scholar]
- 26.Brantl S, Wagner EG. Dual function of the copR gene product of plasmid pIP501. J Bacteriol. 1997;179:7016–7024. doi: 10.1128/jb.179.22.7016-7024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavorgna G, et al. In search of antisense. Trends Biochem Sci. 2004;29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 28.Giordano E, et al. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 30.Fincham JR, Sastry GR. Controlling elements in maize. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- 31.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. [DOI] [PubMed] [Google Scholar]
- 32.Druker R, et al. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 34.Nigumann P, et al. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 35.Nasser W, et al. Transcriptional regulation offis operon involves a module of multiple coupled promoters. EMBO J. 2002;21:715–724. doi: 10.1093/emboj/21.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saha S, et al. The cox protein of bacteriophage P2 inhibits the formation of the repressor protein and autoregulates the early operon. EMBO J. 1987;6:3191–3199. doi: 10.1002/j.1460-2075.1987.tb02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rijn PA, et al. Regulation of phage Mu repressor transcription by IHF depends on the level of the early transcription. Nucleic Acids Res. 1989;17:10203–10212. doi: 10.1093/nar/17.24.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Koudelka GB. Mutually exclusive utilization of P(R) and P(RM) promoters in bacteriophage 434 O(R) J Bacteriol. 2000;182:3165–3174. doi: 10.1128/jb.182.11.3165-3174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M, Yamaguchi I. Convergent transcription units and their promoters at both ends of pot2, an inverted repeat transposon from the rice blast fungus. J Biochem (Tokyo) 1998;124:268–273. doi: 10.1093/oxfordjournals.jbchem.a022106. [DOI] [PubMed] [Google Scholar]
- 40.Simons RW, et al. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983;34:673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- 41.Adhya S, Gottesman M. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell. 1982;29:939–944. doi: 10.1016/0092-8674(82)90456-1. [DOI] [PubMed] [Google Scholar]
- 42.Gafny R, et al. Isolated P2 rRNA promoters of Escherichia coli are strong promoters that are subject to stringent control. J Mol Biol. 1994;243:152–156. doi: 10.1006/jmbi.1994.1641. [DOI] [PubMed] [Google Scholar]
- 43.Hirschman JE, et al. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4608–4615. doi: 10.1128/mcb.8.11.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Irniger S, et al. The yeast actin intron contains a cryptic promoter that can be switched on by preventing transcriptional interference. Nucleic Acids Res. 1992;20:4733–4739. doi: 10.1093/nar/20.18.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corbin V, Maniatis T. Role of transcriptional interference in the Drosophila melanogaster Adh promoter switch. Nature. 1989;337:279–282. doi: 10.1038/337279a0. [DOI] [PubMed] [Google Scholar]
- 46.Salgado H, et al. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 2004;32 (Database issue):303–306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sneppen K, et al. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J Mol Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- 48.Tjaden B, et al. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marin A, et al. Short-range compositional correlation in the yeast genome depends ontranscriptional orientation. Gene. 2004;333:151–155. doi: 10.1016/j.gene.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 50.Bajic VB, et al. Promoter prediction analysis on the whole human genome. Nat Biotechnol. 2004;22:1467–1473. doi: 10.1038/nbt1032. [DOI] [PubMed] [Google Scholar]
- 51.Chiaromonte F, et al. Gene length and proximity to neighbors affect genome-wide expression levels. Genome Res. 2003;13:2602–2608. doi: 10.1101/gr.1169203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen BA, et al. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 53.Portin P. Historical development of the concept of the gene. J Med Philos. 2002;27:257–286. doi: 10.1076/jmep.27.3.257.2980. [DOI] [PubMed] [Google Scholar]
- 54.Takai D, Jones PA. Origins of bidirectional promoters: computational analyses of intergenic distance in the human genome. Mol Biol Evol. 2004;21:463–467. doi: 10.1093/molbev/msh040. [DOI] [PubMed] [Google Scholar]
- 55.Yamada K, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 56.Stolc V, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 57.Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 58.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 59.Cawley S, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 60.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 61.Ward DF, Murray NE. Convergent transcription in bacteriophage lambda: interference with gene expression. J Mol Biol. 1979;133:249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- 62.Horowitz H, Platt T. Regulation of transcription from tandem and convergent promoters. Nucleic Acids Res. 1982;10:5447–5465. doi: 10.1093/nar/10.18.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elledge SJ, Davis RW. Position and density effects on repression by stationary and mobile DNA-binding proteins. Genes Dev. 1989;3:185–197. doi: 10.1101/gad.3.2.185. [DOI] [PubMed] [Google Scholar]
- 64.Conte C, et al. Promoter competition as a mechanism of transcriptional interference mediated by retrotransposons. EMBO J. 2002;21:3908–3916. doi: 10.1093/emboj/cdf367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Choi OR, Engel JD. Developmental regulation of β-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 66.Liang S, et al. Activities of constitutive promoters in Escherichia coli. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 67.Greger IH, et al. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:8415–8420. doi: 10.1073/pnas.140217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bateman E, Paule MR. Promoter occlusion during ribosomal RNA transcription. Cell. 1988;54:985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- 69.Roberts J, Park JS. Mfd, the bacterial transcription repair coupling factor: translocation, repair and termination. Curr Opin Microbiol. 2004;7:120–125. doi: 10.1016/j.mib.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 70.Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- 71.West S, et al. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 72.Teixeira A, et al. Autocatalytic RNA cleavage in the human β-globin pre-mRNA promotes transcription termination. Nature. 2004;432:526–530. doi: 10.1038/nature03032. [DOI] [PubMed] [Google Scholar]
- 73.Nagaich AK, et al. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 74.Remenyi A, et al. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]