Abstract
Protein S-nitrosation has been argued to be the most important signaling pathway mediating the bioactivity of NO. This post-translational modification of protein thiols is the result of chemical nitrosation of cysteine residues. The term NO-donors covers very different chemical classes: from clinical therapeutics to probes of routine use in chemical biology; their different chemistry is predicted to result in distinctive biology regulated by protein S-nitrosation. To measure the extent of protein S-nitrosation by NO-donors, a proteomic mass spectrometry method was developed, which quantitates free thiol versus nitrosothiol for each modified cysteine residue, coined d-Switch. This method is adapted from the biotin switch (BST) method, used extensively to identify S-nitrosated proteins in complex biological mixtures, however, BST does not quantitate free thiol. Since glutathione-S-transferase P1-1 (GST-P1) has been proposed to be a biological “NO carrier”, GST-P1 was used as a reporter protein. The 5 different chemical classes of NO-donors compared by d-Switch demonstrated very different profiles of protein S-nitrosation and response to O2 and cysteine, although, all NO-donors were oxidants towards GST-P1. The low limits of detection and the ability to use established MS database searching allowed facile generalization of the d-Switch method, therefore after incubation of neuronal cell cultures with nitrosothiol, it was possible not only to quantitate S-nitrosation of GST-P1, but also many other proteins, including novel targets such as ubiquitin carboxyl-terminal esterase L1 (UCHL1), moreover d-Switch also allowed identification of non-nitrosated proteins and quantitation of degree of nitrosation for individual protein thiols.
Keywords: Nitric oxide, Oxidation, Nitrosation, Nitrosylation, Cysteine residue, Cysteome
INTRODUCTION
Cysteine residues in proteins play many important roles including in protein quaternary structure, cofactor binding, and as catalysts at enzyme active sites. Since the primary cysteine thiol is a good biological nucleophile and can attain multiple oxidation states, protein thiol residues represent natural targets for electrophiles and redox active agents including nitrosating and oxidizing species generated from NO. Cysteine modification is manifested in post-translational modification leading to modulation of protein function and regulation of biological processes (1, 2); and in triggering of cellular response to xenobiotics, (3). S-Nitrosation, the chemical reaction of a thiol functional group with a nitrosonium equivalent, likely underlies the protein post-translational modification commonly referred to as protein S-nitrosylation. Such protein nitrosylation has been argued to rival phosphorylation in importance as a cell signaling process (4).
A large number of protein thiols have been reported to undergo regulatory nitrosation, for example: S-nitrosation of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is proposed to facilitate nuclear translocation and apoptosis (5). It has even been proposed that many, if not most, biological effects of NO are mediated by protein S-nitrosation (6). The proclivity of the active site thiol in cysteine proteases towards nitrosothiol formation is considered as providing a mechanism of negative regulation of function by endogenous NO and exogenous NO-donors (7), and NO production may itself be regulated by nitrosation of the Zn-tetrathiolate structure of nitric oxide synthase 2 (iNOS) (8). Glutathione S-transferase P1-1 (GST-P1), found at elevated levels in many solid and chemoresistant tumors (9), is a further protein reported to be nitrosated, in this case at the most reactive cysteine (Cys47) and proposed to act as a biological NO carrier (10–12). GST-P1 was therefore a reasonable target to compare and to quantitate protein S-nitrosation in response to 5 different classes of NO-donors.
The term “NO-donor” is widely used in the literature for: i) diazeniumdiolates that are true NO-donors, releasing predictable fluxes of NO; ii) the endogenous nitrosothiols, S-nitrosocysteine (CysNO) and S-nitrosoglutathione (GSNO) that function as NO+ donors; iii) organic nitrites that are in clinical use and are efficient NO+ donors; iv) clinical organic nitrates that release NO2□ and require reductive bioactivation to give NO; and, v) oxadiazole-N-oxides that are in preclinical studies and again require bioactivation, possibly to NO and HNO. It would be more accurate to term these compounds NO-mimetics, since they result in NO bioactivity, including via protein S-nitrosation, which is not derived solely from donation of nitrogen monoxide (NO). The clinical importance of these collective “NO-donors”, and their different chemistries, demands a quantitative comparison of their ability to effect protein S-nitrosation; towards this end a novel adaptation of the biotin switch (BST) method was developed, coined d-Switch.
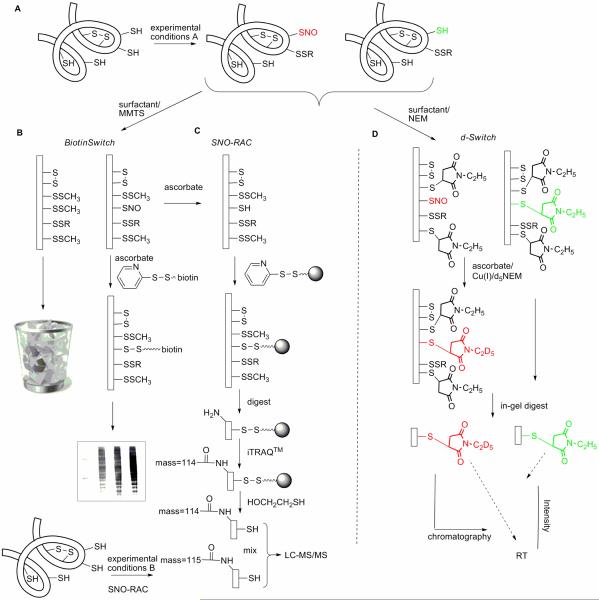
The BST methodology introduced by Jaffrey et al. has been central to reports of protein S-nitrosation (13). This semi-quantitative method consists of three principal steps: blocking of free thiols with a thiol reactive reagent; reducing nitrosothiol to free thiol with ascorbate; and, labeling the resulting free thiol with biotin (Figure 1). A number of sources of artifacts have been identified in the BST approach, resulting in publication of a number of modifications and improvements; most recently, the SNO-RAC methodology was reported, which incorporates iTRAQ™ quantitative proteomic techniques (Figure 1) (14). In concert with LC-MS/MS chromatography, both the BST and SNO-RAC methodologies have the potential to identify nitrosated proteins, the number of which detected may run to the hundreds in cell culture studies. The question remains, however, as to whether the extent of cysteine nitrosation in these proteins is sufficiently high to lead to altered protein function. The objective of the present research was to answer the question: can we quantitate the percentage of a critical protein thiol that is nitrosated by different NO-donor classes? The d-Switch method developed to answer this question is complementary to BST, does not use proprietary reagents and is generalizable in cell culture. Protein S-nitrosation measured by BST and d-Switch was compared for five classes of NO-donors in purified protein and cell cultures, demonstrating the ability of d-Switch to quantify nitrosation and revealing the quite different protein S-nitrosation properties of NO-donors, routinely used in chemical biology and drug discovery.
Fig 1. Schematics for biotin switch (BST), SNO-RAC, and d-Switch methods.
A) The proteome may contain cysteines with varied reactivity and accessibility and intramolecular disulfide bonds; BST and modified BST methods discard non-nitrosated proteins. B) The standard BST method visualizes biotinylated proteins by western blot (shown); or may identify proteins by LC-MS/MS chromatography after of pull-down of biotinylated proteins prior to protein digestion. C) The SNO-RAC is a modified BST methodology that uses activated-disulfide beads in place of biotin to isolate nitrosated proteins in combination with iTRAQ™ quantitative proteomic analysis. D) The d-Switch modification is designed to quantify the absolute ratio of nitrosated versus non-nitrosated, reduced cysteine at individual sites in key proteins; therefore, non-nitrosated proteins must be islolated and quantified, not discarded.
Results & Discussion
There is no doubt that under certain cellular conditions, selected cysteine residues in proteins undergo nitrosation. There is also convincing evidence that S-nitrosation can influence protein function, most simplistically in enzyme inhibition by nitrosation and blocking of catalytic, active site cysteine residues such as in caspases and protein phosphatases (7, 15). In such cases, the percentage of non-nitrosated, free cysteine thiol will directly correlate with enzyme activity and hence biological function. Evidence has been provided that protein thiol nitrosation mediates cell signaling by NO in parallel with signaling via NO/cGMP. Termed protein S-nitrosylation, this signaling mechanism is proposed to represent a broad-based post-translational regulation of protein function (16). Much of the evidence provided for S-nitrosation employs BST immunoassay analysis, followed by elegant studies mutating the relevant cysteine, leading to loss of biological response accompanied by loss of protein S-nitrosation visualized by BST. One example of the use of BST in this way is S-nitrosation of the primary cellular target of NO, soluble guanylyl cyclase (sGC) that has been proposed to be regulated through S-nitrosation by the “NO-donors”, CysNO, GSNO, and nitroglycerin (GTN) (17, 18). This work led to the proposal that inhibition of sGC through S-nitrosation by GTN was the cause of clinical nitrate tolerance (17). This work has been queried recently (19), demonstrating the importance of both a quantitative technique for measuring S-nitrosation and a fuller understanding of agents commonly referred to as NO-donors. A simple method that quantified amounts of protein Cys-SNO relative to the non-nitrosated free protein thiol (Cys-SH) would be invaluable to support a functional role for protein S-nitrosation.
The term NO-donors has come to include newer compounds designed to deliver defined fluxes of NO under specific conditions, in addition to nitrite and nitrate nitrovasodilators that have been in clinical use for well over a century (20–23). There is evidence in the literature that the very different chemistry of these NO-donors inevitably delivers differing biological responses, however, in many reports NO-donors classes are treated as equivalent. Selected for this study were: the endogenous low molecular weight nitrosothiols, GSNO and CysNO; the clinical nitrite vasodilator, isoamyl nitrite (IAN); the clinical nitrate, GTN; and a novel oxadiazole-N-oxide (furoxan), ODZ1. The latter is a drug class that has been the subject of recent intensive studies in NO-donor drug discovery (24, 25). The fast release diazeniumdiolate (DEA/NO) was also selected for study because of the extensive use of diazeniumdiolates in chemical biology.
Human GST-P1 was chosen as a target, in part because of multiple reports of its S-nitrosation. GST-P1, the most widely distributed of the GST phase 2 detoxifying enzymes (26, 27), is found at elevated levels in solid tumors (e.g. breast, lung colon, pancreas) and in drug resistant tumors (9), and both detoxifies and is adducted by electrophilic xenobiotics (28, 29). The 4 cysteine residues in GST-P1, are modified with reactivity, Cys47 > Cys101 > Cys14 >> Cys169; although not being part of the active site, modification does not necessarily lead to loss of enzyme activity. In 2001, Lo Bello et al using ESI-MS analysis reported that in absence of GSH, GST-P1 was nitrosated by the endogenous nitrosothiol, GSNO, and proposed GST-P1 as a cellular “NO carrier” (10). Evolution of this hypothesis viewed GST-P1 as an important endogenous carrier of a dinitrosyl-iron complex (11, 30, 31), however, S-nitrosation of Cys47 by GSNO was still claimed to be a likely physiological scenario (11, 12).
GST-P1 nitrosation by NO-donors
Previous reports on GST-P1 nitrosation using 2 mM GSNO estimated a half-life of 5 min for Cys47 nitrosation (10, 11). Although the concentration of GSNO was supraphysiological, the studies confirmed S-nitrosation. Organic nitrites, such as IAN, are excellent nitrosating agents; the observed rate of reaction with glutathione (GSH; k ~ 2 M−1s−1) indicates that, as for the nitrosothiols, extensive nitrosation of GST-P1 by IAN should be anticipated (32). GST-P1 nitrosation was assayed by BST for all six NO-donors (500 μM) demonstrating qualitatively that the diazeniumdiolate, nitrite, and nitrosothiols were effective nitrosating agents, whereas GTN and ODZ1 were at best, weak nitrosating agents. The increase in GST-P1 nitrosation by DEA/NO, GSNO, CysNO and IAN was significant, but BST was unable to show any significant differences between these four NO-donors (Figure 2A).
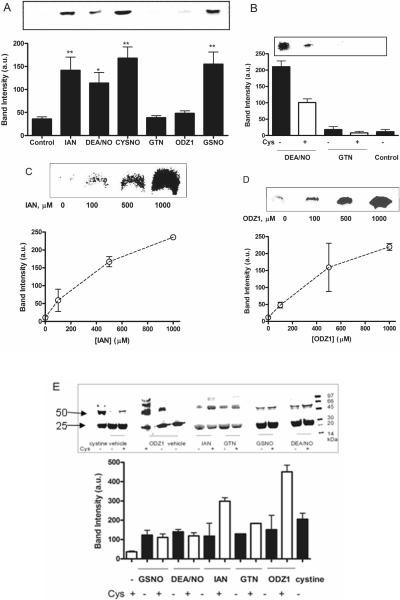
Fig. 2. Comparison of GST-P1 nitrosation and oxidation by NO-donors.
(A) BST analysis of GST-P1 (20 μM) treated with NO-donors (500 μM) at 37 °C for 30 min. ANOVA analysis with Dunnett's post test relative to vehicle control: ** p<0.001; * p<0.005. Response to IAN, GSNO, CysNO and DEA/NO was not significantly different. (B) BST analysis of GST-P1 (20 μM) untreated (control) or incubated with NO-donor (500 μM) in the presence and absence of Cys (2 mM). Quantitation was by image densitometry from blots. (C) BST analysis of GST-P1 (20 μM) nitrosation after incubation with varying concentrations of IAN at 37 °C for 30 min. (D) BST analysis of GST-P1 (20 μM) nitrosation after incubation with varying concentrations of ODZ1. (E) Non-reducing SDS-PAGE analysis of GST-P1 oxidation to dimer (and higher oligomers) by NO-donors (500 μM) visualized by Coomassie blue staining. GST-P1(20 μM) was incubated with NO-donors (500 μM) or cystine (1 mM) in the presence and absence of Cys (2 mM) at 37 °C for 30 min. Oxidation of GST-P1 was significantly increased in the presence of cysteine for IAN, GTN, and ODZ1, but not other NO-donors. Quantitation was by image densitometry from blots; upper panels show representative blots. Data show mean and s.d. in arbitrary units for triplicate experiments, which for oxidation/dimerization were normalized to the untreated control as 0%.
Both nitrates and furoxans may require reductive bioactivation to function as NO-donors; although, likely mediated by multiple enzymes, bioactivation mediated by free thiol has frequently been proposed (21, 23, 25, 33). Addition of cysteine (2 mM) to incubations of GST-P1 with GTN did not cause S-nitrosation, whereas cysteine significantly reduced GST-P1 nitrosation by the spontaneous NO-donor DEA/NO (Figure 2B). The observed loss of signal for nitrosated protein is compatible with accelerated denitrosation by the added cysteine (34), moreover cysteine can compete with GST-P1 for nitrosating species generated from NO autoxidation. The next step was to use BST to examine concentration dependent protein S-nitrosation, as frequently employed in the literature. IAN, which gave significant nitrosation at 500 μM, and the marginal nitrosating agent ODZ1 were studied, the resulting blots apparently showing that both NO-donors were able to elicit protein S-nitrosation (Figure 2C,D). This experiment illustrates a potential pitfall with BST, because as we shall see from d-Switch analysis, GST-P1 nitrosation by ODZ1 is real, but low: 1.2 ± 0.3 % nitrosation of Cys47.
GST-P1 oxidation by NO-donors
GST-P1 is readily oxidized to form oligomers with intermolecular and intramolecular disulfide bonds (27) and thiols are known to undergo oxidation to disulfides in nitrosating solutions and on reaction with GTN (35–37). Oxidative dimerization of GST-P1 was observed for all NO-donors studied by non-reducing SDS-PAGE: GTN was a potent oxidant, and oxidation was dependent on NO-donor concentration (Supplemental Figure 1). Again significant differences were observed between the chemical classes in their response to added cysteine: oxidation was not significantly enhanced for GSNO or DEA/NO, but was enhanced by cysteine (2 mM) for ODZ1 > IAN > GTN (Figure 2E). Cystine (1 mM) was also observed to enhance GST-P1 oxidative dimerization and likely contributed to the enhanced oxidation observed when cysteine is added to IAN, GTN, and ODZ1. Cu/ascorbate treatment used to convert RS-NO to R-SH in BST and d-Switch methods did not reduce protein disulfide bonds, since allowing GST-P1 to dimerize followed by treatment with Cu/ascorbate was without effect (Supplemental Figure 6).
GST-P1 nitrosation by d-Switch
The d-Switch method depicted in Figure 1, is a proteomics method adapted from the BST assay, in which NEM is used in place of MMTS for Cys-SH blocking, and d5-NEM is used in place of biotin-HPDP for Cys-SNO labeling (Figure 1). The use of CuCl/ascorbate was chosen for the denitrosation/labeling step, because of the enhanced signal (Supplemental Figure 2) (38). The d-Switch method relies upon protein digestion to yield a peptide containing the protein-Cys of interest that is amenable to analysis by LC-MS/MS; in studies on GST-P1, this is Cys47. In-gel tryptic digest of GST-P1 monomer followed by LC-MS/MS analysis revealed the desired decapeptide at 12.8–13.4 min in the total ion current chromatogram, the identity of which was confirmed by MS/MS analysis of the doubly charged peptide (Supplemental Figure 2). Reaction times and temperatures and reagent concentrations were optimized at each step in the d-Switch methodology yielding an assay in which the NEM and d5-NEM labeled peptides were readily quantified in extracted ion chromatograms (m/z 603.3 (d0) versus 605.6 (d5) for z = 2; Figure 3A). Both NEM and d5-NEM labeled peptides have almost identical retention time and identical ionization efficiency, hence calculating the peak area ratios of the two peptides from the extracted ion chromatogram, allows for quantitative analysis of Cys47 nitrosation. The final output of d-Switch analysis is the peak area ratio of Cys-SNO labeling (d5) to the summed Cys-SNO plus Cys-SH labeling (d0 + d5), reported as percentage nitrosation: %[d5/(d0 + d5)].
Fig 3. Concentration and time dependent nitrosation of GST-P1 by IAN quantified by dSwitch.
GST-P1 (20 μM) was treated with IAN at various concentrations and the reaction was quenched at various time points by addition of NEM. (A) Representative extracted ion chromatograms showing increasing intensity of d5NEM-labeled peptide ASC[d5NEM]LYGQLPK2+ (m/z = 605.6) derived from GST-P1 Cys47 nitrosation versus NEM-labeled peptide ASC[NEM]LYGQLPK2+ (m/z = 603.3) derived from Cys47 free thiol after 30 min treatment with IAN (0, 0.1, 0.5, 1 mM). The retention time of each of the 4 chromatogram excerpts is 12.6–13.6 min. The ratio of peak areas was used to calculate % nitrosation: A(Cys47-SNO)/[A(Cys47-SNO) + A(Cys47-SH)]. (B) Percentage nitrosation of GST-P1 Cys47 was dependent on NO-donor concentration. Data show mean and s.e.m. of triplicate experiments. (C) Percentage nitrosation of GST-P1 Cys47 by IAN (1 mM) was dependent on incubation time as calculated from extracted ion chromatogram peak areas. Data show mean and s.e.m. of triplicate experiments.
The precision of the d-Switch analysis compared to BST was demonstrated in the measurement of concentration and time dependent S-nitrosation of GST-P1 by IAN (Figure 3 B,C). Measured at 37 °C and 30 min, % nitrosation of Cys47 was linear in IAN concentration, however, at 10 μM IAN, Cys47 was only 0.43 ± 0.04 % nitrosated. The rate constant for hydrolysis of IAN has been measured spectrophotometrically in aqueous phosphate buffer at 37 °C to be 1.18 × 10−3 s−1, corresponding to a half life of approximately 8 min and a second order rate constant for reaction with water of 2 × 10−5 M−1s−1 (39, 40). The rate constant for the nitrosation of cysteine by IAN in phosphate buffer (pH 7.0 and 25 °C) has been reported (k = 1.05 M−1s−1), as has the rate constant for the fast reaction with GSH in an aqueous biological compartment (k ~ 2 M−1s−1) (41). The observed time course for trapping of IAN by Cys47 to give protein nitrosothiol was therefore surprisingly slow (Figure 3C), suggesting that reactions in addition to hydrolysis and transnitrosation determine the % nitrosation observed. For example, intermolecular disulfide formation, observed to be induced by all NO-donors (Supplemental Figure 1), consequent to Cys47 nitrosation, is one such secondary reaction that leads to loss of Cys47-SNO.
Comparison of NO-donors using d-Switch and effect of O2
The concentration dependence of Cys47 nitrosation was measured for six NO-donors (Figure 4A). The two nitrosothiols, CysNO and GSNO, were the most efficient nitrosating agents, outperforming the true NO-donor DEA/NO that theoretically yields twice the amount of NO. The nitrite, an efficient and direct nitrosating agent, was in fact less efficient than the thionitrites (nitrosothiols), because of competing hydrolysis. At lower concentrations of CysNO and GSNO (10 μM) less than 5% nitrosation of GST-P1 was observed. Under high physiological fluxes of NO, the estimated [GSNO] is 1–5 μM.(42), therefore the prediction from extrapolation of these data is that significant S-nitrosation of GST-P1 is unlikely to be observed under physiological conditions. Cys47 nitrosation by GTN was not observed and the levels of nitrosation induced by ODZ1 were only significantly above background at millimolar concentration. The d-Switch technique combines sensitivity with precision, as evidenced by the low variance and the ability to discriminate quantitatively between NO-donors in Cys47 nitrosation, which was not possible using BST analysis. S-nitrosation by NO is expected to be dependent on O2, since oxidation of NO to nitrosating N2O3 is the dominant aerobic mechanism (43). Assuming that diazeniumdiolates are the only true NO-donors of the chemical classes studied herein, the prediction was that Cys47 nitrosation by DEA/NO would require O2; which was borne out by observations in 20% versus 1% O2 where nitrosation was reduced from 11% to 1.8% (Figure 4B). All other NO-donor classes showed no dependence on O2, demonstrating that Cys47 nitrosation is not mediated via release of NO from these agents (Scheme 1).
Fig. 4. d-Switch comparison of GST-P1 nitrosation by six NO-donors in pure form and in human cells.
(A) Concentration dependent Cys47 nitrosation after incubation of GST-P1 (20 μM) with the indicated concentrations of various NO-donors at 37 °C for 30 min from d-Switch analysis. Data showing mean and s.e.m. of 3 experiments for IAN, DEA/NO and ODZ1 are linearly fit with slopes of 0.031, 0.021, and 0.0014, respectively. (B) Effect of O2 pressure on GST-P1 nitrosation at Cys47 by NO-donors (500 μM) analyzed by d-Switch. Data show mean and s.d. of 3 experiments (C) Concentration dependant Cys47 nitrosation after incubation of human neuroblastoma SH-SY5Y cells with CysNO for 20 min and analysis by d-Switch (solid line; open square); data for recombinant protein incubation with CysNO are shown for comparison (dashed line; solid square).
Scheme 1.
Reaction schemes for GST-P1 S-nitrosation by CysNO, IAN and DEA/NO, showing oxygen dependence for NO; and for GST-P1 oxidation by GTN (RONO2) and subsequent to nitrosation.
S-nitrosation induced by NO-donors from five different chemical classes was studied using GST-P1 as a model protein, measuring the extent of nitrosation of Cys47, and examining the influence of O2 and added cysteine. Only 3 of the 5 drug classes gave significant S-nitrosation. Of these 3, only the true NO-donor diazeniumdiolate required aerobic conditions to effect nitrosation via generation of N2O3. In contrast, all NO-donors oxidized GST-P1 to give dimeric protein disulfides. The combined analysis demonstrated that the different chemistry of NO-donor classes is reflected in differing profiles of protein nitrosation and oxidation. Since oxidation of protein thiols to disulfides by NO-donors is concomitant or dominant over nitrosation, cysteome oxidation may be responsible for many actions of endogenous NO and NO-donors. Theoretical and experimental studies have reported thiol oxidation, rather than nitrosation, as the dominant modification induced by NO (44, 45), although this is often overlooked when considering mechanisms of NO bioactivity. A variety of oxidative modifications of protein thiols have been reported for NO and NO-donors, including disulfides, sulfenic and sulfinic acids (46–48).
Transnitrosation of GST-P1-Cys47 by nitrosothiols was observed not to reach completion. The d-Switch analysis indicated that at millimolar nitrosothiol, significant amounts of free thiol persisted; the ratio of Cys47SH to Cys47SH+Cys47SNO was 45%. The extent of protein nitrosation is expected to be determined by the balance of nitrosation versus denitrosation rates; the apparent saturation of Cys47 nitrosation by nitrosothiols at 55% might be in accord with this equilibrium in the transnitrosation reaction between small molecule and protein thiol (Scheme 1). Another possibility was that Cys47 was “hidden” from small molecule nitrosothiol in the interface of GST-P1 homodimers, since this protein is known to dimerize non-covalently. To test these hypotheses, an experiment was performed incubating GST-P1 at 10-fold lower concentration with CysNO, however, the degree of Cys47 nitrosation was not increased (Supplemental Figure 12). Mitchell et al. reported on S-nitrosation of caspase-3, a protein reported to be basally nitrosated in nature, using GSNO and a peptide-SNO complementary to the active site, and rigorous use of BST and mass spectrometry (49). Comparison of comparable GSNO and peptide-SNO esters (0.5 mM) in cell culture showed almost threefold less S-nitrosation of caspase-3 by GSNO. Furthermore, GSNO treatment of pure protein gave protein-disulfide formation. For GST-P1, S-nitrosation did not increase above 0.5 mM CysNO (Figure 4A), whereas SDS-PAGE assay of the concentration dependence of GST-P1 oxidative dimerization showed that oxidation increased threefold with increasing [CysNO], from 0.5 mM to 2 mM (Supplemental Figure 13). The importance of thiol oxidation as a competing and consequent reaction to nitrosation is apparent from these data. Further mechanistic analysis clearly requires parallel measurement of disulfide formation. Indeed, this was the primary impetus for development of an alternative methodology to BST, which unlike BST is not dependent on formation of disulfides in the analytical steps. Adaptation of d-Switch using (tris(2-carboxyethyl)phosphine) in place of Cu/ascorbate is currently under study.
GST-P1 nitrosation in cells
Evidence for GST-P1 as an “NO-carrier” was reported in E. coli cells used to overexpress GST-P1, by exposing cells to 2 mM GSNO or 0.5 mM DEA/NO (11). Herein, under similar conditions, GST-P1 nitrosation was quantified by d-Switch in living cells. E. coli cells overexpressing GST-P1 were exposed to NO-donor, however very low levels of nitrosation were observed and in order to measure 8% Cys47 nitrosation, combinations of bolus injections of DEA/NO were required to give total delivery of 40 mM NO (Supplemental Figure 3C). Unsurprisingly, the same experiment performed with GTN as NO-donor yielded no signal corresponding to nitrosation. More interesting was the measurement of protein nitrosation in human neuronal cells.
SH-SY5Y neuroblastoma cells were treated with different concentration of CysNO (0.1, 1, 2 mM) and divided into two sets: i) for BST analysis; and, ii) for d-Switch analysis. BST analysis showed the expected concentration dependent increases in multiple S-nitroso-proteins (Supplemental Figure 8). After d-Switch treatment, the gel region corresponding to GST-P1 was subjected to in-gel tryptic digestion and analyzed by nLC-LTQ-FT-ICR. The resulting MS and MS/MS data were searched using MassMatrix against the IPI human v3.65 database (50). GST-P1 was identified with excellent sequence coverage of 66%. The MS spectrum for ASCLYGQLPK, the GST-P1 peptide containing Cys47, was searched using the scan number generated from the database search and comparing with the observed full scan MS spectrum. The MS spectrum shown in Figure 5A, showing unlabelled (Cys47-SH; green) and d5-labelled (Cys47-SNO; red) peptide after CysNO treatment of cells, vividly displays the capability of the d-Switch methodology. The GST-P1 peptide fragment, labeled at Cys47 with NEM and/or d5-NEM was rigorously identified using MS/MS (Supplemental Figure 7A). The concentration dependent nitrosation of GST-P1 Cys47 was quantified using the peak height of the most intense isotope peak in the d5-NEM-peptide compared to the corresponding peak in the NEM-peptide. Concentration dependent nitrosation of GST-P1 was again observed by CysNO (Figure 4C, solid line; open square). At 2 mM CysNO, the extent of GST-P1 S-nitrosation in cells was the same as that observed in recombinant protein at 1mM CysNO; whereas at 100 μM CysNO, Cys47 nitrosation was 5-fold lower in cell culture. The extent of GST-P1 Cys47 nitrosation was less than 5% at 100 μM CysNO. In comparable experiments, treatment of SH-SY5Y cells with 5 mM DEA/NO resulted in less than 5% nitrosation of GST-P1 at Cys47; and treatment with GTN gave no nitrosation. BST analysis of cell cultures incubated for up to 24 h with GTN (1 mM) showed no time dependent increase in signal and no significant differences with untreated control blots (Supplemental Figure 10).
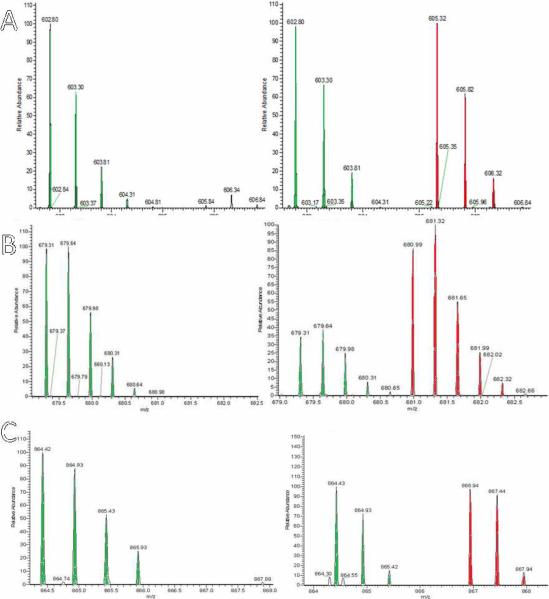
Fig 5. Nitrosation of Proteins in SH-SY5Y cells analyzed by LC-MS/MS.
LTQ-FTMS spectra of NEM and d5-NEM labeled peptides from d-Switch analysis of CysNO treated SH-SY5Y cells. On the left, the spectrum is of the control experiment without CysNO. On the right, treatment of intact cells with CysNO (2 mM) gave spectra showing d0-NEM and d5-NEM labeled peptides. (A) Cys47 peptide of GST-P1; the green signals are natural isotope distribution for the peptide corresponding to the non-nitrosated peptide and the red peaks are the similar isotope signals corresponding to peptide from GSTP1 nitrosated at Cys47. As displayed in figure 4C, under these conditions GST-P1 Cys47 is 51% nitrosated. (B) Cys 152 peptides of UCHL1. (C) Cys53 peptides of DJ-1.
The efficient nitrosothiol nitrosating agents at close to physiological concentrations (10 μM) gave only 3.4 ± 0.5 % Cys47 nitrosation; and nitrosation was never observed to exceed 55% even in the presence of nitrosothiols at millimolar concentrations. These observations are not compatible with a proposed physiological role as a “NO carrier”, but certainly do not rule out the proposal of GST-P1 as a carrier of dinitrosyl-Fe complex (10–12, 30, 31). Previous application of SNO-RAC and SNOCAP methods indicated two pools of S-nitrosated proteins, GSH stable, and GSH labile, the latter defined by rapid denitrosation by GSH (14, 51); of 102 S-nitrosated proteins detected in rat brain homogenates by SNOCAP after incubation with GSNO (40 μM) for 20 min, 10 were in the stable pool, however, GST-P1 Cys47 nitrosation was not detected.
Human GST-P1 has been assigned key roles in response to NO and oxidative stress (10–12, 30, 31). Cytoprotective roles include inhibition of c-Jun N-terminal kinase (JNK) mediated apoptosis (52) and coordinated regulation of stress kinases in response to oxidative stress (53). A role for GST-P1 has recently been proposed in mediating glutathionylation following oxidative and nitrosative stress, induced by exogenous NO or disulfide (54), wherein GST-P1 was reported to be glutathionylated at Cys47 and Cys101 impacting the binding of JNK. NO and ROS induced disulfide formation at Cys47 is also likely to play an important role in regulation of peroxiredoxins through formation of heterodimers (55, 56). The observation that all NO-donors induced Cys47 oxidation to give GST-P1 homodimers confirms that GST-P1 may respond to nitrosative stress by protein and glutathione disulfide formation.
Cellular protein nitrosation beyond GST-P1
The original BST technique has frequently been modified, including the addition of LC-MS to identify S-nitrosated proteins. The SNOSID (SNO site identification) technique was introduced to identify S-nitrosated proteins in complex biological mixtures using LC-MS after tryptic digest of BST treated proteins followed by avidin capture of biotinylated peptides (57). In another study His-tag idoacetamide was used for affinity capture of peptides obtained by trypsinolysis (58). Other laboratories have reported combinations of BST with 1D and 2 D PAGE and in-gel trypsinolysis to identify nitrosated proteins by LC-MS (59). The SNOCAP (S-nitrosothiol capture) modified BST procedure introduced in 2008 bears some similarity to d-Switch in that quantitation is achieved by heavy and light isotope labeling, but uses biotinylated disulfide reagents (51). Recently, Stamler reported the SNO-RAC methodology, a modification of the BST method, which incorporates iTRAQ™ quantitative proteomic techniques to map the “cysteome” (Figure 1) (14). From our perspective, there was a need for a technique, complementary to BST, that was rigorously quantitative for protein S-nitrosation, which did not introduce disulfide bonds, the latter being a hallmark of the BST methodologies, including SNO-RAC. The need to avoid the introduction of disulfides rests upon the desire to extend d-Switch to parallel quantitation of endogenous disulfides. In contrast to BST and SNO-RAC methods, d-Switch carries through both labeled and unlabeled proteins, which is of course essential to quantitate nitrosated versus free thiol for any cysteine residue (Figure 1). Moreover in SNO-RAC, parallel samples are mixed and quantitated using iTRAQ reagents, a relative quantitation approach that reduces precision; d-Switch uses absolute quantitation obtained from single sample analysis.
SNOSID, SNO-RAC, and other BST modifications employ avidin or similar affinity columns or beads to concentrate the labeled peptides that were S-nitrosated in the test sample (14, 57–59). Although peptide loss occurs through inefficient digestion and retention on solid supports, affinity concentration has the potential to identify low abundance proteins, whereas d-Switch relies on the limit of detection of the mass spectrometer. If nitrosated proteins can be quantified at low nano to picomolar levels with modern MS instruments, nitrosated proteins can be unambiguously identified by comparing the non-deuterated tandem mass spectra to the deuterated spectra. d-Switch peptides are readily identified and quantified by adapting currently available database search methods used in established methods such as SILAC (60, 61).
To test the potential of d-Switch, the gel band excised for GST-P1 analysis, from incubation of SH-SY5Y cells with CysNO (2 mM), was further interrogated using Mass Matrix, yielding approximately 50 proteins with acceptable coverage. 20 Peptides derived from these proteins showed NEM modification only, representing non-nitrosated cysteine residues (Supplemental Table S1); 8 were unambiguously identified by d-Switch to be nitrosated after treatment with CysNO (Table 1); 3 of which showed < 10% nitrosation. Of the remaining 5 proteins, ubiquitin carboxyl-terminal esterase L1 (UCHL1) and PARK7 protein DJ-1, both nitrosated at two cysteine residues, deserve comment (Figure 5 B,C). UCHL-1 has been identified as a major target of oxidative stress contributing to Parkinson's and Alzheimer's diseases, at least in part by oxidation of cysteine residues, but nitrosation has not been previously noted (62–64). DJ-1 has also been implicated in mediating oxidative stress in Parkinson's disease, furthermore nitrosation of key cysteine residues has been reported (65). A combination of cysteine mutants and BST was not able to differentiate between Cys46 and Cys53 nitrosation, however, d-Switch quantitatively revealed the degree of nitrosation: 29% and 56%, respectively. The generalization of d-Switch to quantitation of nitrosated and non-nitrosated cysteine residues in cell culture makes this method a valuable complement to published BST methods. It should also be emphasized that in the absence of exogenous NO-donor, the proteins identified in Tables 1 and S1 were identified and observed not to be nitrosated.
Table 1.
Nitrosated proteins detected by d-Switch analysis of ~20 kDa gel band from cell lysates after incubation of SH-SY5Y neuroblastoma cells with CysNO.
| Ave. mass | Sequence coverage | Peptides | % Nitrosation | |
|---|---|---|---|---|
| Glutathione-S-transferase P1-1 (GST-P1) | 23724.66 | 69% | ASC47LYGQLPK | 50.5 |
| Cofilin 1(CFL1) | 18871.37 | 59% | AVLFC39LSEDKK | 37.5 |
| Ubiquitin carboxyl-terminal esterase L1 (UCHL1) | 25443.28 | 53% | QTIGNSC90GTIGLIHAVANNQDK NEAIQAAHDAVAQEGQC152R |
66.6 48.0 |
| PARK7 Protein DJ-1 | 20134.87 | 64% | VTVAGLAGKDPVQC46SR DVVIC53PDASLEDAKK |
28.6 56.2 |
| Chromobox protein homolog 3 (CBX3) | 21055.23 | 38% | GFTDADNTWEPEENLDC69PELIE AFLNSQ |
16.6 |
| Phosphatidylethanolamine binding protein 1(PEB1) | 21175.52 | 72% | APVAGTC168YQAEWDDYVPK | < 10 |
| Transgelin-2 (TAGLN2) | 21329.69 | 58% | QYDADLEQILIQWITTQC38R DGTVLC63ELINALYPEGQAPVKK |
< 10 |
| Ribosomal protein S5 (RPS5) | 23120.19 | 43% | TIAEC172LADELINAAK | < 10 |
In summary, the simple d-Switch proteomic technique is introduced, using commercially available thiolalkylating isotopologues, which identifies and quantitates protein targets of S-nitrosation by measuring the relative amounts of each peptide-SNO versus peptide-SH. The method was used to measure GST-P1 nitrosation induced by 5 classes of NO-donors that were seen to be differentiated in extent of nitrosation and sensitivity to cysteine and oxygen. The methodology was further extended to neuronal cell culture, wherein proteomic analysis identified protein thiols that were either sensitive or insentitive to nitrosation by exogenous nitrosothiol; the in-sample quantitation of both nitrosated and non-nitrosated cysteines providing an advantage over other BST methods. Since all NO-donors induced significant thiol oxidation, parallel or consequent to S-nitrosation, the extension of the d-Switch method to allow quantitation of protein disulfide is important and is in progress.
METHODS
Materials
All chemicals and reagents were purchased from Sigma (St Louis, MO) or Aldrich (Milwaukee, WI) unless otherwise mentioned. d5-NEM was purchased from Cambridge Isotopes (Andover, MA). CysNO (66) and GTN (67) were synthesized as described in the literature. CysNO, GSNO, and DEA/NO were freshly prepared just before experiment and concentrations were determined spectrophotometrically prior to experiment according to the literature (68). Synthesis of ODZ1 is described in Supplemental Information.
GST-P1 expression and purification
Standard recombinant DNA techniques were followed as described by Sambrook et al (69) and detailed in the Supplemental Information as are detailed methods for NO treatment of intact E.coli cells and d-Switch analysislemental Information.
Biotin-switch (BST) technique
The Jaffrey et al. BST technique was followed with some modifications (13) as described in Supplemental Information. The reaction buffer used in BST and GST-P1 oxidation experiments was HEPES (pH 7.4, 250 mM), EDTA (1 mM) and neocuproine (0.1 mM).
Analysis of GST-P1 oxidation
For analysis of oxidation induced by NO-donors, GST-P1 was incubated with desired concentration of NO-donors at 37 °C for 30 min. SDS-PAGE was performed under non-reducing conditions followed by Coomassie blue staining.
d-Switch method for quantitation of nitrosation
All steps were performed in the dark in amber colored vials. Purified GST-P1 protein storage buffer was exchanged with reaction buffer containing 40 mM ammonium bicarbonate, 1mM EDTA and 0.1 mM neocuproine at pH 7.4. Additional use of DTPA in the reaction buffer did not perturb the degree of nitrosation observed on reaction of GST-P1 with CysNO (Supplemental Information). After incubation with NO-donor for 30 mins, the unreacted thiols were blocked with 20 mM NEM and 5% SDS by alternating between sonicating and shaking in a 55 °C water bath for 30 min. The unreacted NEM was removed with Microcon YM-10 filter device and the protein was eluted with 40 mM ammonium bicarbonate. For the labeling step, a saturated solution of CuCl (100 μM) was prepared as described (38). The labeling reaction was carried out at 25 °C for 60 mins with 5 mM sodium ascorbate, 1 μM CuCl and 2 mM d5-NEM. The fresh samples were then run on SDS-PAGE and subjected to trypsinolysis using pierce in-gel digestion kit (cat # 89871). The samples were then analyzed using an Agilent 1100 series LC coupled to an Agilent 6310 ESI Ion Trap mass spectrometer in positive ion mode. The precision of the analysis is high for replicate experiments on the same batch of recombinant protein as reported.
Cell culture and extract preparation for MS analysis
SH-SY5Y cells were treated with CysNO for 20 min. After the treatment, cells were washed, lysed, and centrifuged. Precipitated protein was collected, washed and treated with labeling solution (1mM biotin HPDP/or 2mM d5-NEM, 5 mM sodium ascorbate, 1μM CuCl) and incubated at 25 °C for 60 min. Samples were run on SDS gel and the GST-P1 containing region was excised and subjected to in-gel digestion.
Mass spectroscopic analysis of tryptic digests
The tryptic digest samples were analyzed by using a hybrid linear ion trap FT-ICR mass spectrometer (LTQ-FT-ICR, Thermo Electron Corp., Bremen, Germany) equipped with a nanospray source, and nano-HPLC with autosampler (Dionex, Sunnyvale, CA). Details for MS analysis and database searching are provided in Supplemental Information.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by NIH grants CA 102590 and CA 121107 and the University of Illinois at Chicago (UIC) Center for Clinical and Translational Science (CCTS), Award Number UL1RR029879. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Rod Davis and the RRC facility at the University of Illinois at Chicago for providing necessary advice whenever needed.
Abbreviations
- GST-P1
glutathione-S-transferase P1-1
- BST
biotin switch
- CysNO
S-nitrosocysteine
- GSNO
S-nitrosoglutathione
- ODZ1
3-phenyl-4-furoxanmethanol
- GTN
glyceryl trinitrate (nitroglycerin)
- IAN
isoamyl nitrite
- DEA/NO
2-(N,N-diethylamino)-diazenolate-2-oxide.diethylammonium salt
- GSH
reduced glutathione
- Cys
cysteine
- DTPA
diethylenetriamine pentaacetic acid
- DTT
dithiothreitol
- Biotin HPDP
N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide
- NEM
N-ethylmaleimide
- SILAC
stable isotope labeling with amino acids in cell culture
- FT-ICR
fourier transform ion cyclotron resonance
- LTQ
linear quadrupole ion trap
- ESI
electrospray ionization
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- CID
collision induced dissociation
- NMDA
N-methyl-d-aspartate
- EDTA
ethylenediamine tetraacetic acid
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- PMSF
phenylmethylsulphonyl fluoride
- LB
Luria-Bertani
- MMTS
S-methyl methanesulfonothioate
- DMEM
Dulbecco's modified eagle media
- iTRAQ™
isobaric tags for relative and absolute quantitation
- ROS
reactive oxygen species
Footnotes
Supporting Information Available. The Supporting Information includes the following: (1) figures and table, (2) experimental procedures, and (3) analytical and spectral characterization data.
References
- 1.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Stamler JS, Singel DJ, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 3.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem Biol. 5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane P, Hao G, Gross SS. S-nitrosylation is emerging as a specific and fundamental posttranslational protein modification: head-to-head comparison with O-phosphorylation. Sci STKE. 2001;2001:RE1. doi: 10.1126/stke.2001.86.re1. [DOI] [PubMed] [Google Scholar]
- 5.Hara MR, Thomas B, Cascio MB, Bae BI, Hester LD, Dawson VL, Dawson TM, Sawa A, Snyder SH. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc Natl Acad Sci U S A. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lima B, Forrester MT, Hess DT, Stamler JS. S-nitrosylation in cardiovascular signaling. Circ Res. 106:633–646. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YM, Talanian RV, Billiar TR. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell DA, Erwin PA, Michel T, Marletta MA. S-Nitrosation and regulation of inducible nitric oxide synthase. Biochemistry. 2005;44:4636–4647. doi: 10.1021/bi0474463. [DOI] [PubMed] [Google Scholar]
- 9.Tew KD. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 1994;54:4313–4320. [PubMed] [Google Scholar]
- 10.Lo Bello M, Nuccetelli M, Caccuri AM, Stella L, Parker MW, Rossjohn J, McKinstry WJ, Mozzi AF, Federici G, Polizio F, Pedersen JZ, Ricci G. Human glutathione transferase P1-1 and nitric oxide carriers; a new role for an old enzyme. J Biol Chem. 2001;276:42138–42145. doi: 10.1074/jbc.M102344200. [DOI] [PubMed] [Google Scholar]
- 11.Cesareo E, Parker LJ, Pedersen JZ, Nuccetelli M, Mazzetti AP, Pastore A, Federici G, Caccuri AM, Ricci G, Adams JJ, Parker MW, Lo Bello M. Nitrosylation of human glutathione transferase P1-1 with dinitrosyl diglutathionyl iron complex in vitro and in vivo. J Biol Chem. 2005;280:42172–42180. doi: 10.1074/jbc.M507916200. [DOI] [PubMed] [Google Scholar]
- 12.Tellez-Sanz R, Cesareo E, Nuccetelli M, Aguilera AM, Baron C, Parker LJ, Adams JJ, Morton CJ, Lo Bello M, Parker MW, Garcia-Fuentes L. Calorimetric and structural studies of the nitric oxide carrier S-nitrosoglutathione bound to human glutathione transferase P1-1. Protein Sci. 2006;15:1093–1105. doi: 10.1110/ps.052055206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 14.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett DM, Black SM, Todor H, Schmidt-Ullrich RK, Dawson KS, Mikkelsen RB. Inhibition of protein-tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J Biol Chem. 2005;280:14453–14461. doi: 10.1074/jbc.M411523200. [DOI] [PubMed] [Google Scholar]
- 16.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 17.Sayed N, Kim DD, Fioramonti X, Iwahashi T, Duran WN, Beuve A. Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circ Res. 2008;103:606–614. doi: 10.1161/CIRCRESAHA.108.175133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayed N, Baskaran P, Ma X, van den Akker F, Beuve A. Desensitization of soluble guanylyl cyclase, the NO receptor, by S-nitrosylation. Proc Natl Acad Sci U S A. 2007;104:12312–12317. doi: 10.1073/pnas.0703944104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riego JA, Broniowska KA, Kettenhofen NJ, Hogg N. Activation and inhibition of soluble guanylyl cyclase by S-nitrosocysteine: involvement of amino acid transport system L. Free Radic Biol Med. 2009;47:269–274. doi: 10.1016/j.freeradbiomed.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas DD, Miranda KM, Espey MG, Citrin D, Jourd'heuil D, Paolocci N, Hewett SJ, Colton CA, Grisham MB, Feelisch M, Wink DA. Guide for the use of nitric oxide (NO) donors as probes of the chemistry of NO and related redox species in biological systems. Methods Enzymol. 2002;359:84–105. doi: 10.1016/s0076-6879(02)59174-6. [DOI] [PubMed] [Google Scholar]
- 21.Thatcher GRJ. Organic nitrates and nitrites as stores of NO. In: van Fassen E, Vanin A, editors. Radicals for Life: the various forms of nitric oxide. Elsevier Science; Amsterdam: 2007. [Google Scholar]
- 22.Thatcher GRJ. An introduction to NO-related therapeutic agents. Curr Top Med Chem. 2005;5:597–601. doi: 10.2174/1568026054679281. [DOI] [PubMed] [Google Scholar]
- 23.Thatcher GRJ, Nicolescu AC, Bennett BM, Toader V. Nitrates and NO release: Contemporary aspects in biological and medicinal chemistry. Free Radic Biol Med. 2004;37:1122–1143. doi: 10.1016/j.freeradbiomed.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 24.Turnbull CM, Marcarino P, Sheldrake TA, Lazzarato L, Cena C, Fruttero R, Gasco A, Fox S, Megson IL, Rossi AG. A novel hybrid aspirin-NO-releasing compound inhibits TNFalpha release from LPS-activated human monocytes and macrophages. J Inflamm (Lond) 2008;5:12. doi: 10.1186/1476-9255-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rai G, Sayed AA, Lea WA, Luecke HF, Chakrapani H, Prast-Nielsen S, Jadhav A, Leister W, Shen M, Inglese J, Austin CP, Keefer L, Arner ES, Simeonov A, Maloney DJ, Williams DL, Thomas CJ. Structure mechanism insights and the role of nitric oxide donation guide the development of oxadiazole-2-oxides as therapeutic agents against schistosomiasis. J Med Chem. 2009;52:6474–6483. doi: 10.1021/jm901021k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyland E, Chasseaud LF. Enzyme-catalysed conjugations of glutathione with unsaturated compounds. Biochem J. 1967;104:95–102. doi: 10.1042/bj1040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang M, Shin YG, van Breemen RB, Blond SY, Bolton JL. Structural and functional consequences of inactivation of human glutathione S-transferase P1-1 mediated by the catechol metabolite of equine estrogens, 4-hydroxyequilenin. Biochemistry. 2001;40:4811–4820. doi: 10.1021/bi002513o. [DOI] [PubMed] [Google Scholar]
- 28.Chandrasena RE, Edirisinghe PD, Bolton JL, Thatcher GR. Problematic detoxification of estrogen quinones by NAD(P)H-dependent quinone oxidoreductase and glutathione-S-transferase. Chem Res Toxicol. 2008;21:1324–1329. doi: 10.1021/tx8000797. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, Li Q, Yang X, van Breemen RB, Bolton JL, Thatcher GR. Analysis of protein covalent modification by xenobiotics using a covert oxidatively activated tag: raloxifene proof-of-principle study. Chem Res Toxicol. 2005;18:1485–1496. doi: 10.1021/tx0501738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen JZ, De Maria F, Turella P, Federici G, Mattei M, Fabrini R, Dawood KF, Massimi M, Caccuri AM, Ricci G. Glutathione transferases sequester toxic dinitrosyl-iron complexes in cells. A protection mechanism against excess nitric oxide. J Biol Chem. 2007;282:6364–6371. doi: 10.1074/jbc.M609905200. [DOI] [PubMed] [Google Scholar]
- 31.De Maria F, Pedersen JZ, Caccuri AM, Antonini G, Turella P, Stella L, Lo Bello M, Federici G, Ricci G. The specific interaction of dinitrosyl-diglutathionyl-iron complex, a natural NO carrier, with the glutathione transferase superfamily: suggestion for an evolutionary pressure in the direction of the storage of nitric oxide. J Biol Chem. 2003;278:42283–42293. doi: 10.1074/jbc.M305568200. [DOI] [PubMed] [Google Scholar]
- 32.Patel HMS, Williams DLH. Nitrosation by alkyl nitrites. Part 3. Reactions with cysteine in water in the pH range 6–13. J. Chem. Soc., Perkin Trans. 1989;2:339–341. [Google Scholar]
- 33.Gasco A, Fruttero R, Sorba G, Di Stilo A, Calvino R. NO donors: focus on furoxan derivatives. Pure Appl Chem. 2004;76:973–981. [Google Scholar]
- 34.Mannick JB, Schonhoff CM. Nitrosylation: the next phosphorylation? Arch Biochem Biophys. 2002;408:1–6. doi: 10.1016/s0003-9861(02)00490-3. [DOI] [PubMed] [Google Scholar]
- 35.Hogg N. The biochemistry and physiology of S-nitrosothiols. Annu Rev Pharmacol Toxicol. 2002;42:585–600. doi: 10.1146/annurev.pharmtox.42.092501.104328. [DOI] [PubMed] [Google Scholar]
- 36.Arnelle DR, Stamler JS. NO+, NO, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch Biochem Biophys. 1995;318:279–285. doi: 10.1006/abbi.1995.1231. [DOI] [PubMed] [Google Scholar]
- 37.Thatcher GRJ, Weldon H. NO problem for nitroglycerin: organic nitrate chemistry and therapy. Chem. Soc. Rev. 1998;27:331–337. [Google Scholar]
- 38.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolescu AC, Zavorin SI, Turro NJ, Reynolds JN, Thatcher GRJ. Inhibition of lipid peroxidation in synaptosomes and liposomes by nitrates and nitrites. Chem Res Toxicol. 2002;15:985–998. doi: 10.1021/tx025529j. [DOI] [PubMed] [Google Scholar]
- 40.Nicolescu AC, Reynolds JN, Barclay LR, Thatcher GRJ. Organic nitrites and NO: inhibition of lipid peroxidation and radical reactions. Chem Res Toxicol. 2004;17:185–196. doi: 10.1021/tx034097p. [DOI] [PubMed] [Google Scholar]
- 41.Patel HMS, Williams DLH. Nitrosation by alkyl nitrites. Part 3. Reactions with cysteine in water in the pH range 6–13. J Chem Soc, Perkin Trans. 1989;2:339–341. [Google Scholar]
- 42.Lim CH, Dedon PC, Deen WM. Kinetic analysis of intracellular concentrations of reactive nitrogen species. Chem Res Toxicol. 2008;21:2134–2147. doi: 10.1021/tx800213b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lancaster JR., Jr. Nitroxidative, nitrosative, and nitrative stress: kinetic predictions of reactive nitrogen species chemistry under biological conditions. Chem Res Toxicol. 2006;19:1160–1174. doi: 10.1021/tx060061w. [DOI] [PubMed] [Google Scholar]
- 45.Keszler A, Zhang Y, Hogg N. Reaction between nitric oxide, glutathione, and oxygen in the presence and absence of protein: How are S-nitrosothiols formed? Free Radic Biol Med. 48:55–64. doi: 10.1016/j.freeradbiomed.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker K, Savvides SN, Keese M, Schirmer RH, Karplus PA. Enzyme inactivation through sulfhydryl oxidation by physiologic NO-carriers. Nat Struct Biol. 1998;5:267–271. doi: 10.1038/nsb0498-267. [DOI] [PubMed] [Google Scholar]
- 47.Ishii T, Sunami O, Nakajima H, Nishio H, Takeuchi T, Hata F. Critical role of sulfenic acid formation of thiols in the inactivation of glyceraldehyde-3-phosphate dehydrogenase by nitric oxide. Biochem Pharmacol. 1999;58:133–143. doi: 10.1016/s0006-2952(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 48.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell DA, Morton SU, Marletta MA. Design and characterization of an active site selective caspase-3 transnitrosating agent. ACS Chem Biol. 2006;1:659–665. doi: 10.1021/cb600393x. [DOI] [PubMed] [Google Scholar]
- 50.Xu H, Freitas MA. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adler V, Yin Z, Fuchs SY, Benezra M, Rosario L, Tew KD, Pincus MR, Sardana M, Henderson CJ, Wolf CR, Davis RJ, Ronai Z. Regulation of JNK signaling by GSTp. EMBO J. 1999;18:1321–1334. doi: 10.1093/emboj/18.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin Z, Ivanov VN, Habelhah H, Tew K, Ronai Z. Glutathione S-transferase p elicits protection against H2O2-induced cell death via coordinated regulation of stress kinases. Cancer Res. 2000;60:4053–4057. [PubMed] [Google Scholar]
- 54.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J Biol Chem. 2009;284:436–445. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diet A, Abbas K, Bouton C, Guillon B, Tomasello F, Fourquet S, Toledano MB, Drapier JC. Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J Biol Chem. 2007;282:36199–36205. doi: 10.1074/jbc.M706420200. [DOI] [PubMed] [Google Scholar]
- 56.Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–372. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 57.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camerini S, Polci ML, Restuccia U, Usuelli V, Malgaroli A, Bachi A. A novel approach to identify proteins modified by nitric oxide: the HIS-TAG switch method. J Proteome Res. 2007;6:3224–3231. doi: 10.1021/pr0701456. [DOI] [PubMed] [Google Scholar]
- 59.Han P, Chen C. Detergent-free biotin switch combined with liquid chromatography/tandem mass spectrometry in the analysis of S-nitrosylated proteins. Rapid Commun Mass Spectrom. 2008;22:1137–1145. doi: 10.1002/rcm.3476. [DOI] [PubMed] [Google Scholar]
- 60.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 61.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Butterfield DA, Gnjec A, Poon HF, Castegna A, Pierce WM, Klein JB, Martins RN. Redox proteomics identification of oxidatively modified brain proteins in inherited Alzheimer's disease: an initial assessment. J Alzheimers Dis. 2006;10:391–397. doi: 10.3233/jad-2006-10407. [DOI] [PubMed] [Google Scholar]
- 63.Castegna A, Thongboonkerd V, Klein J, Lynn BC, Wang YL, Osaka H, Wada K, Butterfield DA. Proteomic analysis of brain proteins in the gracile axonal dystrophy (gad) mouse, a syndrome that emanates from dysfunctional ubiquitin carboxyl-terminal hydrolase L-1, reveals oxidation of key proteins. J Neurochem. 2004;88:1540–1546. doi: 10.1046/j.1471-4159.2003.02288.x. [DOI] [PubMed] [Google Scholar]
- 64.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 65.Ito G, Ariga H, Nakagawa Y, Iwatsubo T. Roles of distinct cysteine residues in S-nitrosylation and dimerization of DJ-1. Biochem Biophys Res Commun. 2006;339:667–672. doi: 10.1016/j.bbrc.2005.11.058. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Y, Hogg N. The mechanism of transmembrane S-nitrosothiol transport. Proc Natl Acad Sci U S A. 2004;101:7891–7896. doi: 10.1073/pnas.0401167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meah Y, Brown BJ, Chakraborty S, Massey V. Old yellow enzyme: reduction of nitrate esters, glycerin trinitrate, and propylene 1,2-dinitrate. Proc Natl Acad Sci U S A. 2001;98:8560–8565. doi: 10.1073/pnas.151249098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu X, Murata LB, Weichsel A, Brailey JL, Roberts SA, Nighorn A, Montfort WR. Allostery in recombinant soluble guanylyl cyclase from Manduca sexta. J Biol Chem. 2008;283:20968–20977. doi: 10.1074/jbc.M801501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.