Abstract
High-throughput screening of National Cancer Institute libraries of synthetic and natural compounds identified the vinylogous ureas 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide (NSC727447) and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (NSC727448) as inhibitors of the ribonuclease H (RNase H) activity of HIV-1 and HIV-2 reverse transcriptase (RT). A Yonetani–Theorell analysis demonstrated that NSC727447, and the active-site hydroxytropolone RNase H inhibitor β-thujaplicinol were mutually exclusive in their interaction with the RNase H domain. Mass spectrometric protein footprinting of the NSC727447 binding site indicated that residues Cys280 and Lys281 in helix I of the thumb subdomain of p51 were affected by ligand binding. Although DNA polymerase and pyrophosphorolysis activities of HIV-1 RT were less sensitive to inhibition by NSC727447, protein footprinting indicated that NSC727447 occupied the equivalent region of the p66 thumb. Site-directed mutagenesis using reconstituted p66/p51 heterodimers substituted with natural or non-natural amino acids indicates that altering the p66 RNase H primer grip significantly affects inhibitor sensitivity. NSC727447 thus represents a novel class of RNase H antagonists with a mechanism of action differing from active site, diva-lent metal-chelating inhibitors that have been reported.
Although an absolute requirement for reverse transcriptase (RT)-associated ribonuclease H (RNase H) activity for human immunodeficiency virus (HIV) replication was documented almost two decades ago (1, 2), development of potent and selective RNase H inhibitors has been surprisingly slow compared with the nucleoside and non-nucleoside DNA polymerase inhibitors currently in clinical use. Recently, however, N-hydroxyimides (3, 4), diketo acids (5, 6), and dihydroxytropolones (7) have shown promise by specifically inhibiting RNase H activity of HIV-1 and HIV-2 RT, and in some instances acting synergistically with clinically approved RT inhibitors. The preliminary crystal structure of an N-hydroxyimide bound to the RNase H domain of HIV-1 RT (4) suggests that it sequesters the divalent metal cofactor, laying the foundation for rational design of improved inhibitors. Increasing the diversity of RNase H inhibitors and structures of their complexes with RT would significantly accelerate these efforts.
We report here the identification and characterization of the vinylogous ureas 2-amino-5,6,7,8-tetra-hydro-4H-cyclohepta[b]thiophene-3-carboxamide (NSC727447) and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (NSC727448) as novel HIV-1 and HIV-2 RNase H inhibitors. NSC727447 failed to inhibit both DNA polymerase and pyrophosphorolysis activities at a concentration of 50 μM, indicating preferential inhibition of RNase H function. NSC727447 and NSC727448 are structurally unrelated to the dihydroxytropolone β-thujaplicinol (NSC18806) that we previously identified by high-throughput screening of libraries of synthetic and natural products (7). A Yonetani– Theorell analysis indicated that NSC727447 and NSC18806 were mutually exclusive RNase H inhibitors.
In order to examine the NSC727447 binding site, we performed mass spectrometric protein footprinting based on biotin modification of exposed lysine residues in the free protein and the protein–inhibitor complex (8–10). Cys280 and Lys281, located in helix I of the thumb subdomain, were protected from modification by inhibitor binding. Proximity between the p51 thumb subdomain and the p66 RNase H domain implies that inhibitor binding adjacent to the catalytic center affects either divalent metal coordination or positioning of the nucleic acid substrate in the active site. Although DNA polymerase activity was less sensitive to inhibition, protein footprinting indicated that the analogous region of the p66 was affected by NSC727447 binding.
Although vinylogous ureas display moderate selectivity for the retroviral enzymes, cellular toxicity prevented selection of drug-resistant virus to locate the inactivating lesion. We therefore exploited structural data suggesting that the RNase H primer grip, located in the vicinity of the catalytic center, promotes catalysis by determining the trajectory of the RNA strand accessing the RNase H active site (11). Within this motif, modifying Tyr501 affects the activity of the purified enzyme in vitro (12) and virus replication kinetics in vivo (13). Tyr501 of HIV-1 RT was replaced with the aromatic residues phenylalanine or tryptophan. In addition, we incorporated the unnatural amino acids azido-phenylalanine (Az-Phe) and benzoyl-phenylalanine (Bp-Phe) by amber codon suppression with orthogonal tRNA:aminoacyl-tRNA synthetase pairs (14). A Tyr501Phe substitution reduced NSC727447 sensitivity 3-fold, while replacement with tryptophan and Az-Phe reduced this ~11- and 14-fold, respectively. A Tyr501Bp-Phe substitution exhibited the strongest effect, reducing potency from an IC50 of 6.6 μM to one of 196 μM. Conversely, replacing primer grip residue Thr473 with cysteine resulted in increased sensitivity to NSC727447.
Collectively, our data suggests that regions within the RNase H domain distinct from the catalytic center can be targeted by small-molecule antagonists. Structural alterations in the RNase H domain conferring resistance to NSC727447 furthermore highlight the advantages of unnatural amino acid mutagenesis in probing protein structure and function. The vinylogous ureas occupy a binding pocket outside the RNase H catalytic center and represent a novel and important class of allosteric RNase H antagonists to complement active-site-directed, metal-chelating inhibitors such as dihydroxytropolones, N-hydroxyimides, and diketo acids.
RESULTS AND DISCUSSION
Inhibition of HIV RNase H Activity by Vinylogous Ureas
NCI libraries of synthetic and natural compounds, totaling ~230,000, were screened robotically for HIV-1 RNase H inhibition as described (7, 15, 16). Primary hits were evaluated against the HIV-2 enzyme, while Escherichia coli and human RNases H were used as a counterscreen. This strategy identified the vinylogous ureas 2-amino-5,6,7,8-tetrahydro-4H-cyclohepta [b]thiophene-3-carboxamide (NSC727447) and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (NSC727448) as moderately potent RNase H inhibitors. The structures of both inhibitors and the sensitivity of retroviral, bacterial, and human RNases H are presented (Figure 1). Sensitivity of HIV-2 RT demonstrates that NSC727447 and NSC727448 do not occupy the non-nucleoside binding site of the DNA polymerase domain and allosterically affect RNase H activity as has been shown for other inhibitors (17).
Figure 1.
Inhibition of retroviral, bacterial, and human RNases H by vinylogous ureas NSC727447 and NSC727448. IC50 values are the average of triplicate analyses and were determined under initial-rate conditions from the slope of the time-dependent increase in fluorescence.
Vinylogous Ureas and Hydroxytropolones are Mutually Exclusive RNase H Inhibitors
To compare the properties of the vinylogous urea NSC727447 and β-thujaplicinol (NSC18806), a hydroxytropolone previously identified as a potent and selective HIV RNase H inhibitor (7), a Yonetani–Theorell analysis was performed (18) to detect the degree of synergy or exclusivity. If the two compounds bind in a mutually exclusive fashion, their effects upon enzyme velocity are additive, yielding a series of parallel lines when the data is plotted as the reciprocal of velocity vs concentration of the titrated inhibitor in the presence of a number of fixed concentrations of the second inhibitor. If, in contrast, two compounds bind independently of one another to the enzyme, a Yonetani–Theorell analysis produces a series of converging lines, as previously demonstrated with β-thujaplicinol and the non-nucleoside calanolide A(7). The data (Figure 2) shows a set of parallel lines, illustrating that the interaction term γ approaches infinity, suggesting that NSC727447 and NSC18806 are mutually exclusive inhibitors.
Figure 2.
Yonetani–Theorell plot for inhibition of HIV-1 RNase H activity in the presence of β-thujaplicinol (NSC18806) and vinylogous urea NSC727447. The inverse of the rate of RNase H cleavage (expressed as the change in relative fluorescence per minute) is plotted as a function of NSC727447 concentration at β-thujaplicinol concentrations of 0, 0.1, 0.2, and 0.4 μM. Concentrations of the titrated inhibitor NSC727447 were 0, 0.25, 0.5, 1, 2, 4, 8, and 16 μM. The assay was performed at fixed concentrations of substrate and enzyme (250 nM and 4 nM, respectively). The parallel set of lines indicates that the two compounds bind in a mutually exclusive fashion.
NSC727447 Affects neither Pyrophosphorolysis nor DNA Polymerase Activity
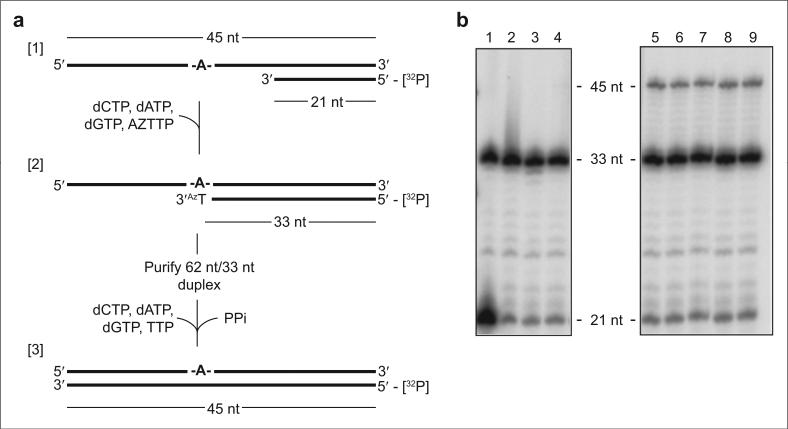
In order to examine the specificity of RNase H inhibition by NSC727447, RNA-dependent DNA polymerase activity was evaluated under conditions requiring prior removal of a chain-terminating nucleotide from the primer by pyrophosphorolysis. To achieve this, the DNA primer 3′ terminus of an RNA/DNA hybrid was blocked by incorporating the nucleoside RT inhibitor azidothymidine monophosphate (AZT-MP). Following purification of the blocked hybrid, DNA synthesis was examined in the presence of both a pyrophosphate donor and deoxynucleoside triphosphates (experimental system, Figure 3, panel a; results, Figure 3, panel b).
Figure 3.
RNA-dependent DNA polymerase activity of HIV-1 RT is not inhibited by NSC727447. a) Substrate was an end-labeled 21 nt DNA oligomer, annealed to a 45 nt RNA template (45R/21D RNA/DNA). In the first step, the DNA oligomer was extended to a 33 nt AZT-monophosphate-terminated primer. The resultant RNA/DNA hybrid (45R/33D) was purified, after which DNA synthesis was examined in the presence of a pyrophosphate donor and NSC727447. b) Lane 1, 21nt/33nt marker mix; lane 2, 45R/33D –RT, +PPi; lane 3, 45R/33D +RT, +PPI, –dNTPs; lane 4, 45R/33D –PPi, +RT, +dNTPs; lane 5, 45R/33D +RT, +PPI, +dNTPs, lanes 6–9, 45R/33D +RT, +PPI, +dNTPs, +NSC727447 at final concentrations of 50.0, 25.0, 12.5, and 6.25 μM, respectively.
In the absence of a pyrophosphate donor, a 33-nt product verifies primer extension and incorporation of AZT-MP as the P + 12 product, after which synthesis terminates. However, a 45-nt primer extension product is evident when the blocked substrate is provided a pyrophosphate donor and dNTPs. Under the same conditions, NSC727447 at a concentration of 50 μM failed to inhibit DNA synthesis. Although not shown here, a similar result was obtained with NSC727448. DNA polymerase activity was also determined in the absence of pyrophosphorolysis and proved likewise insensitive to inhibition at a concentration of 50 μM (data not shown). Under equivalent conditions, we determined an IC50 of ~10 nM for efavirenz, a nonnucleoside RT inhibitor currently used in combination antiretroviral therapy (data not shown).
Footprinting of NSC727447 on the p51 RT Subunit by Mass Spectrometry
Recently, we applied mass spectrometry to identify ligand binding sites on both HIV-1 RT (8) and integrase (9, 10). For RT, this involved (a) modification of surface-exposed lysines by N-hydroxysuccinimidyl-biotin (NHS-biotin) in the absence or presence of ligand, (b) electrophoretic separation of the p66 and p51 subunits, (c) in-gel proteolysis, and (d) resolution of peptide mixtures by mass spectrometry to differentiate lysines susceptible to modification in the protein–ligand complex from those shielded upon complex formation (Figure 4).
Figure 4.
Mass spectrometric protein footprinting strategy. Biotinylation reactions of free heterodimeric HIV-1 RT and of RT in complex with NSC727447 are performed in parallel, allowing accessible lysine residues to be covalently modified by NHS-biotin. RT subunits p66 and p51 are then separated by SDS-PAGE and subsequently subjected to proteolysis by trypsin. Comparative analysis of the generated peptide fragments by MALDI-TOF MS identifies lysine residues shielded from biotinylation in the RT/inhibitor complex.
The MALDI-TOF mass spectrum for tryptic fragments of the p51 subunit was examined (Figure 5). In the unmodified protein (Figure 5, panel c), peaks p3 (1389.8 Da) and p4 (1395.8 Da) represent peptides comprising residues 263–275 of the thumb subdomain (-L-N-W-AS-Q-I-Y-P-G-I-K-) and 144–154 of its fingers/palm junction (-Y-Q-Y-N-V-L-P-Q-G-W-K-), respectively. NHS-biotin treatment resulted in new modified peaks including p1 and p2 (Figure 5, panel a). In the RT/NSC727447 complex, p1 was fully shielded from modification while p2 remained susceptible to biotinylation (Figure 5, panel b). The mass of p2 (1342.7 Da) corresponded to the fingers subdomain peptide comprising amino acids 66–72 (-K-D-G-T-K-W-R-), within which Lys66 and Lys70 contained a biotin adduct. Peak p1 (1325.65 Da) corresponded to the tryptic peptide from thumb residues 278–284 (-Q-L-C-K-L-L-R-) plus two biotin molecules. Given that NHS-ester has been reported to be highly specific for primary amines and that there is only one lysine in p1, we subjected the peptide to postsource decay analysis to identify the sites of modification. The fragmentation pattern (Figure 5, panel d) confirmed the sequence of the peptide and revealed that Cys280 and Lys281 contained a biotin adduct. Equivalent results were confirmed by ESI-TOF MS/MS analysis (data not shown). Such a result was surprising, since to our knowledge there is no precedent indicating reactivity of cysteine to NHS-biotin. It has, however, been reported that for the reaction of NHS-esters of agarose derivatives, sulfhydryl groups can compete with free amino groups (19). To study this in more detail, a peptide comprising residues Val275– Arg284 was examined. Following NHS-biotin treatment and trypsin digestion, a species of mass 1325.65 Da was identified, corresponding to the tryptic product -QL-C-K-L-L-R- containing a biotin adduct on cysteine and lysine (Supplementary Figure 1). These results were fully consistent with the modification pattern of full length RT and confirmed that under our reaction conditions cysteine was reactive to NHS-biotin.
Figure 5.
MALDI-TOF analysis of RT tryptic peptides. a–c) Representative segments of the mass spectra. a) Free RT was treated with NHS-biotin. b) The RT–inhibitor complex was treated with NHS-biotin. c) Unmodified RT. The indicated peaks correspond to the following peptides: p1 = aa 278–284 containing the modified residues Cys280 and Lys281; p2 = aa 66–72 containing modified lysines Lys66 and Lys70; p3 = aa 264–275; p4 = aa 144–154. Comparison of the modification patterns of free RT (a) with the RT–inhibitor complex (b) reveals that p1 was specifically shielded in the preformed complex. Unmodified peptide fragments p3 and p4 (a, b, and c) serve as internal controls. d) Postsource decay data for p1. The fragmentation pattern of the parent ion (1326.5 Da) confirms the following sequence for the modified peptide: QL(C+biot.)(K+biot.)LLR. The “b” and “y” ions are derived from fragmentation of the peptide bonds and provide amino acid sequence information read from the peptide N-terminus toward C-terminus and the peptide C-terminus toward its N-terminus, respectively. During fragmentation of tryptic peptides the loss of ammonia from “b” and “y” ions is frequently observed. The respective peaks in our spectrum are accordingly labeled. We also detected the “a4 – NH3” ion, which resulted from the combined loss of the CO and NH3 groups from the “b4” ion. In addition to the conventional fragmentation pattern, PSD analyses commonly yield other internal fragmentation reactions. We did not attempt to identify these additional ion peaks because the assigned “b” and “y” ions provided unequivocal identification of the peptide sequence and modified residues. The spectrum in the range of 0–1300 m/z has been magnified to better visualize the fragmented ions.
Of those residues readily accessible to biotinylation in free RT, only Cys280 and Lys281 in the thumb subdomain were selectively protected upon binding of NSC727447 (region highlighted in Figure 6, together with RNase H primer grip residues Gln500 and Tyr501 and the active site carboxylates, Asp443, Glu478 and Asp498). Crystallography of HIV-1 RT containing duplex DNA (20, 21) or an RNA/DNA hybrid (11) indicates that the p51 subunit contacts the p66 RNase H domain. Subunit-selective mutagenesis experiments (22) have demonstrated that modifying residue Trp266 of p51 α-helix H affects RNase H activity, and C-terminal truncations of the p51 RT subunit are linked with defects in RNase H activity (23). Such lines of evidence support the notion that ligand binding to the p51 thumb can directly affect activity of the p66 RNase H domain, possibly by altering positioning of the scissile bond of an RNA/DNA hybrid at the active site. However, our data does not indicate whether the p51 peptide identified here by mass spectrometry is the primary ligand binding site or whether binding at one site induces a local conformational change affecting neighboring residues. In order to determine whether Cys280 of the p51 RT subunit was directly involved in NSC727447 binding, we examined NSC727447 sensitivity of an HIV-1 RT variant from which Cys38 and Cys280 had been removed from both the p66 and p51 subunits (Supplementary Figure 4). This mutant enzyme retained sensitivity to NSC727447, excluding its direct binding to Cys280.
Figure 6.
Structure of HIV-1 RT depicting, in magenta, residues of the p66 and p51 thumb subdomains whose biotinylation is affected by NSC727447 binding. p66 subdomains have been color-coded blue (fingers), red (palm), green (thumb), yellow (connection), and gold (RNase H), For simplicity, only the p51 thumb is color-coded in green. Catalytic carboxylates of the RNase H domain are in cyan and Gln500 and Tyr501 of the RNase H primer grip in white. This figure was generated based on PDB entry 1RTD using Discovery Studio 2.0 software (Accelrys).
Footprinting the NSC727447 Binding Site on the p66 Subunit
When mass spectrometric protein footprinting was applied to the p66 RT subunit, an equivalent peptide of mass 1325.65 Da (i.e., peak p1) was detected and protected from biotinylation in the presence of NSC727447 (Supplementary Figure 3). Since (a) the C-terminal tryptic fragment of p51 was not identified during our analysis of the p66 subunit and (b) the p51 and p66 subunits were electrophoretically separated prior to proteolytic digestion, we believe it unlikely that such a result reflects contamination of p66. Footprinting was also performed in a 20-fold excess of inhibitor to account for possible differences in affinity for p66 and p51. Our data therefore suggests that a second and equivalent NSC727447 binding site is located within the p66 thumb. Since the overall geometry of the p51 and p66 thumb subdomains is similar, such a result might not be entirely unexpected. Alternatively, we cannot rule out an allosteric effect induced by NSC727447 binding in the vicinity of the RNase H domain, which results in protection of Cys280 and Lys281 of p66 from chemical modification. However, insensitivity of DNA polymerase activity to NSC727447 (Figure 3), together with decreased sensitivity to inhibition upon introducing mutations into the RNase H domain (see below) suggests the p66 binding site does not contribute significantly to the inhibitory mechanism of NSC727447.
Altering Tyr501 of the RNase H Primer Grip Confers NSC727447 Resistance
Tyr501 of the RNase H primer grip of p66 RT has been implicated in controlling nucleic acid geometry to position the scissile phosphodiester bond at the RNase H catalytic center (11). Mutagenesis of Tyr501 interferes with RNase H activity in vitro (12), as well as virus replication (13) and AZT sensitivity (24) in vivo. Given the intimate interaction between the p51 thumb subdomain and p66 RNase H domain, we next examined whether introducing novel functionality or steric bulk into the RNase H primer grip affected NSC727447 activity. In addition to introducing the natural amino acids phenylalanine and tryptophan, we exploited genetically engineered E. coli containing orthogonal tRNA/aminoacyl tRNA synthetase pairs (25–28) to site-specifically introduce the non-natural phenylalanine analogs Az-Phe and Bp-Phe via nonsense suppression. Reconstituted p66/p51 heterodimers (29) were purified and examined for inhibitor sensitivity.
NSC727447 inhibited wild type RT with an IC50 of 6.6 μM (Figure 7). Replacing Tyr501 with phenylalanine increased the IC50 approximately 3-fold (20.0 μM), indicating that removing the hydroxyl function was relatively benign. However, Tyr501Trp and Tyr501Az-Phe mutations induced an ~11-fold and 14-fold decrease in inhibitor potency (70.6 and 89.6 μM, respectively). Steric bulk introduced by replacing Tyr501 with tryptophan or rigidity introduced by the azido group of Az-Phe may subtly alter inhibitor orientation or restrict its access to the binding site. Although we cannot differentiate between these possibilities, the consequences of increasing steric bulk in the RNase H primer grip were further augmented by p66501BpF/p51 RT, which was almost 30-fold resistant to NSC727447 (196.7 μM). A similar trend was noted with NSC727448, which inhibited wild-type RT with an IC50 of ~5.5 μM and Bp-Phe-substituted enzyme with an IC50 of ~220 μM (data not shown). Together with the biotinylation data (Figure 6), we suggest that substituting Bp-Phe for Tyr501 of the RNase H primer grip establishes a “gate” at the door of the inhibitor binding cleft. Moreover, the effect of RNase H primer grip mutations on inhibitor sensitivity supports our contention that their major effect is associated with the p51 thumb subdomain.
Figure 7.
Sensitivity of HIV-1 RT Tyr501 variants to NSC727447 inhibition. Each IC50 value reported is the average of three independent experiments.
NSC727447 Destabilizes Nucleic Acid Binding to RNase H Primer Grip Mutants
Modifying RNase H primer grip residue Thr473 reduces affinity for nucleic acid in vitro (12) and also impairs virus replication kinetics (13). We therefore examined the affinity of a Thr473Cys HIV-1 RT mutant for nucleic acid in the presence of NSC727447. Substrate for these experiments was a 14 nt RNA–8 nt DNA chimera hybridized to a 22 nt DNA, which mimics removal of the tRNA primer during reverse transcription (30). In order to avoid substrate degradation, gel-mobility shift experiments were performed in a Glu478Gln RNase H– background mutation (1). Under the conditions employed here, the affinity of both wild-type RT and the Glu478Gln mutant for our substrate was unaffected in the presence of 50 μM NSC727447 (Supplementary Figure 2, panels a and b). Although replacing primer grip residue Thr473 with cysteine did not affect nucleic acid binding in the absence of inhibitor, dissociation of the nucleoprotein complex was evident at NSC727447 concentrations as low as 1.6 μM and virtually complete at 50 μM (Supplementary Figure 2, panel c). Thr473Cys RT was also 50-fold more sensitive toward inhibition by vinylogous ureas than the wild-type enzyme (Supplementary Figure 5). Tyr501 and Thr473 contact the primer backbone at position –13 and –14, respectively, that is, 3 bp from the scissile bond. The effect of two RNase H primer grip mutations on inhibitor sensitivity support our contention that vinylogous ureas affect RNase H activity by interacting with the p51 thumb.
In summary, targeting regions of RT in the vicinity of the RNase H domain represents a novel and attractive means of allosterically inhibiting activity, in a manner formally analogous to non-nucleoside inhibition of DNA polymerase function (31). Jochmans et al. (32) have designated indolopyridone-1 (INDOPY-1) a nucleotide-competing RT inhibitor (NcRTI) in that it competes at the active site with the incoming dNTP. In addition, Sluis-Cremer et al., have shown that N3-substituted TSAO-T derivatives disrupt dimerization (33). Together with our report of an RNase H inhibitor targeting a site outside the catalytic center, these studies illustrate that additional inhibitor binding “pockets” on HIV-1 and HIV-2 RT remain to be exploited. This is of particular importance for the RNase H domain of RT, for which so far no potent and selective antagonists are known. The vinylogous ureas reported here represent a novel class of RNase H inhibitors distinct from previously reported active-site directed, metal-chelating antagonists such as dihydroxytropolones, N-hydroxyimides, and diketo acids. Remarkably, while uncovering an inhibitor binding site in the p51 subunit was unexpected, recent data suggests that acyl hydrazones likewise inhibit RNase H function through an interaction with the p51 thumb and also possess a second binding site in the p66 thumb that is unrelated to RNase H inhibition (M. Parniak, University of Pittsburgh, personal communication).
METHODS
Materials
All chemicals were from Sigma unless otherwise stated. The bacterial expression vector pRSET was obtained from Invitrogen and vector pPR-IBA-2 from IBA. Strep-Tactin superflow Sepharose and desthiobiotin were from IBA. dNTPs were purchased from Roche. The Dual Media Set for protein expression by autoinduction was obtained from Zymo Research. Synthetic oligonucleotides were from IDT and oligonucleotides for the fluorescence-based RNase H assay from TriLink Biotechnologies. Sulfo-NHS-biotin was obtained from Pierce. The synthetic peptide NH2-VRQLCKLLR-CO2H was from GenScript Corporation. Plasmids for incorporating p-benzoyl-l-phenylalanine (Bp-Phe) and p-azido-l-phenylalanine (Az-Phe) (14) were provided by Dr. Peter G. Schultz, Scripps Research Institute, La Jolla, CA. Bp-Phe was purchased from Bachem and Az-Phe from Chem-Impex. Vinylogous ureas NSC727447 and NSC727448 were obtained from ChemNavigator.
Expression and Purification of HIV-1 RT Variants
The coding region of RT p66 was cloned into the vector pRSET to introduce a C-terminal (His)6 affinity tag and that of p51 RT into the bacterial expression vector pPR-IBA2, introducing an N-terminal Streptag. For incorporation of non-natural amino acids, the Tyr501 codon was mutated to TAG, generating plasmid pRSET-p66His-501Stop, which was cotransformed with either pSup-BpaRS-6TRN or pSup-pAzPheRS-6TRN into bacterial strain BL21(DE3) (Novagen/EMD Biosciences). Transformants were selected on LB agar containing 80 μg mL–1 ampicillin and 50 μg mL–1 chloramphenicol. A single colony was used to inoculate 250 mL of Expansion Broth containing 50 μg mL–1 ampicillin, 50 μg mL–1 chloramphenicol, and 1 mM of the unnatural amino acid and grown with shaking at 230 rpm. Overexpression Broth (750 mL) containing 50 μg mL–1 ampicillin and 50 μg mL–1 chloramphenicol was then added, and the culture was grown in baffled flasks for 18 h at 37 °C.
Mutations Tyr501Trp and Tyr501Phe of p66 were introduced using the QuikChange site-directed mutagenesis kit. For the expression of wild-type p51 and p66 mutants, BL21(DE3) cells were transformed with the respective plasmids. Protein expression was performed according to the autoinduction protocol described above, except that media contained only 50 μg mL–1 ampicillin and chloramphenicol was withheld. Enzyme purification was as previously described (34).
FRET-Based RNase H Assay
RNaseH assays were performed as described (16) using an 18-nucleotide 3′-fluorescein-labeled RNA annealed to a complementary 18-nucleotide 5′-dabcylconjugated DNA. The increase in fluorescence as a result of RNase H hydrolysis was monitored with a Spectramax Gemini EM fluorescence spectrometer. Data were analyzed using the instrument manufacturer's SoftMax pro software. To determine IC50 values, the slope of the curve representing the time-dependent increase in fluorescence was determined at 10 min, when initial rate conditions are met and substrate depletion is not significant (16). The slope values were plotted against the logarithm of the inhibitor concentrations, and IC50 values were determined using SigmaPlot software. The two previously characterized RNase H inhibitors β-thujaplicinol and manicol were used as positive controls, yielding IC50 values in agreement with previously published data (250 ± 23 nM for thujaplicinol and 0.6 ± 0.09 μM for manicol). For experiments comparing the inhibitory potency of NS727447 toward different RNases H (Figure 1), enzyme amounts were adjusted so that reactions without inhibitor resulted in comparable hydrolysis: HIV-1 RNase H (specific activity toward FRET substrate = 5.9 × 10–3 μmol mg–1 min–1), 7.5 nM; HIV-2 RNase H (specific activity = 2.02 × 10–3 μmol mg–1 min–1), 22 nM; human RNase H (specific activity = 37.5 × 10–3 μmol mg–1 min–1), 1.6 nM; E.coli RNase H (specific activity = 320 × 10–3 μmol mg–1 min–1), 0.9 nM. Substrate concentration for these assays was 0.16 μM.
Yonetani – Theorell Analysis
To determine whether the binding site for vinylogous ureas overlaps with that of the RNase H inhibitor β-thujaplicinol (7), RNase H activity was measured in the presence of varying concentrations of NSC727447 (0.25–16 μM) at fixed concentrations of β-thujaplicinol (0.1–0.4 μM) (18). The assay was conducted at fixed concentrations of RNA/DNA substrate (250 nM) and enzyme (4 nM). Hydrolysis was initiated by adding RNA/DNA hybrid and monitored for 60 min. Data analysis was performed with SigmaPlot (SPSS Inc.). The following equation was used for data an analysis:
where νij = enzyme velocity in the presence of both compounds at concentrations [I] and [J] and γ is the interaction term that defines the degree to which binding of one compound perturbs binding of the other.
RNA-Dependent DNA Polymerase Activity
A 33 nt AZT-monophosphate-terminated primer was generated by extending a 21 nt end-labeled DNA oligomer (5′-GAC AGG GAT GGA AAG GAT CAC-3′) hybridized to a 45 nt RNA template (5′-AUU CCC GUU UAC GCC UCU CCU GGU GAU CCU UUC CAU CCC UGU CCC-3′) in a reaction that used 300 nM primer/template, 2 mM dATP, dCTP, and dGTP, 2.5 mM AZTTP, and 75 nM RT in 50 mM Tris, pH 8.0, 80 mM NaCl, 8 mM MgCl2, and 5 mM DTT for 60 min at 37 °C, followed by phenol/chloroform extraction and dNTP removal via Sephadex G 50 chromatography. AZT-terminated primer was next extended for 20 min at 37 °C in the above buffer using 25 nM primer/template, 2 mM dNTPs, 1 mM NaPPi (pH 7.5), 21 nM RT, and a full complement of dNTPs, in the presence of 50, 25, 12.5, and 6.25 μM NSC727447. Reaction products were resolved by high-voltage gel electrophoresis.
Mass Spectrometric Protein Footprinting
NHS-biotin reacts with accessible primary amines, resulting in the covalent addition of a biotin molecule (adding a mass of 226.3 Da) and release of N-hydroxysuccinimide. Due to better solubility, we used Sulfo-NHS-biotin (Pierce). In the course of this study, we found that this reagent labels not only lysines and the protein N-terminus but, to a lesser extent, also accessible cysteine residues. In a 30 μL reaction volume, 7 μM RT and 1 μL of DMSO or inhibitor solution was incubated in the absence or presence of 600 μM sulfo-NHS-biotin in 50 mM HEPES, pH 8.0, for 30 min at room temperature. We have demonstrated previously that under these conditions, structural integrity of RT is preserved (8). Reactions for free RT and RT in complex with the inhibitor were carried out in parallel, using an inhibitor concentration 20 times above the IC50. The reactions were quenched by adding excess lysine in its free amino acid form. Protein was denatured by adding 2 μL of 20% SDS and 0.2 μL of 1 M DTT and incubating at 70 °C for 20 min. Free cysteine residues were then alkylated with 2 μL of 1 M iodoacetamide and incubated for 45 min at room temperature. The reaction was stopped with 100 mM DTT, and samples were concentrated in a SpeedVac and subjected to SDS-PAGE on 4–12% Bis-Tris gels. RT was visualized with Coomassie blue stain, and the p51 and p66 subunits were excised. Gel slices were destained with 50% acetonitrile/50% 50 mM NH4HCO3, followed by two washes with 50 mM NH4HCO3. The slices were then dehydrated with 100% acetonitrile, dried under vacuum, and subjected to proteolysis using 0.5 μg of trypsin (Roche) in 50 mM NH4HCO3 buffer for 16 h. Peptide spectra were recorded using a Kratos Axima-CFR mass spectrometer equipped with curved field reflectron (Kratos Analytical Instruments) and α-cyano-hydroxy-cinnamic acid as matrix. For quantitative analysis of biotinylated peptide peaks, at least two adjacent unmodified peptide peaks were used as controls. To confirm that sulfo-NHS-biotin also labels cysteine, we analyzed the synthetic peptide VRQLCKLLR as described above. After reduction and alkylation, the samples were desalted using ZipTip C18 tips (Millipore), prior to treatment with 0.2 μg of trypsin in 50 mM NH4HCO3.
Electrophoretic Mobility Shift Analyses
The stability of wild-type and mutant RTs on a model RNA/DNA hybrid in the presence of NSC727447 was evaluated by electrophoretic mobility shift analysis. Enzyme (250 nM) and RNA/DNA hybrid (50 nM (30)) were incubated in a buffer of 50 mM Tris/HCl, pH 7.8, and 50 mM NaCl and in the presence of NSC727447 at concentrations ranging from 0.6–50 μM. Nucleoprotein complexes were resolved by nondenaturing electrophoresis and visualized by autoradiography. RT mutant Thr473Cys was evaluated in the context of the RNase H-inactivating mutation Glu478Gln, the latter of which does not affect complex stability.
Supplementary Material
Acknowledgment
S. Le G. is supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, M.G. by a grant from the Canadian Foundation for AIDS Research, and M.K. by NIH Grant AI062520. We thank A. Wamiru for technical assistance. This project was supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Supporting Information Available: This material is free of charge via the Internet.
REFERENCES
- 1.Schatz O, Cromme FV, Gruninger-Leitch F, Le Grice SF. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 1989;257:311–314. doi: 10.1016/0014-5793(89)81559-5. [DOI] [PubMed] [Google Scholar]
- 2.Schatz O, Mous J, Le Grice SFJ. Inactivation of the RNase H domain of HIV-1 reverse transcriptase blocks viral infectivity. In: Papas T, editor. Oncogenesis and AIDS. Portfolio Publishing; Houston, TX: 1990. pp. 293–303. [Google Scholar]
- 3.Klumpp K, Hang JQ, Rajendran S, Yang Y, Derosier A, Wong Kai In P, Overton H, Parkes KE, Cammack N, Martin JA. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klumpp K, Mirzadegan T. Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr. Pharm. Des. 2006;12:1909–1922. doi: 10.2174/138161206776873653. [DOI] [PubMed] [Google Scholar]
- 5.Shaw-Reid CA, Feuston B, Munshi V, Getty K, Krueger J, Hazuda DJ, Parniak MA, Miller MD, Lewis D. Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry. 2005;44:1595–1606. doi: 10.1021/bi0486740. [DOI] [PubMed] [Google Scholar]
- 6.Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R, Olsen DB, Carroll SS, Embrey M, Wai JS, Miller MD, Cole JL, Hazuda DJ. Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J. Biol. Chem. 2003;278:2777–2780. doi: 10.1074/jbc.C200621200. [DOI] [PubMed] [Google Scholar]
- 7.Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA, Crouch RJ, McMahon JB, Beutler JA, Le Grice SF. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 2005;33:1249–1256. doi: 10.1093/nar/gki268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvaratskhelia M, Miller JT, Budihas SR, Pannell LK, Le Grice SF. Identification of specific HIV-1 reverse transcriptase contacts to the viral RNA:tRNA complex by mass spectrometry and a primary amine selective reagent. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15988–15993. doi: 10.1073/pnas.252550199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Jr., Le Grice SF, Kvaratskhelia M. Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6894–6899. doi: 10.1073/pnas.0400873101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams KL, Zhang Y, Shkriabai N, Karki RG, Nicklaus MC, Kotrikadze N, Hess S, Le Grice SF, Craigie R, Pathak VK, Kvaratskhelia M. Mass spectrometric analysis of the HIV-1 integrase-pyridoxal 5′-phosphate complex reveals a new binding site for a nucleotide inhibitor. J. Biol. Chem. 2005;280:7949–7955. doi: 10.1074/jbc.M413579200. [DOI] [PubMed] [Google Scholar]
- 11.Sarafianos SG, Das K, Tantillo C, Clark AD, Jr., Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001;20:1449–1461. doi: 10.1093/emboj/20.6.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rausch JW, Lener D, Miller JT, Julias JG, Hughes SH, Le Grice SF. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry. 2002;41:4856–4865. doi: 10.1021/bi015970t. [DOI] [PubMed] [Google Scholar]
- 13.Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J. Virol. 2003;77:8548–8554. doi: 10.1128/JVI.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 15.Chan KC, Budihas SR, Le Grice SF, Parniak MA, Crouch RJ, Gaidamakov SA, Isaaq HJ, Wamiru A, McMahon JB, Beutler JA. A capillary electrophoretic assay for ribonuclease H activity. Anal. Biochem. 2004;331:296–302. doi: 10.1016/j.ab.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 16.Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA. A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal. Biochem. 2003;322:33–39. doi: 10.1016/j.ab.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Das K, Lewi PJ, Hughes SH, Arnold E. Crystallography and the design of anti-AIDS drugs: conformational flexibility and positional adaptability are important in the design of nonnucleoside HIV-1 reverse transcriptase inhibitors. Prog. Biophys. Mol. Biol. 2005;88:209–231. doi: 10.1016/j.pbiomolbio.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Yonetani T. The Yonetani-Theorell graphical method for examining overlapping subsites of enzyme active centers. Methods Enzymol. 1982;87:500–9. doi: 10.1016/s0076-6879(82)87028-6. [DOI] [PubMed] [Google Scholar]
- 19.Cuatrecasas P, Parikh I. Adsorbents for affinity chromatography. Use of N-hydroxysuccinimide esters of agarose. Biochemisry. 1972;11:2291–2299. doi: 10.1021/bi00762a013. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Harrison SC, Verdine GL. Trapping of a catalytic HIV reverse transcriptase*template:primer complex through a disulfide bond. Chem. Biol. 2000;7:355–364. doi: 10.1016/s1074-5521(00)00113-7. [DOI] [PubMed] [Google Scholar]
- 21.Jacobo-Molina A, Ding J, Nanni RG, Clark AD, Jr., Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao HQ, Boyer PL, Arnold E, Hughes SH. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J. Mol. Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 23.Jacques PS, Wohrl BM, Howard KJ, Le Grice SF. Modulation of HIV-1 reverse transcriptase function in “selectively deleted” p66/p51 heterodimers. J. Biol. Chem. 1994;269:1388–1393. [PubMed] [Google Scholar]
- 24.Delviks-Frankenberry KA, Nikolenko GN, Barr R, Pathak VK. Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J. Virol. 2007;81:6837–6845. doi: 10.1128/JVI.02820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Summerer D, Chen S, Wu N, Deiters A, Chin JW, Schultz PG. A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9785–9789. doi: 10.1073/pnas.0603965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 27.Xie J, Schultz PG. A chemical toolkit for proteins–an expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Alfonta L, Tian F, Bursulaya B, Uryu S, King DS, Schultz PG. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Grice SF, Naas T, Wohlgensinger B, Schatz O. Subunit-selective mutagenesis indicates minimal polymerase activity in heterodimer-associated p51 HIV-1 reverse transcriptase. EMBO J. 1991;10:3905–3911. doi: 10.1002/j.1460-2075.1991.tb04960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JS, Roth MJ. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3) J. Biol. Chem. 1992;267:15071–15079. [PubMed] [Google Scholar]
- 31.Zhou Z, Lin X, Madura JD. HIV-1 RT nonnucleoside inhibitors and their interaction with RT for antiviral drug development. Infect. Disord. Drug Targets. 2006;6:391–413. doi: 10.2174/187152606779025833. [DOI] [PubMed] [Google Scholar]
- 32.Jochmans D, Deval J, Kesteleyn B, Van Marck H, Bettens E, De Baere I, Dehertogh P, Ivens T, Van Ginderen M, Van Schoubroeck B, Ehteshami M, Wigerinck P, Gotte M, Hertogs K. Indolopyridones inhibit human immunodeficiency virus reverse transcriptase with a novel mechanism of action. J. Virol. 2006;80:12283–12292. doi: 10.1128/JVI.00889-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sluis-Cremer N, Hamamouch N, San Felix A, Velazquez S, Balzarini J, Camarasa MJ. Structure–activity relationships of [2′,5′-bis-O-(tert-butyldimethylsilyl)-β-d-ribofuranosyl]-3′-spiro-5″-(4″-amino-1″,2″-oxathiole-2″,2″-dioxide)thymine derivatives as inhibitors of HIV-1 reverse transcriptase dimerization. J. Med. Chem. 2006;49:4834–4841. doi: 10.1021/jm0604575. [DOI] [PubMed] [Google Scholar]
- 34.Klarmann GJ, Eisenhauer BM, Zhang Y, Sitaraman K, Chatterjee DK, Hecht SM, Le Grice SF. Site- and subunit-specific incorporation of unnatural amino acids into HIV-1 reverse transcriptase. Protein Expression Purif. 2004;38:37–44. doi: 10.1016/j.pep.2004.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.