Abstract
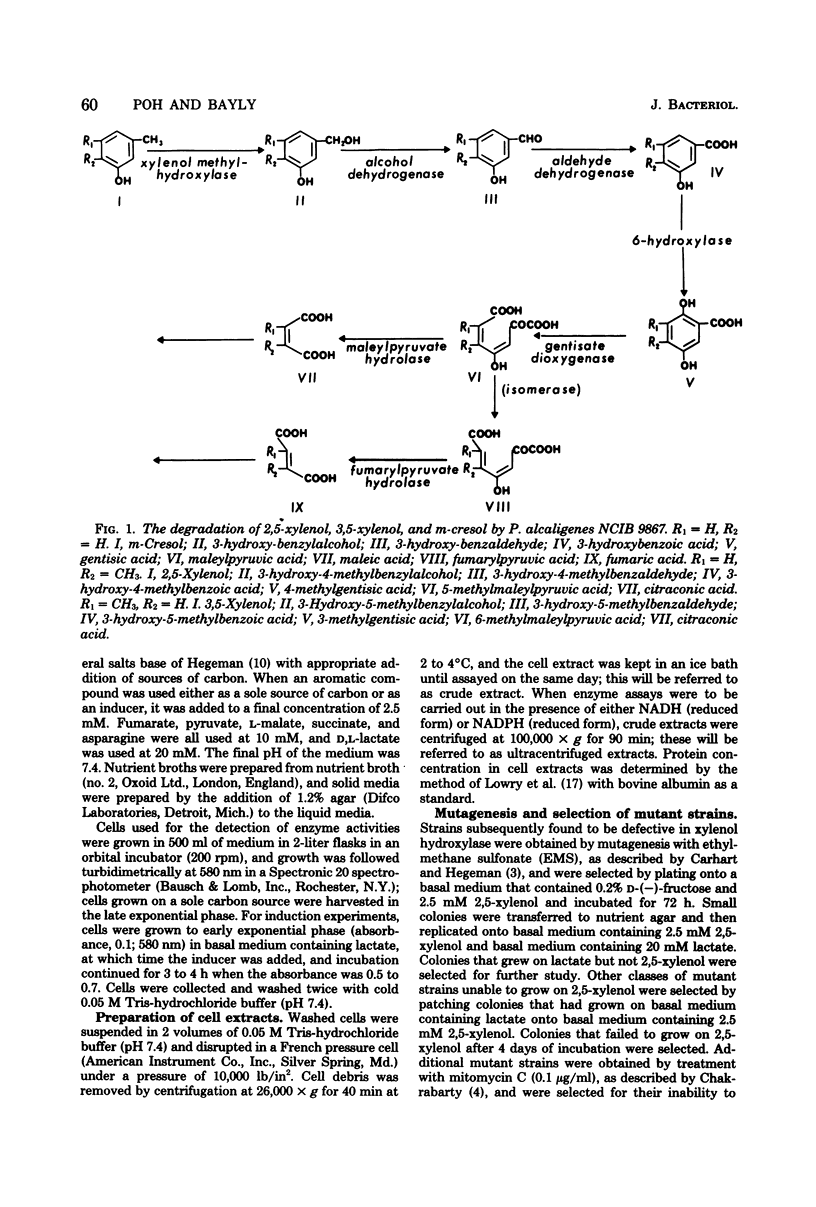
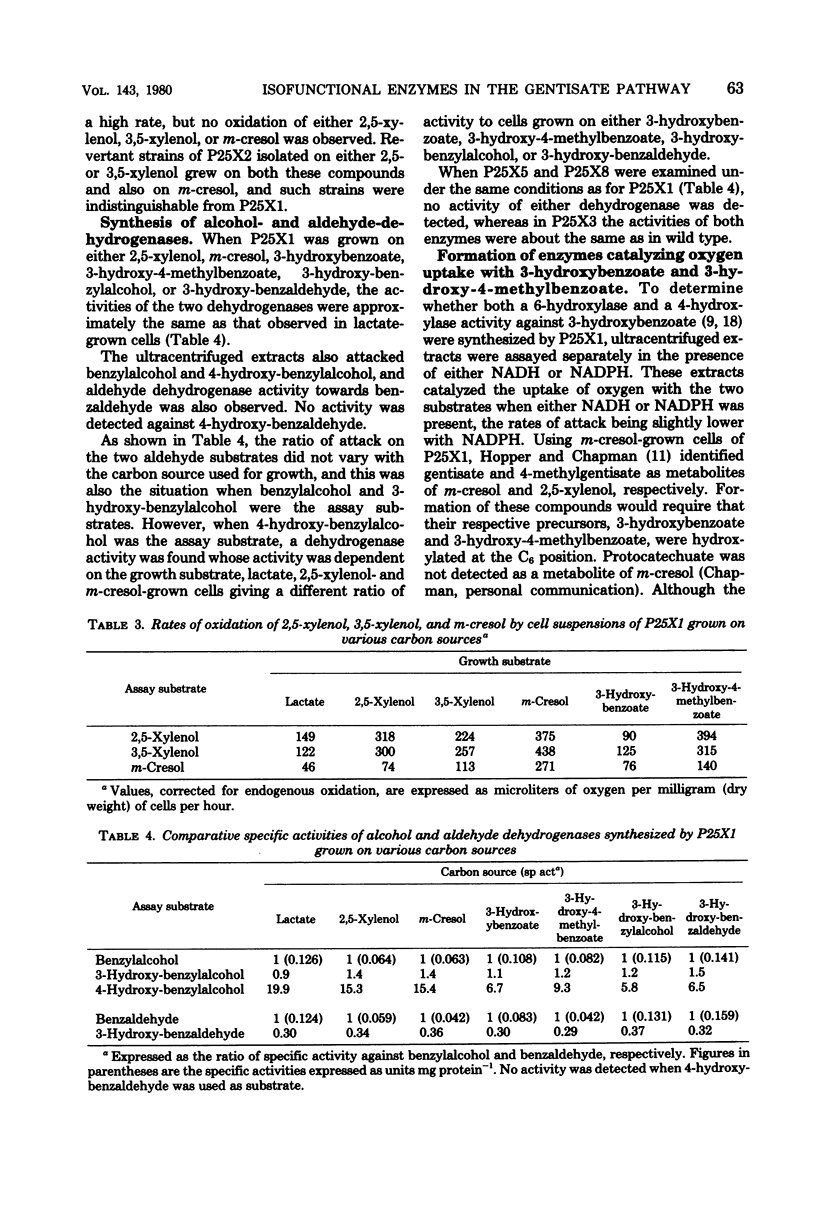
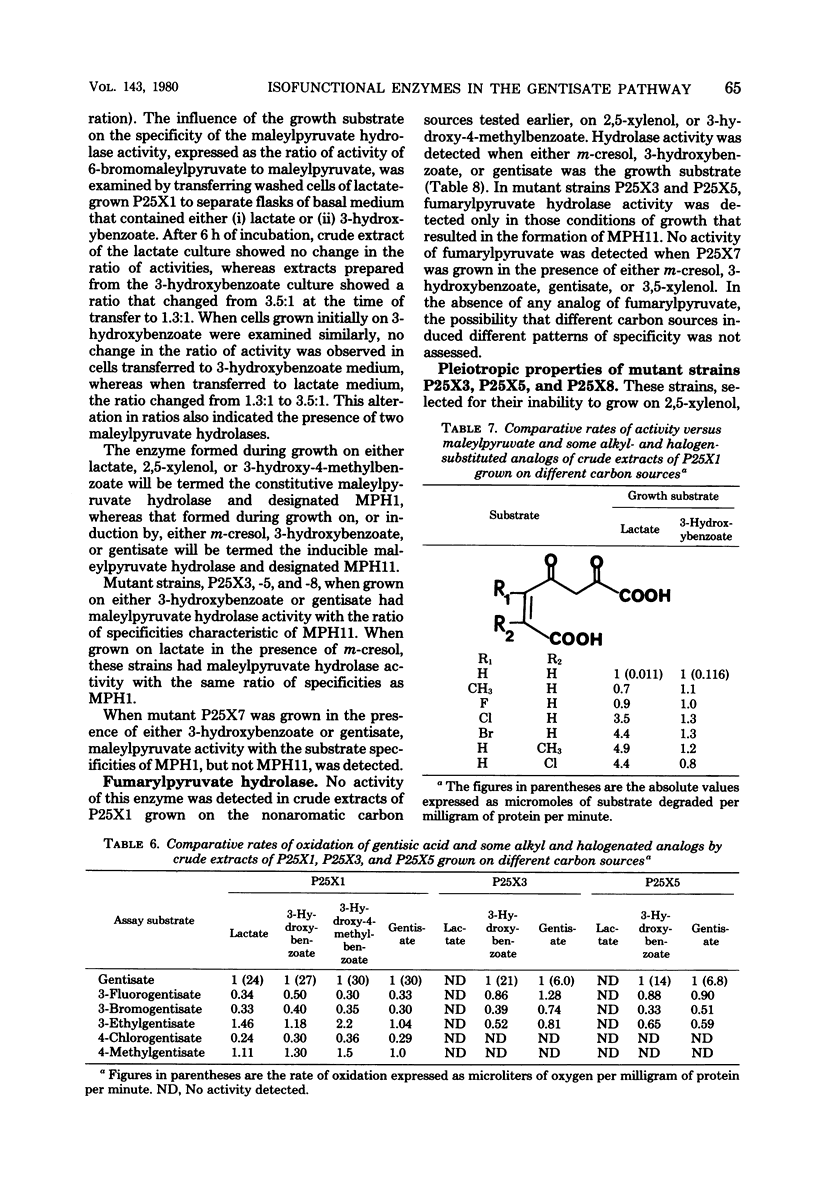
Study of the reaction sequence by which Pseudomonas alcaligenes (P25X1) and derived mutants degrade m-cresol, 2,5-xylenol, and their catabolites has provided indirect evidence for the existence of two or more isofunctional enzymes at three different steps. Maleylpyruvate hydrolase activity appears to reside in two different proteins with different specificity ranges, one of which (MPH1) is expressed constitutively; the other (MPH11) is strictly inducible. Two gentisate 1,2-dioxygenase activities were found, one of which is constitutively expressed and possesses a broader specificity range than the other, which is inducible. From oxidation studies with intact cells, there appear to be two activities responsible for the 6-hydroxylation of 3-hydroxybenzoate, and again a broadly specific activity is present regardless of growth conditions; the other is inducible by 3-hydroxybenzoate. Three other enzyme activities are also detected in uninduced cells, viz., xylenol methylhydroxylase, benzylalcohol dehydrogenase, and benzaldehyde dehydrogenase. All apparently possess broad specificity. Fumarylpyruvate hydrolase was also detected but only in cells grown with m-cresol, 3-hydroxybenzoate, or gentisate. Mutants, derived either spontaneously or after treatment with mitomycin C, are described, certain of which have lost the ability to grow with m-cresol and 2,5-xylenol and some of which have also lost the ability to form the constitutive xylenol methylhydroxylase, benzylalcohol dehydrogenase, benzaldehyde dehydrogenase, 3-hydroxybenzoate 6-hydroxylase, and gentisate 1,2-dioxygenase. Such mutants, however, retain ability to synthesize inducibly a second 3-hydroxybenzoate 6-hydroxylase and gentisate 1,2-dioxygenase, as well as maleylpyruvate hydrolase (MPH11) and fumarylpyruvate hydrolase; MPH1 was still synthesized. These findings suggest the presence of a plasmid for 2,5-xylenol degradation which codes for synthesis of early degradative enzymes. Other enzymes, such as the second 3-hydroxybenzoate 6-hydroxylase, gentisate 1,2-dioxygenase, maleylpyruvate hydrolase (MPH1 and MPH11), and fumarylpyruvate hydrolase, appear to be chromosomally encoded and, with the exception of MPH1, strictly inducible.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayly R. C., Chapman P. J., Dagley S., Di Berardino D. Purification and some properties of maleylpyruvate hydrolase and fumarylpyruvate hydrolase from Pseudomonas alcaligenes. J Bacteriol. 1980 Jul;143(1):70–77. doi: 10.1128/jb.143.1.70-77.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly R. C., McKenzie D. I. Catechol oxygenases of Pseudomonas putida mutant strains. J Bacteriol. 1976 Sep;127(3):1098–1107. doi: 10.1128/jb.127.3.1098-1107.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carhart G., Hegeman G. Improved method of selection for mutants of Pseudomonas putida. Appl Microbiol. 1975 Dec;30(6):1046–1047. doi: 10.1128/am.30.6.1046-1047.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. L. Degradation of 3-hydroxybenzoate by bacteria of the genus Bacillus. Appl Microbiol. 1975 Sep;30(3):439–444. doi: 10.1128/am.30.3.439-444.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. L., Frick T. D. Rapid spectrophotometric differentiation between glutathione-dependent and glutathione-independent gentisate and homogentisate pathways. Appl Environ Microbiol. 1977 Aug;34(2):170–174. doi: 10.1128/aem.34.2.170-174.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford R. L., Hutton S. W., Chapman P. J. Purification and properties of gentisate 1,2-dioxygenase from Moraxella osloensis. J Bacteriol. 1975 Mar;121(3):794–799. doi: 10.1128/jb.121.3.794-799.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseclose E. E., Ribbons D. W., Hughes H. 3-Hydroxybenzoate 6-hydroxylase from Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1973 Dec 10;55(3):897–903. doi: 10.1016/0006-291x(73)91228-x. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966 Mar;91(3):1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Chapman P. J., Dagley S. Enzymic formation of D-malate. Biochem J. 1968 Dec;110(4):798–800. doi: 10.1042/bj1100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Chapman P. J., Dagley S. The enzymic degradation of alkyl-substituted gentisates, maleates and malates. Biochem J. 1971 Mar;122(1):29–40. doi: 10.1042/bj1220029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Chapman P. J. Gentisic acid and its 3- and 4-methyl-substituted homologoues as intermediates in the bacterial degradation of m-cresol, 3,5-xylenol and 2,5-xylenol. Biochem J. 1971 Mar;122(1):19–28. doi: 10.1042/bj1220019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. P-cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem J. 1978 Nov 1;175(2):649–658. doi: 10.1042/bj1750649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. The aromatic alcohol dehydrogenases in Pseudomonas putida N.C.I.B. 9869 grown on 3,5-xylenol and p-cresol. Biochem J. 1978 Nov 1;175(2):659–667. doi: 10.1042/bj1750659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LACK L. Enzymic cis-trans isomerization of maleylpyruvic acid. J Biol Chem. 1961 Nov;236:2835–2840. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Ornston L. N., Ornston M. K., Chou G. Isolation of spontaneous mutant strains of Pseudomonas putida. Biochem Biophys Res Commun. 1969 Jul 7;36(1):179–184. doi: 10.1016/0006-291x(69)90666-4. [DOI] [PubMed] [Google Scholar]
- Wigmore G. J., Bayly R. C., Di Berardino D. Pseudomonas putida mutants defective in the metabolism of the products of meta fission of catechol and its methyl analogues. J Bacteriol. 1974 Oct;120(1):31–37. doi: 10.1128/jb.120.1.31-37.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]



