Summary
With the decipherment of the human genome, new questions have moved into the focus of today's research. One key aspect represents the discovery of DNA variations capable to influence gene transcription, RNA splicing, or regulating processes, and their link to pathology. Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS) is a powerful tool for the qualitative investigation and relative quantification of variations like single nucleotide polymorphisms, DNA methylation, microsatellite instability, or loss of heterozygosity. After its introduction into proteomics, efforts were made to adopt this technique to DNA analysis. Initially intended for peptide/protein analysis, it held several difficulties for application to nucleic acids. Today, MALDI-TOF-MS has reached worldwide acceptance and application in nucleic acid research, with a wide spectrum of methods being available. One of the most versatile approaches relies on primer extension to genotype single alleles, microsatellite repeat lengths or the methylation status of a given cytosine. Optimized methods comprising intelligent primer design and proper nucleotide selection for primer extension enabled multiplexing of reactions, rendering the analysis more economic due to parallel genotyping of several alleles in a single experiment. Laboratories equipped with MALDI-TOF-MS possess a universal technical platform for the analysis of a large variety of different molecules.
Key Words: MALDI-TOF, Mass spectrometry, Single nucleotide polymorphism, Methylation, Microsatellite instability, Genotyping
Zusammenfassung
Mit der Entzifferung des humanen Genoms sind neue Fragestellungen in den Mittelpunkt heutiger Forschung getreten. Eine zentrale Rolle spielt die Erforschung von Sequenzvariationen der DNA, die Einfluss auf die Transkriptionsrate, das Spleißen oder die Regulation verschiedener Gene haben und zu pathologischen Veränderungen führen können. Die Matrix-assistierte Laser-De-sorption-und-Ionisation-Flugzeit-Massenspektrometrie (MALDI-TOF-MS) hat sich als ein geeignetes Werkzeug nicht nur zur qualitativen Untersuchung, sondern auch zur relativen Quantifizierung von Einzelbasenaustauschen, DNA-Methylierung, Mikrosatelliteninstabilitäten oder dem Verlust der Heterozygotie erwiesen. Nach Einführung der ursprünglich für die Untersuchung von Peptiden und Proteinen entwickelten Methode wurde diese Technik für die Nukleinsäureanalyse weiterentwickelt und zur Anwendung gebracht. Nach anfänglichen Schwierigkeiten bei der Anpassung an die Nukleinsäu-reanalytik hat MALDI-TOF-MS mittlerweile eine große Akzeptanz und eine breite Anwendung in der Analyse dieser Molekülklasse erreicht. Die Hauptanwendungsgebiete basieren auf einer Primer-Verlängerungsreaktion die zur Genotypisierung einzelner Allele und zur Bestimmung der Mikrostellitenlänge oder des Cytosin-Methy-lierungsgrads verwendet werden. Derzeit stehen optimierte Methoden mit einem intelligenten Primer-Design und genauer Nukleotidzusammensetzung zur Primer-Verlängerung zur Verfügung, die eine Vervielfältigung der Reaktionen in einem Ansatz ermöglichen und somit durch parallele Genotypisierung mehrerer Allele Kosten einsparen. Insgesamt verfügen Labors mit einer MALDI-TOF-MS-Ausstattung über eine Plattform zur breiten Analyse von Makromolekülen verschiedenster Art.
Introduction
During the last 50 years, DNA research has unravelled many mysteries of this pivotal molecule, culminating in the complete sequencing of the human genome. With this knowledge, new interesting questions emerged concerning DNA variations as these are considered to play an important role in different types of diseases such as cancer [1, 2] or psychiatric disorders [3, 4]. Key variations of DNA like single nucleotide polymorphisms (SNPs), insertions and deletions (indels), microsatellites (MSs), and differences in the methylation status of important regions (e.g. CpG islands) moved into the focus of today's research. One of the most powerful alternatives to conventional Sanger or pyrosequencing addressing these aspects is matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). MALDI-TOF-MS was invented in the 1980s by different groups [5, 6]. Initially intended for peptide/protein analysis, it was predominantly used in proteomic approaches for protein identification and characterization, e.g. in combination with two-dimensional polyacrylamide gel electrophoresis [7, 8]. Its usability in genomic research, however, became obvious by the work of Tang and co-workers [9] and Tolson and Nicholson [10] who investigated DNA and RNA, respectively. In the following years, MALDI-TOF-MS was used for other molecules such as oligosaccharides and glycoproteins [11], lipids [12], and synthetic polymers [13]. Today, the most common application for MALDI-TOF-MS in genomic research is SNP genotyping. In combination with primer extension and polymerase chain reaction (PCR), this technique allows high-throughput screening for allelic variants. Although there are alternative methods for SNP analysis, like the Affymetrix 10 or 100 K chips [14], the Illumina bead arrays [15] or the ParAllele chips [16], these approaches are most efficient when several thousand SNPs in one sample are to be investigated in parallel. The relative cost per SNP on these platforms rises disproportionately if they are not used at full capacity. In contrast, the TaqMan™ assay (Applied Biosystems, Foster City, CA, USA) [17] or the MALDI-TOF-MS approaches [18] are most reasonable for genotyping a limited number of SNPs in a large number of different samples.
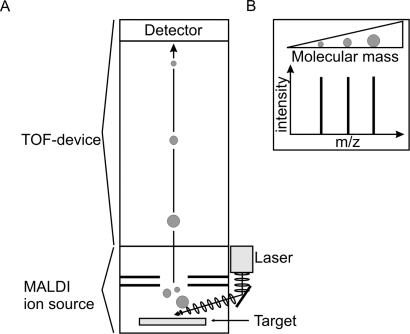
In general, for MALDI-TOF-MS analysis of single-stranded oligonucleotides the sample is spotted onto an appropriate matrix (e.g. 3-hydroxy-picolinic acid) on a MALDI target plate. Irradiation of the matrix-oligonucleotide-cocrystal with a laser beam of defined wavelength ultimately results in desorption and ionization of the oligonucleotide which then can be accelerated in an electrical field into the time-of-flight (TOF) device. The TOF device serves to separate the accelerated analyte ions of different mass-to-charge (m/z) ratios by providing a field-free drift tube of defined length (fig. 1A). After having passed the tube, ions are detected and every signal is assigned to a specific molecular mass calculated from the TOF (fig. 1B). Applying external calibration and/or internal standards, a mass deviation of as low as 5 ppm can be achieved. The resolution, however, largely depends on the size of the analyte, and therefore small molecules can be determined more exactly than large ones.
Fig. 1.
MALDI-TOF mass spectrometry. A Schematic drawing of a MALDI-TOF mass spectrometer. Analyte ions are generated by laser irradiation of the matrix-analyte-cocrystal within the ion source and accelerated into the TOF device. The detector transforms the electrical information of the striking analyte ions into a signal suitable for computational processing. B Schematic MALDI-TOF-MS mass spectrum of three different analytes. Their assignment to discrete m/z rations is calculated from their time of flight based on constant acceleration energy. Smaller ions are detected first, larger ones later.
SNP Analysis
SNPs are subtle variations of the DNA sequence with a prevalence of at least 1 in 100 within a given population, potentially causing phenotypic alterations in gene transcription or protein function. On average, they are found every 500-1,000 base pairs in the human genome. Generally, coding and non-coding SNPs are being discriminated. Depending on the position within the codon and the base exchange, the variation leads to a synonymous or non-synonymous amino acid sequence. Furthermore, SNPs being within the non-coding regions may affect the transcription of a gene by disrupting or creating regulatory sequence elements such as binding sites for transcription factors, enhancers, silencers, or splicing factors. In addition, synonymous coding SNPs are no longer termed silent variations as they may also affect splice enhancers or silencers. Thus, the screening for specific known SNPs in the course of association studies provides valuable information to determine a linkage to a given disorder, or to identify potential markers for clinical diagnosis, prognosis, monitoring, and forensic applications.
Besides the identification of variations directly involved in the susceptibility to a disorder, the comparative analysis of a set of well-known SNPs in affected and control persons can be used to investigate the association of a specific haplotype to a disorder. This is due to the observation that neighboring SNPs are most often transmitted as a block to the next generation. Therefore, only a subset of all known SNPs has to be analyzed to decipher the haplotype of the accompanying ones.
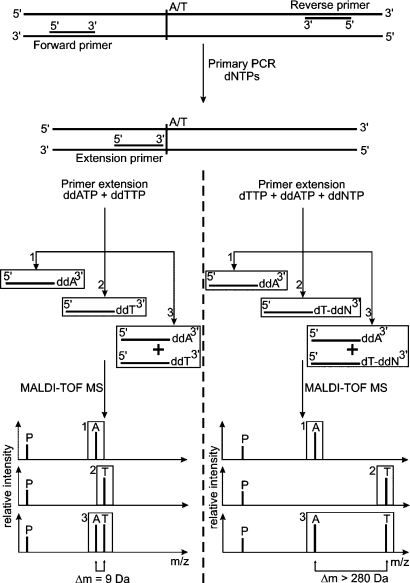
SNP genotyping is one of the most common applications for MALDI-TOF-MS in genomic research. Based on the method of primer extension [19], allelic variants of known SNPs can readily be identified. For this purpose, a genomic DNA template is used to amplify the region of interest containing the variable base. This is achieved by a regular PCR creating a fragment of usually about 300-400 base pairs. The double-stranded DNA is purified subsequently to avoid undesirable side products in the second step, which constitutes the primer extension reaction. The primer to be extended is designed in a way that its 3'-terminal base anneals directly in front of the investigated SNP. For the primer extension PCR, two strategies are applied (fig. 2). In the GOOD assay [20], the PinPoint assay [21] or the GenoSNIP assay [22], terminating dideoxynucleotides (ddNTPs) are added to the reaction mixture, interrupting primer extension immediately after the first nucleobase (fig. 2A). MALDI-TOF-MS determination of the extended primer molecular mass allows the identification of the allelic variant of the respective SNP. In this approach, however, the major drawback is the mostly small mass difference between the differentially extended primers, thus hampering analysis of the data. Especially low-molecular-weight adducts, such as sodium or potassium ions binding to the negatively charged DNA molecules, can lead to unde-sired mass shifts (+Na+ = M + 23 Da; +K+ = M + 39 Da) and thereby interfere with the informative mass signals. These circumstances raised the need for efficient sample purification. To space the alleles further apart and thereby enhance clear assignment of the mass peaks, other methods like the PROBE assay [23], the VSET assay [24], or the MassEXTEND assay [25] are used. Here, mixtures of deoxynucleotides (dNTPs) and appropriate ddNTPs are added to the primer extension reaction. Termination of the extension occurs at the first base of the template complementary to the ddNTP (fig. 2B). This results either in the addition of only the respective ddNTP or of at least two nucleotides (dNTP and ddNTP), thus spacing the two allelic variants about 300 Da apart. Special care has to be taken since the extension by dATP and ddGTP lead to the same mass shift. Primers extended by either dCTP or ddTTP are spaced by only 1 Da. Thus, discrimination of ddGTP or incomplete dATP extension is impossible, eventually leading to false results. The same holds true for the combination of dCTP and ddTTP. This problem can be circumvented by extension of one allele by three nucleobases or by designing a proper reverse extension primer. For molecular masses of nucleotides and the induced mass shift of an oligonucleotide upon their incorporation see table 1.
Fig. 2.
SNP genotyping by MALDI-TOF-MS. After primary PCR amplification of a DNA fragment harboring the SNP of interest, primer extension is carried out with either ddNTPs (left of dotted line) or a proper mixture of dNTP and ddNTP (right of dotted line). The first method results in an extension of a single nucleobase per genotype, resulting in a small difference (here 9 Da) of the respective primer molecular mass. The second method spaces one allele further apart (by at least 280 Da) by incorporation of two nucleobases as compared to a single one. This strategy facilitates data interpretation. For internal calibration, the mass of residual non-extended primer (P) can be used.
Table 1.
Molecular masses of nucleotidesa
| Nucleotide | Formula | Molecular mass, Da | Mass shift, Da |
|---|---|---|---|
| dATP | C10H16N5O12P3 | 491.2 | 313.2 |
| dTTP | C10H17N2O14P3 | 482.2 | 304.2 |
| dCTP | C9H16N3O13P3 | 467.2 | 289.2 |
| dGTP | C10H16N5O13P3 | 507.2 | 329.2 |
| ddATP | C10H16N5O11P3 | 475.2 | 297.2 |
| ddTTP | C10H17N2O13P3 | 466.1 | 288.2 |
| ddCTP | C9H16N3012P3 | 451.1 | 273.2 |
| ddGTP | C10H16N5O12P3 | 491.2 | 313.2 |
Chemical composition of deoxy- as well as dideoxynucleotides and their molecular masses. The value for the mass shift refers to the respective increase in molecular mass when incorporated into a single stranded oli-gonucleotide.
For large-scale SNP genotyping in a more economic way, the strategy of sample multiplexing was employed, which allows parallel detection of several allelic variants in only one reaction mix [26]. In recent years, much effort has been invested to optimize conditions for multiplexing MALDI-TOF-MS-based SNP genotyping. Routinely, it is possible to analyze simultaneously 3-5 SNPs in one experiment, thus lowering both working time and costs per SNP. Multiplexing to a higher degree is difficult due to the necessary increase in the number of extension primers. This raises the probability of primer interaction such as dimerization, leading to less efficient reactions or false-positive results due to unspecific primer extension. A major step towards solving these problems was the recent generation of algorithms for automated primer design and BLAST searches ensuring primer specificity [27]. These efforts resulted in the possibility of detecting as many as 20-30 SNP variations in a single experiment, although suitability for routine use remains to be shown. Reliable data interpretation, however, is dependent on both excellent mass accuracy as well as high resolution since molecular mass signals move closer together within spectra of highly multiplexed samples. Furthermore, the large number of molecular species may lead to suppression effects, and thus informative mass signals, although present in the sample, might not be visible. In general, with high number of simultaneously investigated SNPs, increased care has to be taken in establishing and validating PCR, e.g. classical gel-based control of the primary PCR product to ensure specificity and purity. Finally, one has to consider unknown SNPs within the primer binding sites, which lower the annealing efficiency and eventually impede either primary amplification or extension. To scale down the initial number of genotyping reactions, pooling of DNA samples has been applied successfully [25]. SNPs, considered to be interesting, can be investigated subsequently in separate experiments. It should be mentioned, however, that allelic variants with low prevalence will most probably be missed within pooled DNA samples. Moreover, when pooling samples, special care has to be taken to ascertain comparable quality as well as concentration of the samples to be pooled.
Several SNPs of diagnostic importance have been genotyped successfully with MALDI-TOF-MS. Among these were the clinically relevant sequence variations within genes for the blood clotting factors II and V [28]. Both factors enhance the risk of thrombosis in case of heterozygosity for the respective mutant allele. In factor V, the A allele of SNP G1691A results in resistance to proteolytic inactivation by activated protein C and therefore enhanced overall activity, concomitant with increased blood coagulation [29, 30]. The A variant of SNP G20210A within the 3'-untranslated region of the factor II gene results in elevated plasma prothrombin levels and thereby increases the risk of thrombosis [31, 32]. In the first genotyping approach, both SNPs were analyzed separately by MALDI-TOF-MS-based primer extension and were confirmed in all cases with classical sequencing. Moreover, it was possible to multiplex the reaction, enabling a simultaneous analysis of both SNPs.
Only recently, multiplexing was also applied by Garritsen and coworkers [33] to genotype six clinically relevant human platelet antigen (HPA) loci (HPA-1, HPA-2, HPA-3, HPA-4, HPA-5 and HPA-15). Genotyping of platelet antigens has nowadays become an important tool supporting diagnosis and therapy of various platelet disorders. Knowledge of the individual genotype of HLA is also valuable concerning improved success in platelet transfusion or stem cell transplantation.
Another evaluation of hematologically relevant SNPs was carried out by Welsby and coworkers [34] who identified genetic polymorphisms that are associated with bleeding after cardiac surgery. For this purpose, they screened 19 candidate genes based on a priori hypotheses about their potential role in postoperative bleeding outcome comprising several platelet glycoproteins, blood clotting factors and fibrinogen ß-chain, tissue factor pathway inhibitor, thrombomodulin plasminogen activator inhibitor type I, methylenetetrahydrofolate reductase, and angiotensin converting factor. Out of these, six turned out to be significantly associated with bleeding (table 2). Screening was performed using the Sequenom MassARRAY™ system (San Diego, CA, USA) combined with MALDI-TOF-MS analysis of the genotypes.
Table 2.
List of selected SNPs partially associated with disorders
| Disorder | Gene | Reference |
|---|---|---|
| Asthma | NOS1, EDN1, GPRA | [65,66] |
| Arrhythmias | KCNQ1 | [67] |
| Biliary cirrhosis | MBL | [68] |
| Bleeding after cardiac surgery | GPIaIIa, GPIbα, FII, FIII, TFPI | [34] |
| Blood pressure | TAF1 | [69] |
| Creutzfeldt-Jakob disease | PRNP | [70] |
| Hemochromatosis | HFE | [71] |
| Idiopathic arthritis | MIF | [72] |
| Lacunar stroke | MMP-2 | [73] |
| Lung cancer | MMP-1, p53 | [74,75] |
| Multiple sclerosis | IL2RA, IL7RA | [76] |
| Ossification | Npps | [77] |
| Systemic lupus erythematosus | Prolactin | [78] |
| Systemic sclerosis | Fibrillin1 | [79] |
| Thrombosis | FV, FII | [28,29] |
| Type II diabetes | Syntaxin 1A | [80] |
| N.d. | HLA-DQA1.2.3.4 | [81] |
| N.d. | HPA-1,-2,-3,-4,-5,-15 | [33] |
EDN1 = Endothelin 1; FII = factor II; FIII = factor III; FV = factor V; GPIaIIa = platelet glycoprotein IaIIa; GPIbα = platelet glycoprotein Ibα; GPRA = G-protein-related receptor for asthma; IL2/7RA = interleukin-2/-7 receptor alpha subunit; KCNQ1 = potassium channel protein; MBL = mannose binding protein; MIF = macrophage inhibitory factor; MMP-1/-2 = matrix metalloproteinase 1/2; N.d. = no specific disorder: genotyping for method evaluation purposes; NOS1 = neuronal nitric oxide synthetase 1; Npps = nucleotide pyrophosphatase; PRNP = prion protein; TAF1 = thrombin-activable fibrinolysis inhibitor; TFPI = tissue factor pathway inhibitor.
Slightly different, but also based on primer extension, was the strategy employed by Misra and colleagues [35] to genotype as many as 40 different SNP variants in the CYP2C9 gene and 50 variants in the CYP2A13 gene. Both gene products belong to the family of cytochrome P450 isoenzymes which are involved in the metabolism of 70-80% of all therapeutically relevant drugs [36]. For CYP2C9, it has been shown that allelic variants influence the activity of the protein for different substrates [37]. As yet, CYP2A13 has been less well characterized but plays a role in carcinogen metabolism [38]. For SNP genotyping, a highly multiplexed approach was chosen. To simplify the MALDI-TOF-MS analysis, extension primers were elongated with biotinylated ddNTPs to label specifically the extension products. Making use of the tight interaction between biotin and streptavidin, purification of the extended primers was carried out by solid-phase affinity chromatography. By this means, incomplete extension products were removed and a higher degree of multiplexing could be achieved.
These are, however, only two examples out of a large number of successfully genotyped SNPs for a broad variety of disorders making use of MALDI-TOF-MS-based techniques (table 2).
Taken together, MALDI-TOF-MS enables high-throughput genotyping of known SNPs. Results obtained with this method feature high reliability and reproducibility which renders it suitable for routine diagnostics in the clinical environment. By multiplexing, the relative costs per SNP can be reduced, but data interpretation becomes more difficult.
Additional Applications of MALDI-TOF-MS in Genomic Research
Microsatellite Instability
Microsatellite instability (MSI) represents another important genetic variation that was demonstrated to be linked to carcinogenesis [39]. MSs are short repetitive sequences present throughout the genome. There are considerable interindividual differences concerning the repeat number of a given microsatellite. Due to this phenomenon, they serve as valuable markers in several genetic applications, e.g. linkage analysis or paternity test. Even healthy individuals display variations especially for long microsatellites as these are hotspots for DNA polymerase slippage and are more difficult for the DNA mismatch repair system to restore. Increased variability within short repetitive MSs points to defects in this DNA mismatch repair system and is referred to as MSI. MSIs within coding MSs (cMSs) of crucial genes contribute to the pathogenesis of DNA mismatch repair-deficient cancers [40]. Particularly affected are MSs associated with the hereditary nonpolyposis colorectal cancer (HNPCC) syndrome [41, 42]. Genotyping of these MSIs is therefore increasingly used to support the molecular diagnosis of HNPCC. Furthermore, several cMS-harboring genes that encode proteins involved in e.g. DNA mismatch repair (hMSH3 and hMSH6) [43] are frequently mutated in MSI cancers.
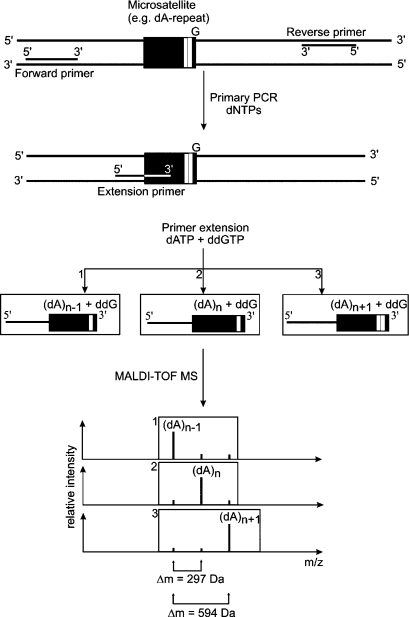
Classical techniques for detection of MSI comprise DNA sequencing or gel electrophoresis, both of which are relatively labor-intensive, time-consuming, and provide only little information on quantitative differences. Making use of MALDI-TOF-MS, the visualization of variability in several repeat lengths in a single experiment is possible (fig. 3). So far, two different approaches have been developed for the assessment of MSI. First, primer extension is employed with primers designed to anneal directly (or with partial overlap) upstream of the short nucleotide repeat. By choosing the proper combination of dNTPs and ddNTPs, the primer is extended according to the length of the repeat and terminated right after the first non-repeat nucleotide. This strategy has been used successfully to genotype five genomic MSIs linked to neoplastic lesions [44]. For the investigation of longer repeats, however, this approach is not suitable because of the restrictions in fragment size for MALDI-TOF-MS analysis. Moreover, including a long 3'-repetitive sequence within the extension primer may lead to partial mispairing and thus might produce heterogeneous results. In addition, one has to take into account the naturally occurring slippage of the DNA polymerase within repetitive sequences such as MSs. As a consequence, even when analyzing homogenous samples without MSI, a small percentage of the extended primers differ from their expected length displaying a population of shorter as well as longer species (fig. 3). This heterogeneity is not only observed with MALDI-TOF-MS analysis but is detected with all polymerase-based methods. By comparing the relative amount of the inherent ‘background’ heterogeneity of controls with unknown samples to be analyzed, an estimation of MSI becomes feasible.
Fig. 3.
MSI genotyping by MALDI-TOF-MS. After PCR amplification of the DNA fragment harboring the microsatellite of interest, primer extension is carried out. Depending on the length of the microsatellite, a defined number of nucleobases is added to the extension primer. Note that due to inherent slippage of the polymerase, a small percentage of heterogeneity can be observed. The signal with the largest area under the peak represents the most abundant microsatellite length. Comparing the peak areas of samples of known genotypes as a reference to unknown samples allows the determination of the microsatellite instability. For internal calibration the mass of remaining unextended primer (P) can be used.
As a second strategy, in vitro transcription resulting in informative RNA fragments has been developed [45]. For this approach, the sequence of interest comprising the MS to be analyzed is transcribed into RNA which then is cut 3' and 5' of the MSI site by specific molecular scissors. In case of MSI, this leads to fragments of different molecular masses as expected for different lengths of the MS. These fragments can be analyzed by MALDI-TOF-MS, allowing the identification of the included repeat length.
DNA Methylation
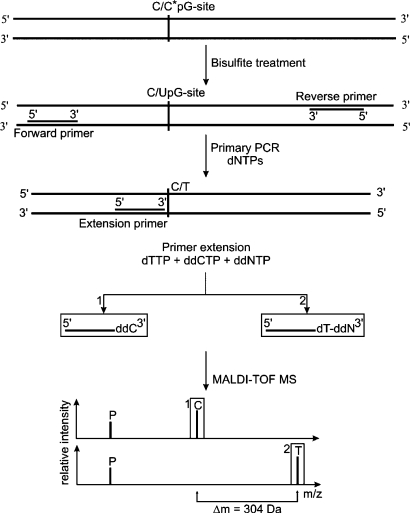
After completion of the human genome project, basic research focuses on understanding the mechanisms of gene function and regulation. Although almost all cells in an organism harbor the same set of genes, their transcription is spatially and temporally regulated. One major epigenetic factor influencing gene activation or silencing is DNA methylation [46]. This modification almost exclusively affects cytosines within the context of CpG dinucleotides. Highly CpG-rich regions, referred to as CpG islands, are found within or near promoter regions in an above-average frequency [47]. In general, highly methylated CpG islands mediate gene silencing, whereas demethylation increases gene transcription. Aberrant methylation is thought to contribute to a variety of human genetic disorders as well as influence the susceptibility of an individual to such disorders [48]. Several techniques have been developed to investigate the methylation status of DNA [49] some of which are eligible for high-throughput screening applications. When investigating the methylation status of a given DNA sequence, the major drawback is the fact that the methylation pattern is lost upon the PCR amplification required for generating sufficient amounts of DNA for the analysis. To overcome this problem, bisulfite treatment of the DNA ‘freezes’ the methylation status by converting free cytosines into uracils by hydrolytic deamination [50]. Cytosines initially methylated are protected from the bisulfite conversion and remain unchanged. Subsequently, the amplified DNA displays thymines at the positions of initially unmethylated cytosines, allowing genotyping of virtual C/T SNPs to assess the native methylation status of the respective cytosines (fig. 4). Bisulfite conversion combined with the classical primer extension-based GOOD assay was successfully applied to determine and quantify the methylation status of cytosines within CpG dinucleotides [51]. An impediment of this approach is the limited suitability of primer extension for the analysis of methylation within long CpG-rich regions: First, repetitive sequences are disadvantageous for primer specificity. Second, potentially methylated cytosines within the primer binding site might inhibit proper annealing and thus efficient detection of the chemically induced SNP.
Fig. 4.
DNA methylation determination by MALDI-TOF-MS. Bisulfite treatment of DNA containing putative methylated cytosines ‘freezes’ their methylation state by converting non-methylated cytosines into uracils while leaving methylated cytosines unaffected. After PCR amplification of the DNA region of interest, initially non-methylated cytosines are displayed as thymines. This chemically induced SNP can then be analyzed using the classical primer extension reaction. For internal calibration the mass of residual non-extended primer (P) can be used.
To overcome these problems, a method (MassCLEAVE™; Sequenom) was developed using T7-promoter-tagged primers for amplification of bisulfite-converted DNA followed by generation of a single-stranded RNA. Treatment with RNase A resulted in base-specific cleavage of the RNA (3' either to rUTP or rCTP) to fragments suitable for analysis by MALDI-TOF-MS [52,53,54]. By further improvement of this method, allele-specific methylation and a quantification of methylation differences as small as ∼5% may be possible. Quantification is the major advantage of the MassCLEAVE method compared to pyrosequencing where only statistically distributed clones are analyzed [55].
Besides the analysis of eukaryotic DNA, the methylation of synthetic oligonucleotide substrates by the Escherichia coli dam DNA methyltransferase has also been investigated successfully using MALDI-TOF-MS [56]. By this approach, the processivity of the enzyme as well as its substrate specificity could be determined directly. Classical techniques are based on the use of either radioactive (methyl-3H)AdoMet [57] or the effect that methylation within the sequence of a restriction site protects the DNA from endonuclease cleavage [58]. MALDI-TOF-MS offers, amongst others, the simultaneous detection of methylated as well as unmethylated samples and thus allows their relative quantification. By this means, more comprehensive and detailed information on the enzymatic characteristics for the methyltransferase could be obtained.
Quantification
A quantification of molecules analyzed by MALDI-TOF-MS is desirable for genotyping of pooled DNAs, for methylation analysis, and for the identification of loss of heterozygosity in non-microdissected tumor samples [59]. Quantification cannot be achieved by measurement of peak height in MALDI-TOF-MS analysis as molecules with higher m/z ratios often display lower peaks within the same sample/spectrum due to suppression effects. An absolute quantification of a solitary signal without a spectrum-internal reference signal is impossible. Nevertheless, relative quantification with regard to a spectrum-internal signal is possible. The area under the curve of this normalized signal in a MALDI-TOF-MS mass spectrum is then proportional to the amount of oligonucleotide that is present in the matrix-analyte-cocrystal and desorbed by the laser beam. In addition to 3-hydroxypicolinic acid, a good matrix to ensure high spot-to-spot reproducibility was found to be a mixture of 2,3,4- and 2,4,6-trihydroxyacetophenone with ammonium citrate [60], allowing a relative quantification of the analytes by comparing the peak area ratios. A relative quantification of peak area ratios was also successfully applied for genotyping of pooled DNAs. Allele frequencies in the samples were determined by normalization of the ratio measured for a single heterozygous DNA, as this was expected to represent a 1:1 ratio between the two allelic variants [61]. The possibility of sample pooling significantly reduces the effort, the cost as well as the time needed for such extensive analyses and renders them more rational. In terms of accuracy, the MALDI-TOF-MS approach for genotyping has turned out to be one of the most accurate techniques, superior to TaqMan, SNaPshot (Applied Biosystems) or pyrosequencing [62], and has already been applied to disease association studies [63]. Nevertheless, pooling of DNA samples, besides all positive aspects mentioned above, is strongly dependent on high DNA quality and exact adjustment of all pools to preserve accurate as well as reproducible results resembling the in vivo distribution of the respective allele frequencies. Thus, generating proper DNA pools out of large numbers of individual DNA samples turns out to be laborious, but this drawback might be overcome by performing a whole-genome amplification step to ensure rather equal amounts of all PCR products within the final sample [64]. For loss of heterozygosity analysis, it was demonstrated that comparative quantification of MALDI-TOF-MS analysis of heterozygous SNPs in non-microdissected normal and tumor tissue resulted in reproducible results to detect the loss of one allele in the tumor sample [59].
Conclusions
Initially invented for the purpose of proteomics, nowadays MALDI-TOF-MS has reached the status of a technical platform for a variety of different applications. Its versatility renders it economic as laboratories equipped with this instrumentation are able to address a variety of questions dealing with different species of molecules. Today, virtually any type of molecule has been investigated using MALDI-TOF-MS, even though with varying success. Starting from proteins, peptides, carbohydrates and nucleic acids, even lipids or inorganic polymers have been analyzed. With the completion of the human genome project, the field of nucleic acid research utilizes DNA analysis to combat disease and improve health. Minor variations within the genomic sequence account for a broad range of disorders or an individual's susceptibility to them. Screening for these variations in a high-throughput manner became more and more important in association studies investigating the influence of defined loci on a given disorder. The most important genomic variations comprise SNPs, MSIs as well as epigenetic factors such as DNA methylation. For analyzing all of these, a variety of different methods has been established, including MALDI-TOF-MS, predominantly relying on primer extension reactions. Making use of this strategy, virtually any allelic variations may be analyzed, including also those of hematological relevance. It has been shown that clinically important genotypes of the hemoglobin ß-chain, eventually causing different forms of thalassemia or sickle cell anemia, could readily and reliably be detected using singleplex as well as multiplex primer extension reactions followed by MALDI-TOF-MS (Humeny, unpublished data). It is conceivable to apply this method also to screen for variations of coagulation factors other than factor V or II. As a prerequisite, however, the exact localization as well as the expected genotypes of the polymorphic bases have to be identified. If this information is not available, the array-based SNP chip is the platform of choice as it covers a large number of allelic variants in one experiment. Nevertheless, with its robustness, high accuracy, versatility and suitability for quantitative studies, MALDI-TOF-MS still experiences growing attention, even though during the last years, break-through developments of innovative methods opening up completely new fields of research have not appeared. Nevertheless, improvement of existing protocols and their adaptation to special applications is still ongoing. Especially multiplexing, allowing investigation of several samples in one reaction, renders mass spectrometry highly cost-effective. For clinical routine, however, multiplexing might hold the drawback in being a multiple-step procedure and hence requiring too much operational time. Also, no mass spectrometry-based DNA assay has yet been approved by the Food and Drug Administration (FDA) which may be one reason why MS-based genotyping is not commonly used for clinical diagnosis. Moreover, the purchase cost for highperformance MALDI-TOF mass spectrometers is relatively high, rendering it more economic for laboratories addressing basic research questions rather than applying it to genotyping of a broad spectrum of polymorphisms in smaller collectives. Nevertheless, MALDI-TOF-MS will continue to be an important tool in research even though, in the meantime, it is being complemented by other mass spectrometric techniques like ESI-MS/MS, which tends to provide advanced mass accuracy but with increased need of sample purity. Taken together, for genomic research in the whole genome age, MALDI-TOF-MS is an important tool that facilitates quick, robust, reliable, and high-throughput analyses of nucleic acids for a broad variety of applications.
Disclosure
The authors declared no conflict of interest.
References
- 1.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, Wen W, Levy S, Deming SL, Haines JL, Gu K, Fair AM, Cai Q, Lu W, Shu XO. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imyanitov EN. Gene polymorphisms, apoptotic capacity and cancer risk. Hum Genet. 2009 doi: 10.1007/s00439-009-0636-7. DOI: 10.1007/s00439-009-0636-7. [DOI] [PubMed] [Google Scholar]
- 3.Wasserman D, Wasserman J, Rozanov V, Soko-lowski M. Depression in suicidal males: genetic risk variants in the CRHR1 gene. Genes Brain Behav. 2009;8:72–79. doi: 10.1111/j.1601-183X.2008.00446.x. [DOI] [PubMed] [Google Scholar]
- 4.Post RM. Role of BDNF in bipolar and unipolar disorder: clinical and theoretical implications. J Psychiatr Res. 2007;41:979–990. doi: 10.1016/j.jpsychires.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:151. [Google Scholar]
- 7.Dhingra V, Gupta M, Andacht T, Fu ZF. New frontiers in proteomics research: a perspective. Int J Pharm. 2005;299:1–18. doi: 10.1016/j.ijpharm.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Canas B, Lopez-Ferrer D, Ramos-Fernandez A, Camafeita E, Calvo E. Mass spectrometry technologies for proteomics. Brief Funct Genomic Proteomic. 2006;4:295–320. doi: 10.1093/bfgp/eli002. [DOI] [PubMed] [Google Scholar]
- 9.Tang K, Fu D, Kotter S, Cotter RJ, Cantor CR, Koster H. Matrix-assisted laser desorption/ionization mass spectrometry of immobilized duplex DNA probes. Nucleic Acids Res. 1995;23:3126–3131. doi: 10.1093/nar/23.16.3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolson DA, Nicholson NH. Sequencing RNA by a combination of exonuclease digestion and uridine specific chemical cleavage using MALDI-TOF. Nucleic Acids Res. 1998;26:446–451. doi: 10.1093/nar/26.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey DJ. Matrix-assisted laser desorption/ionisation mass spectrometry of oligosaccharides and glycoconjugates. J Chromatogr A. 1996;720:429–446. doi: 10.1016/0021-9673(95)00307-x. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs B, Schiller J. MALDI-TOF MS analysis of lipids from cells, tissues and body fluids. Subcell Biochem. 2008;49:541–565. doi: 10.1007/978-1-4020-8831-5_21. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, He M, Pei J, He H. Quantitative analysis of synthetic polymers using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2003;75:6531–6535. doi: 10.1021/ac0344034. [DOI] [PubMed] [Google Scholar]
- 14.Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R. Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res. 2004;14:414–425. doi: 10.1101/gr.2014904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. BioTechniques. 2002;(suppl):56–58. 60-61. [PubMed] [Google Scholar]
- 16.Hardenbol P, Baner J, Jain M, Nilsson M, Nam-saraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, Landegren U, Davis RW. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]
- 17.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′—3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer S, Gut IG. Genotyping single-nucleotide polymorphisms by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:73–87. doi: 10.1016/s1570-0232(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 19.Syvanen AC, Aalto-Setala K, Harju L, Kontula K, Soderlund H. A primer-guided nucleotide incorporation assay in the genotyping of apolipoprotein E. Genomics. 1990;8:684–692. doi: 10.1016/0888-7543(90)90255-s. [DOI] [PubMed] [Google Scholar]
- 20.Sauer S, Lechner D, Berlin K, Plancon C, Heuermann A, Lehrach H, Gut IG. Full flexibility genotyping of single nucleotide polymorphisms by the GOOD assay. Nucleic Acids Res. 2000;28:E100. doi: 10.1093/nar/28.23.e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haff LA, Smirnov IP. Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res. 1997;7:378–388. doi: 10.1101/gr.7.4.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenzel T, Elssner T, Fahr K, Bimmler J, Richter S, Thomas I, Kostrzewa M. Genosnip: SNP genotyping by MALDI-TOF MS using photocleavable oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1579–1581. doi: 10.1081/NCN-120023038. [DOI] [PubMed] [Google Scholar]
- 23.Braun A, Little DP, Koster H. Detecting CFTR gene mutations by using primer oligo base extension and mass spectrometry. Clin Chem. 1997;43:1151–1158. [PubMed] [Google Scholar]
- 24.Sun X, Ding H, Hung K, Guo B. A new MALDITOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Res. 2000;28:E68. doi: 10.1093/nar/28.12.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buetow KH, Edmonson M, MacDonald R, Clifford R, Yip P, Kelley J, Little DP, Strausberg R, Koester H, Cantor CR, Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaderali L. Primer design for multiplexed genotyping. Methods Mol Biol. 2007;402:269–286. doi: 10.1007/978-1-59745-528-2_13. [DOI] [PubMed] [Google Scholar]
- 27.Rachlin J, Ding C, Cantor C, Kasif S. MuPlex: multi-objective multiplex PCR assay design. Nucleic Acids Res. 2005;33:W544–547. doi: 10.1093/nar/gki377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humeny A, Bonk T, Berkholz A, Wildt L, Becker CM. Genotyping of thrombotic risk factors by MALDI-TOF mass spectrometry. Clin Biochem. 2001;34:531–536. doi: 10.1016/s0009-9120(01)00264-8. [DOI] [PubMed] [Google Scholar]
- 29.Bertina RM, Koeleman BP, Koster T, Rosendaal FR, Dirven RJ, de Ronde H, van der Velden PA, Reitsma PH. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–67. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 30.Page MJ. Factor V Leiden mutation: a nursing perspective. J Vasc Nurs. 1998;16:73–77. doi: 10.1016/s1062-0303(98)90010-3. [DOI] [PubMed] [Google Scholar]
- 31.Poort SR, Rosendaal FR, Reitsma PH, Bertina RM. A common genetic variation in the 3'-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–3703. [PubMed] [Google Scholar]
- 32.Rosendaal FR, Siscovick DS, Schwartz SM, Psaty BM, Raghunathan TE, Vos HL. A common prothrombin variant (20210 G to A) increases the risk of myocardial infarction in young women. Blood. 1997;90:1747–1750. [PubMed] [Google Scholar]
- 33.Garritsen HS, Fan AX, Bosse N, Hannig H, Kelsch R, Kroll H, Holzgreve W, Zhong XY. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for genotyping of human platelet-specific antigens. Transfusion. 2009;49:252–258. doi: 10.1111/j.1537-2995.2008.01953.x. [DOI] [PubMed] [Google Scholar]
- 34.Welsby IJ, Podgoreanu MV, Phillips-Bute B, Mathew JP, Smith PK, Newman MF, Schwinn DA, Stafford-Smith M, Perioperative Genetics and Safety Outcomes Study (PEGASUS) Investigative Team Genetic factors contribute to bleeding after cardiac surgery. J Thromb Haemost. 2005;3:1206–1212. doi: 10.1111/j.1538-7836.2005.01337.x. [DOI] [PubMed] [Google Scholar]
- 35.Misra A, Hong JY, Kim S. Multiplex genotyping of cytochrome p450 single-nucleotide polymorphisms by use of MALDI-TOF mass spectrometry. Clin Chem. 2007;53:933–939. doi: 10.1373/clinchem.2006.080739. [DOI] [PubMed] [Google Scholar]
- 36.Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Xie HG, Prasad HC, Kim RB, Stein CM. CYP2C9 allelic variants: ethnic distribution and functional significance. Adv Drug Deliv Rev. 2002;54:1257–1270. doi: 10.1016/s0169-409x(02)00076-5. [DOI] [PubMed] [Google Scholar]
- 38.Su T, Bao Z, Zhang QY, Smith TJ, Hong JY, Ding X. Human cytochrome P450 CYP2A13: predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 2000;60:5074–5079. [PubMed] [Google Scholar]
- 39.Brouwer JR, Willemsen R, Oostra BA. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 41.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 42.Peltomaki P, Lothe RA, Aaltonen LA, Pylkkanen L, Nystrom-Lahti M, Seruca R, David L, Holm R, Ryberg D, Haugen A. Microsatellite instability is associated with tumors that characterize the hereditary nonpolyposis colorectal carcinoma syndrome. Cancer Res. 1993;53:5853–5855. [PubMed] [Google Scholar]
- 43.Malkhosyan S, McCarty A, Sawai H, Perucho M. Differences in the spectrum of spontaneous mutations in the hprt gene between tumor cells of the microsatellite mutator phenotype. Mutat Res. 1996;316:249–259. doi: 10.1016/s0921-8734(96)90007-7. [DOI] [PubMed] [Google Scholar]
- 44.Bonk T, Humeny A, Gebert J, Sutter C, von Knebel Doeberitz M, Becker CM. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry-based detection of microsatellite instabilities in coding DNA sequences: a novel approach to identify DNAMismatch repair-deficient cancer cells. Clin Chem. 2003;49:552–561. doi: 10.1373/49.4.552. [DOI] [PubMed] [Google Scholar]
- 45.Sasayama T, Kato M, Aburatani H, Kuzuya A, Komiyama M. Simultaneous genotyping of indels and SNPs by mass spectroscopy. J Am Soc Mass Spectrom. 2006;17:3–8. doi: 10.1016/j.jasms.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 46.Attwood JT, Yung RL, Richardson BC. DNA methylation and the regulation of gene transcription. Cell Mol Life Sci. 2002;59:241–257. doi: 10.1007/s00018-002-8420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 48.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 49.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. BioTechniques. 2002;33:636–649. doi: 10.2144/02333rv01. 632, 634. [DOI] [PubMed] [Google Scholar]
- 50.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tost J, Schatz P, Schuster M, Berlin K, Gut IG. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31:e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D. Quantitative high-throughput analysis of DNA methylation patterns by basespecific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schatz P, Dietrich D, Schuster M. Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF. Nucleic Acids Res. 2004;32:e167. doi: 10.1093/nar/gnh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gut IG. DNA analysis by MALDI-TOF mass spectrometry. Hum Mutat. 2004;23:437–441. doi: 10.1002/humu.20023. [DOI] [PubMed] [Google Scholar]
- 55.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humeny A, Beck C, Becker CM, Jeltsch A. Detection and analysis of enzymatic DNA methylation of oligonucleotide substrates by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Anal Biochem. 2003;313:160–166. doi: 10.1016/s0003-2697(02)00568-7. [DOI] [PubMed] [Google Scholar]
- 57.Roth M, Jeltsch A. Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol Chem. 2000;381:269–272. doi: 10.1515/BC.2000.035. [DOI] [PubMed] [Google Scholar]
- 58.Gowher H, Jeltsch A. Molecular enzymology of the EcoRV DNA-(adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J Mol Biol. 2000;303:93–110. doi: 10.1006/jmbi.2000.4127. [DOI] [PubMed] [Google Scholar]
- 59.Reuland A, Humeny A, Magener A, Becker CM, Schiebel K. Detection of loss of heterozygosity by matrix-assisted laser desorption/ionization timeof-flight mass spectrometry-based analysis of singlenucleotide polymorphisms. Clin Chem. 2005;51:636–639. doi: 10.1373/clinchem.2004.040899. [DOI] [PubMed] [Google Scholar]
- 60.Zhu YF, Chung CN, Taranenko NI, Allman SL, Martin SA, Haff L, Chen CH. The study of 2,3,4-trihydroxyacetophenone and 2,4,6-trihydroxyacetophenone as matrices for DNA detection in matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:383–388. doi: 10.1002/(SICI)1097-0231(199602)10:3<383::AID-RCM485>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 61.Downes K, Barratt BJ, Akan P, Bumpstead SJ, Taylor SD, Clayton DG, Deloukas P. SNP allele frequency estimation in DNA pools and variance components analysis. BioTechniques. 2004;36:840–845. doi: 10.2144/04365RR01. [DOI] [PubMed] [Google Scholar]
- 62.Shifman S, Pisante-Shalom A, Yakir B, Darvasi A. Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol Cell Probes. 2002;16:429–434. doi: 10.1006/mcpr.2002.0440. [DOI] [PubMed] [Google Scholar]
- 63.Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Beckmann JS, Yakir B, Risch N, Zak NB, Darvasi A. A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet. 2002;71:1296–1302. doi: 10.1086/344514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao ZZ, Nyholt DR, James MR, Mayne R, Treloar SA, Montgomery GW. A comparison of DNA pools constructed following whole genome amplification for two-stage SNP genotyping designs. Twin Res Hum Genet. 2005;8:353–61. doi: 10.1375/1832427054936718. [DOI] [PubMed] [Google Scholar]
- 65.Immervoll T, Loesgen S, Dutsch G, Gohlke H, Herbon N, Klugbauer S, Dempfle A, Bickeboller H, Becker-Follmann J, Ruschendorf F, Saar K, Reis A, Wichmann HE, Wjst M. Fine mapping and single nucleotide polymorphism association results of candidate genes for asthma and related phenotypes. Hum Mutat. 2001;18:327–336. doi: 10.1002/humu.1194. [DOI] [PubMed] [Google Scholar]
- 66.Kormann MS, Carr D, Klopp N, Illig T, Leupold W, Fritzsch C, Weiland SK, von Mutius E, Kabesch M. G-protein-coupled receptor polymorphisms are associated with asthma in a large German population. Am J Respir Crit Care Med. 2005;171:1358–1362. doi: 10.1164/rccm.200410-1312OC. [DOI] [PubMed] [Google Scholar]
- 67.Kubota T, Horie M, Takano M, Yoshida H, Takenaka K, Watanabe E, Tsuchiya T, Otani H, Sasayama S. Evidence for a single nucleotide polymorphism in the KCNQ1 potassium channel that underlies susceptibility to life-threatening arrhythmias. J Cardiovasc Electrophysiol. 2001;12:1223–1229. doi: 10.1046/j.1540-8167.2001.01223.x. [DOI] [PubMed] [Google Scholar]
- 68.Matsushita M, Miyakawa H, Tanaka A, Hijikata M, Kikuchi K, Fujikawa H, Arai J, Sainokami S, Hino K, Terai I, Mishiro S, Gershwin ME. Single nucleotide polymorphisms of the mannose-binding lectin are associated with susceptibility to primary biliary cirrhosis. J Autoimmun. 2001;17:251–257. doi: 10.1006/jaut.2001.0538. [DOI] [PubMed] [Google Scholar]
- 69.Koschinsky ML, Boffa MB, Nesheim ME, Zinman B, Hanley AJ, Harris SB, Cao H, Hegele RA. Association of a single nucleotide polymorphism in CPB2 encoding the thrombin-activable fibrinolysis inhibitor (TAF1) with blood pressure. Clin Genet. 2001;60:345–349. doi: 10.1034/j.1399-0004.2001.600504.x. [DOI] [PubMed] [Google Scholar]
- 70.Vollmert C, Windl O, Xiang W, Rosenberger A, Zerr I, Wichmann HE, Bickeboller H, Illig T, KORA group, Kretzschmar HA. Significant association of a M129V independent polymorphism in the 5′ UTR of the PRNP gene with sporadic Creutzfeldt-Jakob disease in a large German case-control study. J Med Genet. 2006;43:e53. doi: 10.1136/jmg.2006.040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim S, Edwards JR, Deng L, Chung W, Ju J. Solid phase capturable dideoxynucleotides for multiplex genotyping using mass spectrometry. Nucleic Acids Res. 2002;30:e85. doi: 10.1093/nar/gnf084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Donn RP, Shelley E, Ollier WE, Thomson W, British Paediatric Rheumatology Study Group A novel 5′-flanking region polymorphism of macrophage migration inhibitory factor is associated with systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 2001;44:1782–1785. doi: 10.1002/1529-0131(200108)44:8<1782::AID-ART314>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 73.Fatar M, Stroick M, Steffens M, Senn E, Reuter B, Bukow S, Griebe M, Alonso A, Lichtner P, Bugert P, Meitinger T, Wienker TF, Hennerici MG. Single-nucleotide polymorphisms of MMP-2 gene in stroke subtypes. Cerebrovasc Dis. 2008;26:113–119. doi: 10.1159/000139657. [DOI] [PubMed] [Google Scholar]
- 74.Zhu Y, Spitz MR, Lei L, Mills GB, Wu X. A single nucleotide polymorphism in the matrix metallopro-teinase-1 promoter enhances lung cancer susceptibility. Cancer Res. 2001;61:7825–7829. [PubMed] [Google Scholar]
- 75.Biros E, Kalina I, Biros I, Kohut A, Bogyiova E, Salagovic J, Stubna J. Polymorphism of the p53 gene within the codon 72 in lung cancer patients. Neoplasma. 2001;48:407–411. [PubMed] [Google Scholar]
- 76.Weber F, Fontaine B, Cournu-Rebeix I, Kroner A, Knop M, Lutz S, Muller-Sarnowski F, Uhr M, Bettecken T, Kohli M, Ripke S, Ising M, Rieckmann P, Brassat D, Semana G, Babron MC, Mrejen S, Gout C, Lyon-Caen O, Yaouanq J, Edan G, Clanet M, Holsboer F, Clerget-Darpoux F, Muller-Myhsok B. IL2RA and IL7RA genes confer susceptibility for multiple sclerosis in two independent European populations. Genes Immun. 2008;9:259–263. doi: 10.1038/gene.2008.14. [DOI] [PubMed] [Google Scholar]
- 77.Koshizuka Y, Kawaguchi H, Ogata N, Ikeda T, Mabuchi A, Seichi A, Nakamura Y, Nakamura K, Ikegawa S. Nucleotide pyrophosphatase gene polymorphism associated with ossification of the posterior longitudinal ligament of the spine. J Bone Miner Res. 2002;17:138–144. doi: 10.1359/jbmr.2002.17.1.138. [DOI] [PubMed] [Google Scholar]
- 78.Stevens A, Ray D, Alansari A, Hajeer A, Thomson W, Donn R, Ollier WE, Worthington J, Davis JR. Characterization of a prolactin gene polymorphism and its associations with systemic lupus erythematosus. Arthritis Rheum. 2001;44:2358–2366. doi: 10.1002/1529-0131(200110)44:10<2358::aid-art399>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 79.Tan FK, Wang N, Kuwana M, Chakraborty R, Bona CA, Milewicz DM, Arnett FC. Association of fibrillin 1 single-nucleotide polymorphism haplotypes with systemic sclerosis in Choctaw and Japanese populations. Arthritis Rheum. 2001;44:893–901. doi: 10.1002/1529-0131(200104)44:4<893::AID-ANR146>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 80.Tsunoda K, Sanke T, Nakagawa T, Furuta H, Nanjo K. Single nucleotide polymorphism (D68D, T to C) in the syntaxin 1A gene correlates to age at onset and insulin requirement in Type II diabetic patients. Diabetologia. 2001;44:2092–2097. doi: 10.1007/s001250100015. [DOI] [PubMed] [Google Scholar]
- 81.Jiang-Baucom P, Girard JE, Butler J, Belgrader P. DNA typing of human leukocyte antigen sequence polymorphisms by peptide nucleic acid probes and MALDI-TOF mass spectrometry. Anal Chem. 1997;69:4894–4898. doi: 10.1021/ac970639u. [DOI] [PubMed] [Google Scholar]