Summary
The Luminex-based human leukocyte antigen (HLA) antibody screening technology is widespread used in laboratories affiliated to kidney transplantation programs and enables both screening (i.e. the definition of positive or negative antibody status) and antibody identification with high sensitivity and specificity. HLA typing at different levels of resolution with Luminex technology uses sequence-specific oligonucleotide probes bound to color-coded microbeads in order to identify HLA alleles encoded by the DNA sample. In general, the Luminex technology for histocompatibility analyses provides rapid sample processing in a 96-well format combined with limited technical complexity which means high cost efficiency.
Key Words: Antibody identification, Antibody screening, Cost effectiveness analysis, HLA, HLA antibodies, PCR
Zusammenfassung
Die Luminex-basierte Technologie des «Human leukocyte antigen»(HLA)-Antikörper-Screenings ist in Laboratorien, die an Nierentransplantationsprogrammen teilnehmen, weit verbreitet. Sie ermöglicht sowohl das Screening (d.h. die Bestimmung eines positiven oder negativen Antikörperstatus) als auch die Antikörperidentifizierung mit hoher Sensitivität und Spezifität. Zur HLA-Typisierung für verschiedene Auflösungslevel mit Hilfe der Luminex-Technologie werden sequenzspezifische Oligonukleotid-Sonden (sequence-specific oligonucleotide probes), die an farbkodierte Mikrobeads gebunden sind, verwendet, um HLA-Allele zu identifizieren, die durch die DNA-Probe kodiert werden. Im Allgemeinen ermöglicht die Luminex-Technologie für Histokompatibilitätsanalysen eine schnelle Probenprozessierung in 96-Well-Platten bei begrenzter technischer Komplexität und geht so mit einer hohen Kosteneffektivität einher.
Introduction
Histocompatibility diagnostics is traditionally performed in medical laboratories which are specialized in the field of analyzing human leukocyte antigens (so called HLA laboratories). However, it has to be realized that the complete range of histocompatibility diagnostics from low to high resolution levels will be concentrated to a smaller number of high-throughput laboratories with diminished laboratory staff in the future. Today, most small- and medium-sized HLA laboratories use commercially available kits on the basis of sequence-specific primers (PCR-SSP technology) for HLA typing which is work-intensive and time-consuming for technical staff. In contrast, a sequence-based HLA typing strategy using capillary electrophoresis sequencers requires large investments in technology and is therefore often used in high-throughput HLA laboratories. The new Luminex (Luminex Corporation, Austin, TX USA) technology combines rapid sample processing in a 96-well format combined with limited technical complexity which means high cost efficiency. Especially the test duration and the costs for acquisition of machines are significantly lower as compared to the establishment of a sequence-based HLA typing strategy. Moreover, the companies providing histocompatibility testing assays for Luminex offer a variety of tests covering different HLA loci and levels of resolution for genotyping and also for antibody diagnostics (commercial kits are LABScreen™ and LABType™ assays provided by One Lambda Inc., Canoga Park, CA, USA, and Lifecodes™ assays by Tepnel Corp., Stamford, CT, USA).
The Luminex Technology
The basic principle of the Luminex technology is the ability to measure multiple analytes simultaneously in a single reaction well. Molecular reactions take place on the surface of plastic microbeads (microspheres) that are color-coded using a blend of different fluorescent intensities of two dyes. When using this method, over 100 distinct microbead sets can be created. The microbeads act as molecular carriers that capture samples and are then tagged with a fluorescence-labeled reporter binding to the captured sample on the microbead. Afterwards, the microbeads are transported into the Luminex analyzer by using a precision sheath fluid stream to align them in single file, where they pass through two lasers, similarly to classical flow cytometry. One laser excites the colors inside the microbeads to identify which microbead is currently being read. The second laser excites the color on the microbead surface, i.e. the labeled reporter tag. Finally, the color signals are detected by an advanced optical system, and the signals are processed into data for each reaction.
HLA Antibody Characterization using Luminex
Despite effective immunosuppression, the presence of anti-HLA antibodies in the recipient's serum prior to renal transplantation is still recognized as a prominent risk factor for irreversible kidney allograft rejection. Screening for such preformed alloreactive antibodies was traditionally performed using the complement-dependent cytotoxicity assay (CDC) which detects complement-activating IgG1/3 and IgM antibodies. This assay uses a panel of HLA-typed lymphocytes to determine so-called panel-reactive antibodies (PRA). For instance, a positive reaction against half of the cells within the panel is expressed as a panel reactivity of 50% PRA. The specificity of the CDC is limited due to positive reactions mediated through cytotoxic antibodies directed against non HLA molecules. In addition, the sensitivity of the CDC is also limi ted because only complement-activating antibodies result in positive reactions. More recently, solid-phase methods such as ELISA and flow cytometry-based techniques (using classical flow cytometry or Luminex) were introduced to overcome the limited sensitivity and specificity of the CDC. An additional advantage of these new techniques is a differential detection of anti-HLA class I and class II antibodies of the IgG isotypes which are generally accepted to be of relevance for kidney transplantation [1].
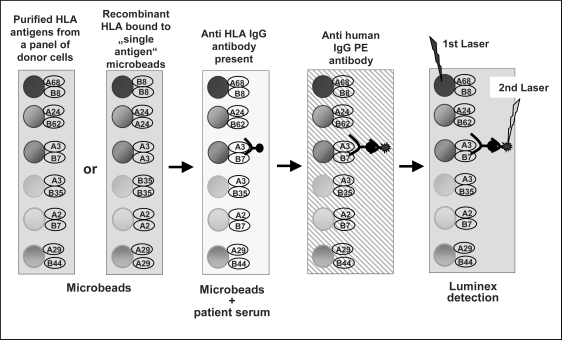
Today the Luminex-based antibody screening technology is widespread used in laboratories affiliated to kidney transplantation programs and enables both screening (i.e. the definition of positive or negative antibody status) and antibody identification with high sensitivity and specificity. The commercially available test kits use purified HLA antigens immobilized directly on a panel of microbeads (fig. 1). These HLA molecules on the microbeads are targets for HLA-specific antibodies in a given patient sample instead of viable cells as targets for the antibodies when the CDC is used. Anti-HLA antibodies of the IgG isotypes which are bound to the microbeads are detected by a secondary IgG-specific antibody which is conjugated with R-phycoerythrin (PE). For washing steps, centrifugation or filter plates combined with vacuum are used. The size of the microbead panel used by a specific test kit differs depending on the type of analysis the kit was composed for: Screening assays for pure detection of antibodies (‘positive’ or ‘negative’ HLA class I or class II antibody status of a serum sample) use only a few microbead populations, whereas a highly specific so-called ‘single antigen’ assay needs almost the complete set of 100 microbeads available to cover each HLA specificity on a distinct microbead population. Unfortunately, the number of microbead populations used for a specific assay is directly reflected in the costs for this assay.
Fig. 1.
HLA antibody screening using Luminex technology. The Luminex technology uses a set if microbeads covered with HLA antigens. Antibodies against HLA bind and are stained by a secondary antihuman IgG antibody conjugated with PE. The microbead population and the tagged sample are detected simultaneously using the Luminex flow analyzer.
Introduction of the Luminex Antibody Screening Technology in Laboratory Routine
In our HLA laboratory we started with Luminex-based antibody screening (LABScreen) in the year 2001. Three years later we performed a comparative analysis which included 458 serum samples analyzed by Luminex for HLA class I- and 347 for HLA class II-specific antibodies from the local transplant program [2]. The results were correlated with the CDC- (inhouse) and ELISA-derived (LATM™, One Lambda Inc.) results. Additionally, in a second study we analyzed ‘difficult’ serum samples from two study groups (i.e. sera that revealed borderline positive results with ELISA in comparison to high PRA sera) using Luminex and classical flow cytometry (FlowPRA™, One Lambda Inc.) ‘single antigen’ microbead assays which were newly available at that time [3].
In 241/458 (53%) sera analyzed in the first study we could identify antibodies directed against HLA class I allotypes both by CDC and Luminex. However, only 121/458 (26%) sera showed fully concordant specificities, whereas with the Luminex approach additional HLA class I specificities were found in 183/458 (40%) sera. In 46/458 (10%) sera we found completely different results in the two assays. We could observe that the CDC (with or without the addition of dithiothreitol (DTT) to eliminate antibodies of the IgM isotype) was able to identify HLA specificities in sera with moderate PRA values (ranging from 10 to 70% PRA). However, in several sera with high PRA values (70–100% PRA) or sera which contain very low antibody titers (below 10% PRA), the CDC is unable to clearly define HLA specificities. In such cases the Luminex method, especially in combination with ‘single antigen’ microbeads, can substantially improve the HLA antibody screening results. Regarding the FlowPRA ‘single antigen’ test, our study revealed difficulties with the interpretation of the results due to insufficient discrimination between positive and negative reactions. False-positive reactions may also occur in Luminex as a consequence of the software analysis when the fixed microbead default background values were used for calculating the final result. An advantage of up to date analysis software is the option to use a background value obtained from an internal laboratory-specific negative control. This procedure provides the benefit of uniform machine conditions, identical reagents, and minimal technical handling problems for all samples including the negative control. Using a laboratory-specific negative control may reduce the percentage of discrepancies of false-positives and false-negatives between Luminex and flow cytometry significantly. Other problems could be patient samples with high background, low positive control values, and low negative control values. These can be resolved when the patient serum is treated with a special adsorption procedure using the reagent AdsorbOut™ (One Lambda Inc.).
Finally, a positive cross-match test, which is still done by CDC in most countries, represents the ultimate evidence for the presence of donor-specific antibodies prior to transplantation [4]. To improve the cross-match sensitivity the HLA laboratory may introduce the flow cytometry cross-match, as we have done in 2004. Today, Luminex technology for HLA antibody screening provides high sensitivity which is comparable to the high sensitivity of the flow cytometry cross-match.
Perspectives of Antibody Characterization using Luminex
The clinical relevance of antibodies detected by highly sensitive solid-phase assays (e.g. Luminex) and also by flow cytometry cross-match has still not been fully elucidated. However, the Luminex technology provides an appropriate platform to study the impact of different isotypes of antibodies on the outcome of transplantation.
To address this issue, we studied the antibodies accumulated directly in transplanted kidney allografts [5]. By acid elution we isolated proteins accumulated in rejected and explanted kidneys and characterized their HLA specificities by CDC, ELISA, and Luminex techniques. In addition, we differentially analyzed non-complement-binding IgG2 and IgG4 anti-HLA antibodies in the eluates using modified ELISA. This study clearly showed that explanted kidney allografts harbor HLA-specific antibodies that can be detected by sensitive Luminex techniques. Moreover, also non-complement-binding anti-HLA antibodies accumulate in renal allografts [5]. Very recently, our group was also able to adapt the Luminex-based antibody detection technology to anti-HLA antibodies of the IgM isotype, which is an improvement in sensitivity compared to the conventionally used CDC for the definition of IgM antibodies [6].
Supplemental to the isotype-specific HLA antibody characterization, Luminex technology also enables the detection and characterization of antibodies against MHC class I-related chain A (MICA). MICA antigens are HLA-related, polymorphic glycoproteins expressed on the surface of human endothelial cells, epithelial cells (especially of the gastrointestinal tract), fibroblasts, and also in certain tumors [7]. AntiMICA antibodies have been detected in the serum of patients awaiting kidney transplantation as well as in kidney allograft recipients, and accumulate in rejected kidney allografts [8]. Presensitization of kidney transplant recipients against MICA antigens is associated with an increased frequency of kidney allograft loss [9]. Today, test kits are commercially available for the detection of anti-MICA antibodies (‘positive’ or ‘negative’ serum status) and also for the characterization of MICA antibody specificities using ‘single antigen’ microbeads, but the clinical relevance of anti-MICA antibodies is currently not fully understood.
In conclusion, the Luminex technology enables up to date HLA antibody detection and specification for daily laboratory routine in high sample throughput, but there is still demand for automation of the whole technical procedure. In addition, the technology appears to be appropriate to answer scientific questions when the impact of antibodies on organ transplantation outcome is particularly analyzed. On the other hand, the clinical relevance of quite a number of HLA antibody specificities detectable by Luminex is still controversially discussed.
HLA Genotyping using Sequence-Specific Oligonucleotide Probe Hybridization in Combination with Luminex Technology
Historically, tissue typing was done by CDC using a defined set of sera containing HLA-specific antibodies. The resolution of HLA typing using CDC is limited by the fact that antibodies have limited access to all epitopes of the HLA molecules on the cell surface. However, since PCR technologies were available, DNA-based tissue typing techniques generating low-resolution (i.e. 2-digits HLA nomenclature) results or high-resolution (i.e. at least 4-digits HLA nomenclature) results have become routine in HLA laboratories. Intermediate HLA resolution comprises a 2-digits HLA nomenclature with supplemental characters to define groups of HLA alleles and was introduced by the National Marrow Donor Program (NMDP) in the USA [10]. In general, the HLA typing resolution accepted as relevant for different clinical applications should be considered [11]. The PCR-SSP technology using a variety of SSPs to cover different HLA alleles at a given HLA locus has become the tissue typing technology mostly used for low-resolution HLA typing. It is easy to perform and needs little technical equipment. However, the PCR-SSP method is not suitable for high sample throughput due to a variety of manual pipetting steps. After PCR-SSP amplification, a single gel electrophoresis gives the information about the HLA alleles present in a DNA sample. In contrast to this, hybridization assays for tissue typing need additional post-PCR amplification steps to discriminate between the different alleles. The commercially available Luminex-based tissue typing assays use sequence-specific oligonucleotide (SSO) probes bound to the color-coded microbeads to identify alleles encoded by the DNA sample.
Similarly to Luminex-based antibody screening, tissue typing using Luminex SSO technology is also widespread in HLA laboratories affiliated to transplant programs or stem cell donor registries. The commercially available test kits enable HLA genotyping on low, intermediate or in parts high resolution level of all HLA loci: HLA-A, HLA-B, HLAC, HLA-DRB1, HLA-DRB3, HLA-DRB4, HLA-DRB5, HLADQA1, HLA-DQB1, and HLA-DPB1. In addition, Luminex-based SSO typing assays are available for the definition of MICA alleles, which are discussed to be of relevance for organ transplantation outcome. In the field of hematopoietic stem cell transplantation genomic variants of the killer-cell immunoglobulin-like receptor (KIR) genes seem to have an impact on success rates. Typing for KIR genes is also possible using Luminex-based SSO technology.
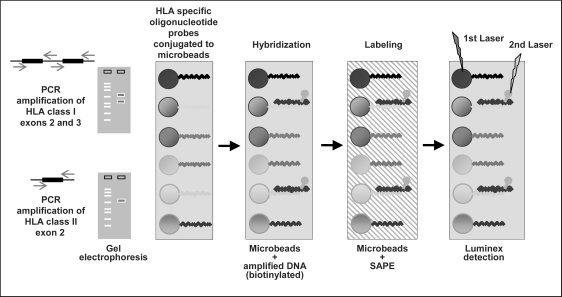
The test procedure starts with PCR amplification of the most polymorphic part of the HLA class II gene using exon 2-specific primers for HLA-DRB1, HLA-DQB1, HLA-DQA1, and HLA-DPB1 (fig. 2). For the HLA-A, HLA-B, and HLAC loci two PCR products covering the most polymorphic exons 2 and 3 of these HLA class I loci are generated. The PCR product is biotinylated, which allows it to be detected by strepavidin-conjugated with PE (SAPE). Subsequently, PCR products are denaturated enabling rehybridization to complementary DNA probes bound to the microbeads. Each microbead mixture includes negative and positive control probes necessary for subtraction of nonspecific background signals and normalization of raw data. For washing steps centrifugation or filter plates combined with vacuum are used. Finally, the Luminex flow analyzer identifies the fluorescent intensity of PE on each microbead. Interpretation software analyzes reaction patterns and assigns the matching HLA alleles. This assignment is based on the HLA sequences listed in the official IMGT/HLA database [12]. In addition, allele codes recently defined by the NMDP will be considered. The combination of a single PCR amplification step with hybridization and detection procedures in a single reaction mixture per sample enables high-throughput tissue typing.
Fig. 2.
HLA class I and class II typing using Luminex technology. Luminex SSO HLA typing uses SSO probes for distinct sequence motifs which are bound to a set of microbeads. A biotinylated PCR product binds and is subsequently stained by SAPE. The microbead population and the tagged sample are detected simultaneously using the Luminex flow analyzer.
Low- and High-Resolution HLA Typing using Luminex Technology
Comparable to the Luminex-based antibody screening tests, the size of the microbead panel used by a specific tissue typing test kit also differs dependent on polymorphism of the different HLA loci: Typing for HLA-DPB1 needs only a few microbead populations, whereas HLA-B typing on an intermediate resolution level needs almost the complete set of 100 microbeads available. In general, this technology involves the danger of losing microbeads when washing of the trays is inadequately done. Microbead loss will result in low microbead count for one or more bead populations. In addition, it is strongly recommended to check complete denaturation and neutralization of the PCR product and to meet the exact hybridization/labeling temperature and duration.
A central question is the manipulation of the cut-offs defined by the analysis software. Although the Luminex SSO typing method is robust in the complete procedure, it may be necessary to modify cut-offs due to variables in DNA quality and laboratory-specific assay performance. It is recommended to make adjustments after comparing the performance of a probe under local laboratory conditions against the manufacturer's quality control panel for that probe.
Recently, both manufacturers providing tissue typing test kits for Luminex introduced assays for high-resolution typing of the DRB1 gene. These tests contain a special probe technology which allows resolution of ambiguities at a level the conventional Luminex SSO method does not offer. The special probe microbeads were added to the panel of conventional beads and are covered with more than a single probe, which means they are specific for several DNA motifs on one DNA strand. The upgrade of the conventional Luminex technology to high-resolution HLA typing requires accurate washing and complex software analysis. Modification of cut-offs is particularly critical when these newly available kits are used. However, the test performance is principally limited due to the initial PCR amplification of only exon 2 of the HLA class II gene which is currently regarded as sufficient for all clinical applications.
However, for high-resolution SSO typing of the highly polymorphic HLA-A or HLA-B genes this strategy had to be extended. For the latest test kits, the companies now use more than 200 different probes on the initial set of 100 Luminex microbeads for high-resolution HLA class I typing. The future challenge is the development of powerful software to differentiate the different probes on the same bead and to analyze the reaction patterns in adequate time and with satisfying reliability.
Finally, some fundamental limitations remain: Similarly to the HLA class II typing, polymorphisms outside the amplified region cannot be resolved, i.e. HLA alleles not amplified by the primers cannot be identified. The same holds true for polymorphisms located at the primer binding sites. Another problem for all high-resolution tissue typing assays is the continuous increase of new HLA alleles in the official database [12]. New alleles require new probes to be permanently designed. Only a sequence-based HLA typing strategy enables the specific analysis of the relevant region within the HLA gene actually present in a given DNA sample. However, HLA sequencing requires large investments in machines and a comprehensive PCR amplification strategy if a typing result superior to the advanced Luminex SSO technology should be generated.
Disclosure
The author declared no conflict of interest.
References
- 1.Terasaki PI, Cai J. Humoral theory of transplantation: further evidence. Curr Opin Immunol. 2005;17:541–545. doi: 10.1016/j.coi.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Heinemann FM, Rebmann V, Arnold ML, Spriewald BM, Lachmann N, Schönemann C, Grosse-Wilde H. HLA antibody characterization in kidney transplantation: development of new technologies and introduction in laboratory routine. In: Terasaki PI, editor. Visuals of the 6th European Clinical Histocompatibility Workshop, Newcastle. Canoga Park: One Lambda Inc.; 2005. pp. 34–35. [Google Scholar]
- 3.Schönemann C, Lachmann N, Heinemann FM, Grosse-Wilde H. Comparison of two sensitive flow cytometry based methods for HLA-antibody detection. In: Terasaki PI, editor. Visuals of the 5th European Clinical Histocompatibility Workshop, Rhodes. Canoga Park: One Lambda Inc.; 2004. pp. 13–14. [Google Scholar]
- 4.Patel R, Terasaki PI. Significance of the positive cross-match test in kidney transplantation. N Engl J Med. 1969;280:735–739. doi: 10.1056/NEJM196904032801401. [DOI] [PubMed] [Google Scholar]
- 5.Heinemann FM, Roth I, Rebmann V, Arnold ML, Spriewald BM, Grosse-Wilde H. Immunoglobulin isotype-specific characterization of anti-human leukocyte antigen antibodies eluted from explanted renal allografts. Hum Immunol. 2007;68:500–506. doi: 10.1016/j.humimm.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Arnold ML, Heinemann FM, Horn PA, Spriewald BM. IgM antibodies and transplantation: auto or allo? Tissue Antigens. 2009;73:389–390. [Google Scholar]
- 7.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou Y, Heinemann FM, Grosse-Wilde H, Sireci G, Wang Z, Lavingia B, Stastny P. Detection of anti-MICA antibodies in patients awaiting kidney transplantation, during the post-transplant course, and in eluates from rejected kidney allografts by Luminex™ flow cytometry. Hum Immunol. 2006;67:230–237. doi: 10.1016/j.humimm.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Zou Y, Stastny P, Süsal C, Döhler B, Opelz G. Antibodies against MICA antigens and kidney-transplant rejection. N Engl J Med. 2007;357:1293–1300. doi: 10.1056/NEJMoa067160. [DOI] [PubMed] [Google Scholar]
- 10.Allele Code Lists of the National Marrow Donor Program (NMDP): http://bioinformatics.nmdp.org/
- 11.IMGT/HLA Nomenclature Databases: www.ebi.ac.uk/imgt/hla/
- 12.Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol. 2008;20:588–593. doi: 10.1016/j.coi.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]