Abstract
Does basal type breast cancer arise from oncogenic transformation of a basal cell type? In this issue Molyneux and colleagues investigate the provenance of the basal-type BRCA1 breast carcinoma and come up with unanticipated results.
Breast cancer is a diverse disease that can be categorized into at least six clinically relevant subtypes based upon molecular gene signatures. The subtypes fit into the broader groupings of either ‘basal’ or ‘luminal’ types due to their molecular similarity to the basal or luminal cells of the normal mammary gland. Thus, basal-type breast cancers express high levels of basal cell markers (cytokeratins 5/6, 14 and 17), while luminal-type breast cancers are defined by high expression of luminal cell markers (estrogen receptor alpha, cytokeratins 8/18 and GATA 3-binding protein) (Sorlie et al., 2003). Such parallels suggest that the biology of the target cell of oncogenic transformation is echoed in the disease that ensues. Put simply, basal-type breast cancers would seem to arise from transformed mammary basal progenitor cells and luminal-type breast cancers from transformed luminal progenitor cells (Petersen and Polyak, 2010).
BRCA1 is a tumor suppressor gene that is often mutated in the germ line, giving rise to greatly elevated risk of developing basal-type breast carcinoma. Previous work had proposed that BRCA1 is an important regulator of mammary stem cell fate, and that breast tissues from women with germ-line BRCA1 mutations had an expansion of BRCA1 mutant mammary stem/progenitor cells (Liu et al., 2008). Furthermore, K14-Cre Brca1f/f p53f/f transgenic mice with targeted deletion of BRCA1 to mammary basal cells developed basal-like tumors with features of BRCA1-mutant breast carcinomas (Liu et al., 2007). As such, BRCA1-mutant breast cancer was thought to arise from a basal progenitor/stem cell.
A recent flow of papers, however, call this notion into question. In this issue, Molyneux and colleagues took a novel approach to define the cell-of-origin for BRCA1-mutant breast cancer. They analyzed a conditional mouse model of BRCA1 deficiency, where Cre recombinase-dependent deletion of exons encoding the C-terminus of the BRCA1 protein combined with p53 heterozygosity, lead to tumour formation. Pertinent to this model, Cre expression was driven by the Beta lactoglobulin (Blg) promoter and is therefore confined to a subpopulation of mammary epithelial cells. Importantly, the Blg-Cre Brca1f/f p53+/− transgenic mice developed mammary tumors that closely resembled human BRCA1-mutant breast cancer. Accordingly, identifying the phenotype of the Blg positive cells in the mouse mammary gland should shed light on the cell-of-origin for BRCA1 basal-type breast carcinoma.
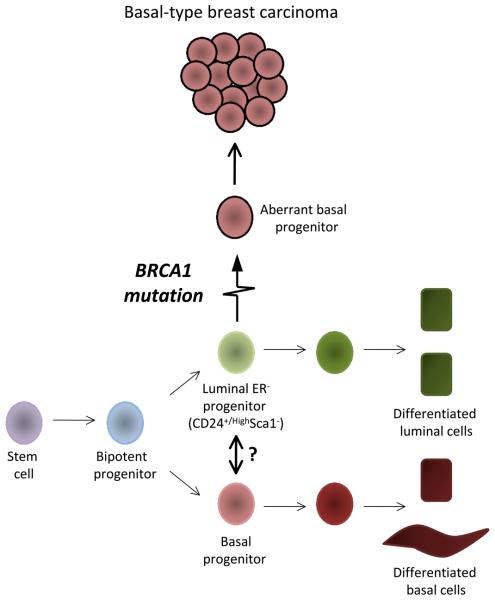
To that end, the authors used cell surface antigen profiles to distinguish three different mammary epithelial populations. They demonstrated that Blg activity is most evident in a CD24+/High Sca-1− ER− cell population, thus defining a luminal ER− progenitor cell as the cell-of-origin for BRCA1-mutant basal-like tumors (Figure 1).
Figure 1. Schematic diagram of a mammary stem cell hierarchy.
In this diagram, a luminal ER- progenitor cell (CD24+/HighSca1-ER-) acquires a BRCA1 mutation and subsequently transitions into an aberrant basal progenitor cell (or directly differentiates into aberrant basal –like cells) and thus gives rise to BRCA1- mutant basal-like breast carcinoma. It is also possible that luminal ER- progenitor cells can transdifferentiate into basal progenitor cells as part of a normal physiological process, either directly (depicted by arrow with a question mark) or first via dedifferentiation to a common progenitor/stem cell
This novel work is at odds with the previous contention that BRCA1-mutant tumors arise from a mammary basal cell (Liu et al., 2007). To further address these conflicting data, Molyneux also presents findings directly comparing tumors arising from Blg-Cre Brca1f/f p53+/− or an independently constructed line of K14-Cre Brca1f/f p53+/− transgenic mice. They arrive at the conclusion that their K14-Cre Brca1f/f p53+/− transgenic mice developed metaplastic carcinoma or malignant adenomyoepithelioma (both rare types of breast cancer), and not a disease reminiscent of BRCA1-mutation. It is possible that the differences in these two studies are due to subtleties in experimental design (e.g., floxed p53 allele versus p53 heterozygote background, different K14-Cre transgenes or floxed Brca1 alleles) which may have lead to oncogenic transformation of different target cell populations within the mammary gland.
In fact, identifying specific target populations in the normal mammary gland remains a daunting challenge, given the dearth of useful stem-cell-specific markers. Thus, current cellular classifications rely heavily upon cell-surface antigen profiling, which results at best in enrichment of target cells rather than isolation of homogenous populations. For example, the CD49fhiCD29hiCD24+Sca1− murine mammary stem cell identifier enriches for a cell population of which less than 5% are actually mammary stem cells (Visvader, 2009). Given these limitations, the most definitive means of assigning mammary cell phenotype is by functionally assessing a cell's ability to repopulate a cleared mouse mammary stromal fat pad. In this assay, mammary stem cells, and to a lesser extent bipotent progenitor cells, generate epithelial outgrowths containing basal and luminal cell lineages. It is therefore somewhat surprising that the luminal ER− progenitor cells defined in the Molyneux study, which seem firmly committed to enter and remain within the luminal differentiation lineage, actually generated epithelial outgrowths containing both luminal and basal cell types in the cleared mammary fat pad.
One interpretation of these data is that luminal ER− progenitor cells can display a capacity for context-dependent multilineage differentiation. While this conclusion is reasonable, it seems equally plausible that the antigen profile used to enrich luminal ER− progenitor cells might also include bipotent progenitor cells. If so, it might be the case that BRCA1 mutations actually occur in a bipotent progenitor cell that subsequently enters exclusively into the committed luminal-progenitor lineage. This possibility would account for the expansion of a luminal–progenitor cell population described in this work by Molyneux and other recent work showing that luminal progenitor numbers are increased in the breast tissue of BRCA1 mutation carriers (Lim et al., 2009).
If this were the case, it is feasible that the earlier work defining the mammary stem/progenitor cell as the BRCA1 breast cancer cell-of-origin could be reconciled with the new findings presented in this issue by concluding that both groups have identified the same cell-of-origin, yet labelled it differently. That conclusion has some support in data reported by Molyneux in directly comparing the gene signatures of Blg-Cre Brca1f/f p53+/− (asserted by Molyneux to target a luminal-cell population) and K14-Cre Brca1f/f p53+/− (asserted by Molyneux to target a basal-cell population) tumors and demonstrating that both tumor types most closely resembled the normal cell type they classified as luminal ER− cells, and thus did not fall into different subgroups.
Perhaps the key to interpreting these novel findings lies in a degree of cellular plasticity that is thought to be largely confined to mammary stem cells. The current depiction of the conventional stem cell hierarchy suggests that stem cells can undergo asymmetric division to create non-stem daughters. Conversely, non-stem cells are portrayed as being unable to move back up the hierarchy and dedifferentiate into a stem-cell state. However, if (as the authors suspect) non-stem cells did indeed have this ability, via either a normal cellular process or a consequence of genetic alterations (such as those associated with tumorigenesis), it is possible that luminal progenitor cells acquiring a BRCA1 mutation might undergo a degree of dedifferentiation, allowing them to revert to a bipotent progenitor or even an oligopotent stem-cell state (both of which states exhibit basal characteristics). In this event, luminal progenitor cells would be able to give rise to carcinomas with a basal phenotype (via conversion to a bipotent progenitor or stem cell).
In accord with the idea presented by Molyneux, another group recently demonstrated that a basal cell might also be the cell-of-origin in a model of prostate cancer, a disease characterized by luminal cell expansion and the absence of basal cells (Goldstein et al., 2010). These findings lend further support to the emerging theme that the histology of cancer does not always reflect the nature of the cell-of-origin. Identifying the target cells of transformation is key to understanding the pathogenesis of these common human tumors. Such information may also prove critical to developing more powerful diagnostic and prognostic tools than are currently available.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, van Vliet MH, Wessels LF, Peterse JL, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen OW, Polyak K. Stem cells in the human breast. Cold Spring Harb Perspect Biol. 2010;2:a003160. doi: 10.1101/cshperspect.a003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]