Abstract
Some of the staphylococcal superantigen-like (SSL) proteins SSL5, SSL7, SSL9, and SSL11 act as immunomodulatory proteins in Staphylococcus aureus. However, little is known about their regulatory mechanisms. We determined the expression levels of ssl5 and ssl8 in seven clinically important S. aureus strains and their regulatory mechanisms in the Newman strain, which had the highest ssl5 and ssl8 expression. Independent comparisons of ssl5 or ssl8 coding and upstream sequences in these strains identified multiple haplotypes that did not correlate with the differential expression of ssl5 and ssl8, suggesting the role of additional regulatory elements. Using knockout mutant strains of known S. aureus global regulators such as Agr, Sae, and SigB in the Newman strain, we showed that both ssl5 and ssl8 were induced by Sae and repressed by Agr, suggesting that Sae and Agr are the positive and the negative regulators, respectively, of these two ssl genes. Moreover, we observed upregulation of sae in the agr mutant and upregulation of agr in the sae mutant compared with the isogenic Newman strain, suggesting that the Agr and Sae may be inhibiting each other. The SigB mutation did not affect ssl5 and ssl8 expression, but they were downregulated in the agr/sigB double mutant, indicating that SigB probably acts synergistically with Agr in their upregulation.
Keywords: ssl5, ssl8, gene expression, Staphylococcus aureus, MRSA, staphylococcal superantigen-like protein
Introduction
Staphylococcus aureus is a significant human pathogen capable of causing a variety of diseases ranging from mild skin and soft tissue infections to bacteremia, pneumonia, endocarditis, and osteomyelitis (Lowy, 1998). The ability of S. aureus to cause a wide range of infections is partly due to the expression of a wide array of virulence factors including, but not limited to, cell wall-associated adhesions, clumping factors, exotoxins, and secreted proteins such as staphylococcal superantigen-like (SSL) proteins (Lowy, 1998; Dinges et al., 2000; Williams et al., 2000; Fitzgerald et al., 2003). The SSL proteins are encoded by a cluster of 11 ssl genes located on S. aureus pathogenicity island-2 (Fitzgerald et al., 2003). These proteins have limited sequence homology to the enterotoxins and toxic shock syndrome toxin 1 and thus represent a novel family of exotoxin-like proteins (Williams et al., 2000). The overall order of ssl genes on an S. aureus chromosome is conserved, and allelic forms of individual ssl genes have been identified in different strains. The sequence homology for individual ssl genes ranges from 85% to 100% in different strains. However, 11 ssl genes within a strain have sequence homology from 36% to 67%, suggesting possible selective pressures encountered during infection (Kuroda et al., 2001; Smyth et al., 2007).
Every strain of S. aureus examined so far carries a cluster of at least seven of the 11 ssl genes, suggesting that they probably have distinct and possibly nonredundant functions (Arcus et al., 2002; Fitzgerald et al., 2003; Smyth et al., 2007). Expression studies of a family of ssl genes in COL, an early methicillin-resistant S. aureus (MRSA) strain, showed that they are upregulated during the stationary phase like other exotoxin genes (Fitzgerald et al., 2003). SSL5 and SSL11 show high structural homology with the chemotaxis inhibitory protein of S. aureus and have been shown to interfere with the interaction between P-selectin glycoprotein ligand-1 and P-selectin, suggesting that S. aureus uses SSL proteins to prevent neutrophil recruitment towards the site of infection (Bestebroer et al., 2007; Chung et al., 2007). The same binding site was also found in SSL2, SSL3, SSL4, and SSL6 (Baker et al., 2007). SSL7 and SSL9 interact with two separate cell surface ligands of human antigen-presenting cells (monocytes and dendritic cells), leading to internalization by these cells, and may thus play a role in the modulation of host immunity against S. aureus (Al-Shangiti et al., 2005). In addition, the ability of SSL5, SSL7, SSL9, and SSL11 to impair the protective immune response against S. aureus (Al-Shangiti et al., 2005; Bestebroer et al., 2007; Chung et al., 2007) suggests that these proteins could represent potential targets for prophylactic or therapeutic agents to treat invasive staphylococcal diseases (Chung et al., 2007). Heme-sensing defective strains of S. aureus have shown enhanced expression of ssl genes, which was associated with the increased S. aureus survival and abscess formation in a host (Torres et al., 2007; Langley et al., 2009). Despite their well-described role in S. aureus pathogenesis, it is not known whether individual SSL proteins are produced in varying amounts in different S. aureus clones or multilocus sequence-based sequence types (ST). It is also not known whether genetic polymorphisms in SSL genes influence their expression levels. The aim of this study was to determine the regulatory mechanism of ssl5 and ssl8 in clinical strains of S. aureus using the Newman as a reference strain.
Materials and methods
Bacterial strains
The S. aureus wild-type and mutant strains used in this study are listed in Table 1. These strains include three ST8 strains (Newman, FPR3757, and RN6390), two ST5 strains (Mu50 and N315), two ST1 strains (MW2 and MSSA476), and one ST250 strain (COL). Epidemiologically, these strains represent two CA-MRSA strains (FPR3757 and MW2), two nosocomial strains (N315 and MSSA476), two laboratory strains (RN6390 and Newman), one vancomycin intermediate resistance strain (Mu50), and an early MRSA (COL) strain. Because COL lacked ssl5 and ssl8 genes, it was used as a negative control in gene expression studies. In addition, the mutant strain of agr (accessory gene regulator) (Δagr::tetM, ALC355) (Wolz et al., 1996); sae (S. aureus exoprotein expression) (sae::Tn917, AS3) (Goerke et al., 2001); sigB (sigma factor B) (ΔrsbUVWsigB::erm(B), IK184) (Kullik et al., 1998); and an agr/sigB double mutant (Δagr::tetM/sigB::kanr) (VKS104, this study) in the Newman background were used to observe the effect of these regulatory genes on ssl5 and ssl8 expression.
Table 1.
Staphylococcus aureus strains used in this study
| S. aureus strains | Source, genotype, and phenotype description | References |
|---|---|---|
| Wild-type strains | ||
| MW2 (NRS123) | CA-MRSA; agr group III; spa type t128; ST1; PVL+ | Baba et al. (2002) |
| FPR3757 (NRS384) | CA-MRSA; agr group I; spa type t008; ST8; PVL+ | Diep et al. (2008) |
| COL (NRS100) | An early MRSA; ST250 | Gill et al. (2005) |
| NCTC8325/RN6390 (NRS147) | Laboratory strain; agr group I; ST8 | Peng et al. (1988) |
| Newman | A clinical isolate, routinely used as a laboratory strain; spa type t008 | Baba et al. (2008) |
| MSSA476 | Nosocomial strain; ST1 | Holden et al. (2004) |
| N315 | Nosocomial strain; ST5 | Kuroda et al. (2001) |
| Mu50 | Vancomycin-intermediate resistant Staphylococcus aureus; ST5 | Hiramatsu et al. (1997) |
| Mutant strains | ||
| ALC355 | Newman Δagr::tetM | Wolz et al. (1996) |
| AS3 | Newman sae::Tn917 | Goerke et al. (2001) |
| IK184 | Newman ΔrsbUVWsigB; Emr | Kullik et al. (1998) |
| VKS104 | Newman, Δagr::tetM/sigB::kanr | This study |
CA-MRSA, community-associated methicillin-resistant S. aureus.
Media and growth conditions
Staphylococcus aureus strains were grown either in tryptic soy broth (TSB) or on tryptic soy agar plates (Beckton Dickinson). For broth culture, an overnight shaking culture, grown at 37 °C in TSB, was used to inoculate 50mL of fresh TSB (1: 200 dilutions). Bacterial growth was subsequently monitored by incubating the flask in a shaking incubator and measuring the turbidity of the culture every 30 min at OD600 nm using a Spectrophotometer (Beckman Coulter Inc., CA) until the culture reached the stationary phase. Cells were collected at the early stationary phase. The MW2, FPR3757, Newman, and MSSA476 reached the early stationary phase (OD600 nm = 4.5) after 4.5 h, whereas strains RN6390, Mu50, N315, and COL reached the early stationary phase after 5.5 h. The transition phase between the late log phase and the stationary phase was considered as the early stationary phase. None of the Newman mutant strains showed any appreciable growth differences from the Newman wild-type strains (data not shown).
Construction of the agr/sigB knockout mutant
For this study, an agr/sigB double mutant was generated by transferring the mutation in the sigB gene to the agr mutant of the Newman strain using a phage transduction procedure as described previously (Singh et al., 2003).
RNA isolation
For gene expression studies, total RNA was isolated at the early stationary phase from all the strains listed in Table 1. Total RNA isolations were performed using a Qiagen RNeasy Mini Kit (Qiagen Inc., Valencia, CA) according to the manufacturer’s recommendations. The extracted RNA concentration was determined using a Bio-Rad SmartSpec Plus Spectrophotometer (Analytical Instruments, LLC, MN). An aliquot of each RNA sample was electrophoresed on a 1.0% agarose gel to assess its integrity and quality.
Relative quantification of ssl5, ssl8, sae, and RNAIII transcripts by real-time reverse transcriptase (RT)-PCR
We quantified the relative transcript ratio of ssl5, ssl8, regulatory genes, sae, and agr (RNAIII) against an endogenous control gene, gmk (guanylate kinase involved in nucleic acid metabolism), in all the strains mentioned in the Table 1. The extracted RNA samples were treated with RNAse-free DNAse using the Turbo DNA-free™ kit (Ambion, Austin, TX) and confirmed to be DNA free by PCR before cDNA synthesis. cDNA synthesis was performed with 2 μg of total RNA using the High-Capacity cDNA Reverse Transcription Kit following the manufacturer’s protocol (Applied Biosystems Inc., Foster City, CA).
From the above reaction mix, ~200 ng of cDNA was mixed with TaqMan Universal PCR Master Mix (2×) (Applied Biosystems Inc.), TaqMan assays containing appropriate PCR primers (900nMμL−1) and a 6-FAM dye-labeled MGB probe (250nMμL−1). The quantitative real-time PCR was performed in a Light cycler (Roche Diagnostics Corp., Indianapolis, IN). The PCR primers and probes are listed in Table 2. Real-time PCR conditions were as follows: one cycle at 50 °C for 2 min is required for optimal AmpErase UNG activity, one cycle of 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min each.
Table 2.
Primers and probes used in this study
| Genes | Primer sequence (50′ → 30′) | Probe sequence (50′ FAM → 30′ NFQ) | References |
|---|---|---|---|
| ssl5 | |||
| Forward | GGTGGTGTCACTAAGAAAAATCAAGAC | ACGCACCAAGATTTC | This study |
| Reverse | CAATACCGTCACCTTCATCTCTCTT | This study | |
| ssl8 | |||
| Forward | GTTCCAAGTGTTTTTATTGGGAAAAGATGA | ACACATGGTTTAGATGTCTTTG | This study |
| Reverse | GTTACACCACTAACACTAAATATTCTTCCATCTA | This study | |
| gmk | |||
| Forward | ACTAGGGATGCGTTTGAAGCTTTAA | AAAGATGACCAATTTATAGAATATG | This study |
| Reverse | CTGGTGTACCATAATAGTTGCCTACAT | This study | |
| sae | |||
| Forward | CAACTGTCGTTTGATGAATTAACACTTATTAACT | ATGGTCACGAAGTCCC | This study |
| Reverse | CCACAATAACTCAAATTCCTTAATACGCAT | This study | |
| RNAIII | |||
| Forward | TCCATTTTACTAAGTCACCGATTGT | ATCTTGTGCCATTGAAATCACTCCTTCCTT | Loughman et al. (2009) |
| Reverse | TGTGATGGAAAATAGTTGATGAGTTGT | Loughman et al. (2009) | |
Relative quantifications of ssl5 and ssl8 and regulatory gene agr (RNAIII) and sae were determined by measuring against the endogenous control, gmk, in the seven clinical and mutant strains (Table 1). Relative quantification was performed using the 2−ΔΔCT calculation according to the manufacturer’s guidelines (Roche Diagnostics Corp.). This method compensates factors such as variability in cDNA synthesis and template concentration and calculates transcript ratios (ssl5/gmk, ssl8/gmk, sae/gmk, and RNAIII/gmk) rather than absolute values. All of the RT-PCR efficiency was ~2 as required for the reliability of 2−ΔΔCT calculation.
In these experiments, gmk was used as a reference gene as its expression levels have been shown to be unchanged under different experimental conditions (Vandecasteele et al., 2001; Nieto et al., 2009). We confirmed that with equal amounts of RNA in our experiments, the gmk transcript levels were the same in the wild-type and the mutant strains. It has been shown that gmk works as well an internal control as gyrA (Eleaume & Jabbouri, 2004). All RT-PCR results were obtained from two independent cultures.
Sequencing of ssl5 and ssl8
Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen Inc.) from all the wild-type and the mutant strains mentioned in Table 1. To amplify the ssl5 and ssl8 upstream and coding sequences primers were designed to cover the 100 bp upstream promoter region and 705 bp ssl5 and 699 bp ssl8 coding regions (Table 3). The amplified products were column purified using the QIAquick PCR Purification Kit (Qiagen Inc.) and sequenced with PCR primers using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems Inc.). Unincorporated dye terminators were removed from the extension products using DyeEx 96 Kit (Qiagen Inc.). Sequences of both strands were analyzed using an ABI Prism 3100 DNA genetic analyzer (Applied Biosystems Inc.). The ssl5 and ssl8 sequences obtained were compared against the DNA sequence database in GenBank to confirm their identity.
Table 3.
Sequencing primers used in this study
| Genes | Target strains | Primer sequence (5′ → 3′) | References |
|---|---|---|---|
| ssl5-F | Mu50 | TCCTCAAATGTGCCAAGTGT | This study |
| ssl5-F | Newman, RN6390, FPR3757, MW2, MSSA476, and N315 | CAGTTTTATTTAACGAACATTATAGATTCC | This study |
| ssl5-R | Newman, RN6390, FPR3757, MW2, MSSA476, Mu50, and N315 | CAACTTATGTTGCCTAACTCCTC | This study |
| ssl8-F | Newman, RN6390, and FPR3757 | TGAAAGTGATGCCCATTGAA | This study |
| ssl8-F | MW2, MSSA476, Mu50, and N315 | CTTCTGAAAGTGATGTCCATTGAA | This study |
| ssl8-R | Newman, RN6390, FPR3757, MW2, MSSA476, Mu50, and N315 | ACATGGGATTATTAAACCGCTTC | This study |
Comparative sequence analysis of ssl5 and ssl8 coding and the putative promoter region
ssl5 coding and its 100 bp upstream sequences in the seven clinical strains were compared with each other. A similar comparison was made for ssl8 alone. The sequence comparison was performed by DNASTAR MEGALIGN program using the CLUSTALW method (LASERGENE, Version 7.2.1, Madison, WI). Allelic forms of the ssl5 and ssl8 present in different strains were identified.
Statistical analysis
Student’s t-test was used to determine the statistical significance for the gene expression data. P values of <0.05 were considered to be statistically significant.
Results
Relative quantification of ssl5 and ssl8 expression in S. aureus strains
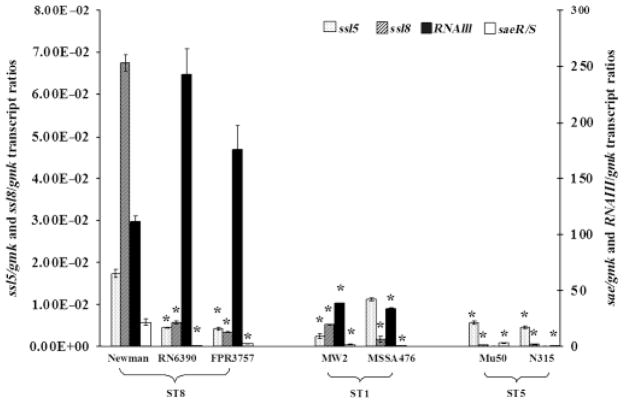
The expression of ssl5 and ssl8 was quantified at the early stationary phase in all the strains listed in Table 1. As expected, the negative control strain, COL, did not show ssl5 or ssl8 expression as it lacked these genes. Both ssl5 and ssl8 had the highest expression in the Newman strain, whereas MW2 and Mu50 strains had the lowest expression, respectively. Both ssl5 and ssl8 expression levels varied in strains within an ST and also when compared among strains with different STs (Fig. 1). The ST8 strains, RN6390 and FPR3757, showed ssl5 levels comparable to each other; however, they had fourfold less expression compared with the Newman strain. In the case of ST1 strains, MSSA476 showed fivefold higher ssl5 expression compared with the MW2 strain. However, MSSA476 and MW2 strains showed 1.5- and 7-fold lower ssl5 expression, respectively, in comparison with the Newman strain. The ST5 strains, Mu50 and N315, showed similar ssl5 expression levels, but showed three- and four-fold less expression, respectively, when compared with the Newman strain (Fig. 1).
Fig. 1.
Transcript ratios of ssl5/gmk, ssl8/gmk, RNAIII/gmk, and sae/gmk quantified at the early stationary phase of seven strains: RN6390, FPR3757, Newman, MW2, MSSA476, Mu50, and N315. The dotted and the hatched bars on the primary axis show ssl5/gmk and ssl8/gmk transcript ratios, respectively. The closed and open bars are relative to the secondary axis and show RNAIII/gmk and sae/gmk transcript ratios. Data represented here show the mean values of two independent measurements. An asterisk (*) denotes that the values are statistically significant (i.e. P<0.05, by Student’s t-test) compared with the Newman strain.
The ssl8 expressions were relatively similar in RN6390 and FPR3757. However, its expression was 12- and 20-fold lower in RN6390 and FPR3757, respectively, compared with the Newman strain. The MW2 strain showed threefold higher ssl8 levels compared with MSSA476; however, these strains showed 13- and 40-fold less ssl8 expression, respectively, compared with the Newman strain. In N315 and Mu50, the ssl8 levels were similar to each other, but in a negligible amount when compared with the Newman strain (Fig. 1). When the expression levels of ssl5 and ssl8 were compared, they were found to be similar in RN6390 and FPR3757, but ssl8 expression was fourfold higher in the Newman strain compared with ssl5. Interestingly, MW2 had twofold higher ssl8 levels compared with ssl5, whereas MSSA476 showed sevenfold higher ssl5 levels compared with ssl8 levels. In contrast, Mu50 and N315 showed 17- and 10-fold higher ssl5 levels, respectively, compared with their ssl8 expression levels (Fig. 1). The differential expression of both ssl5 and ssl8 in different strains prompted us to see whether different haplotypes of ssl5 and ssl8 are present in these strains and whether they correlated with their differential expression.
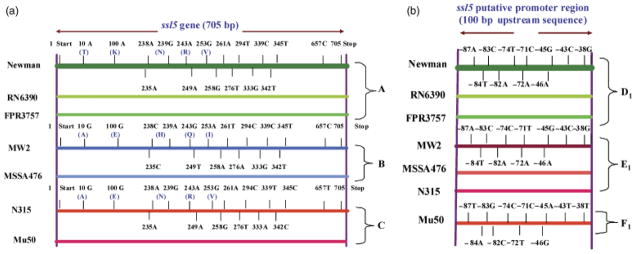
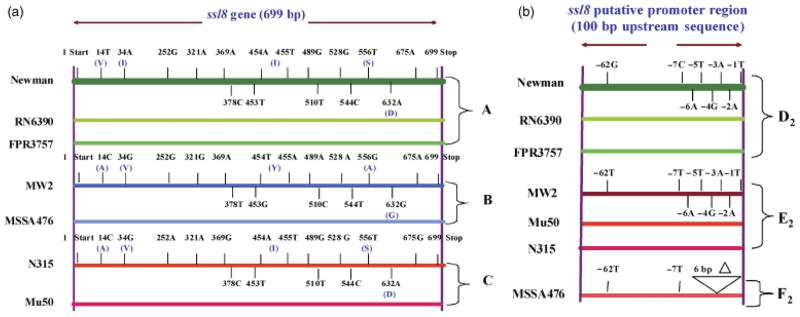
Allelic differences within ssl5 and ssl8 in the clinical strains tested
We sequenced ssl5, ssl8 and their 100 bp upstream regions from the seven clinical strains and various Newman mutant strains used in this study. Because the Newman strain had the highest expression of both ssl5 and ssl8 compared with the other clinical strains tested, the ssl5, ssl8 and their 100 bp upstream sequences obtained were compared with the respective genes of this strain to determine any allelic differences. Based on the respective comparison of ssl5 and ssl8 coding sequences of the seven strains tested (Table 1), three haplotypes emerged. Haplotype A included Newman, FPR3757, and RN6390 strains; haplotype B included MW2 and MSSA476 strains; and haplotype C included Mu50 and N315 strains (Figs 2a and 3a). For the ssl5 or ssl8 upstream sequence comparative analysis, three allelic forms were identified for each one. For both ssl5 and ssl8, allelic type A included the same three strains: Newman, FPR3757, and RN6390. However, for ssl5, allelic type B included MW2, MSSA476, and N315, whereas allelic type C included Mu50 (Fig. 2b). For ssl8, allelic type B included MW2, Mu50, and N315, whereas allelic type C included MSSA476 (Fig. 3b).
Fig. 2.
(a) Schematic comparison of the ssl5 gene sequences found in seven Staphylococcus aureus strains mentioned in Fig. 1. Different-colored horizontal lines represent different haplotypes (A, B, and C) as indicated on the right side. Only differences in the SNPs and the corresponding amino acid (in parentheses) with respect to the Newman strain are shown. The thick green horizontal line at the top represents the ssl5 sequence from the Newman strain. Short vertical lines above and below the horizontal lines indicate the relative positions of SNPs on the gene. The purple long vertical lines at two ends indicate the 5′ and 3′ ends of ssl5. T, threonine; K, lysine; N, asparagine; R, arginine; V, valine; A, alanine; E, glutamic acid; H, histidine; Q, glutamine; and I, isoleucine. (b) The SNPs in the 100 bp upstream sequences of the ssl5 putative promoter region from the strains mentioned in Fig. 1. The SNPs among seven strains are shown with reference to the Newman strain. The thick green horizontal line at the top represents the ssl5 putative promoter sequence from the Newman strain. Short vertical lines above and below the horizontal lines indicate the relative positions of SNPs. The right vertical line indicates the beginning of the upstream sequence. Three allelic subtypes (D1, E1, and F1) are indicated on the right side and strain names on the left side.
Fig. 3.
(a) Schematic diagram of the ssl8 gene sequences from the seven Staphylococcus aureus strains mentioned in Fig. 1. Different-colored horizontal lines represent different S. aureus strains as indicated on the left side. Haplotype groups (A, B, and C) are indicated on the right side. The SNPs and the corresponding amino acid (in parentheses) changes with respect to the Newman strain are shown. The thick green horizontal line at the top represents the ssl8 sequence from the Newman strain. Short vertical lines above and below the horizontal lines indicate the relative positions of SNPs on the gene. The purple long vertical lines at two ends indicate the 5′ and 3′ ends of ssl8. V, valine; I, isoleucine; S, serine; A, alanine; Y, tyrosine; G, glycine; and D, aspartic acid. (b) The SNPs in the 100 bp upstream sequences of the ssl8 putative promoter sequence from strains mentioned in Fig. 1. The deletion and SNPs among seven strains with reference to the Newman strain are shown. The thick green horizontal line at the top represents the ssl8 putative promoter sequence from the Newman strain. Short vertical lines above and below the horizontal lines indicate the relative positions of SNPs. The right vertical line indicates the beginning of the upstream sequence. Symbol ‘Δ’ denotes the deletion. Three allelic subtypes (D2, E2, and F2) are indicated on the right side and strain names on the left side.
The ssl5 and ssl8 coding and promoter sequences showed several single nucleotide polymorphisms (SNPs) (Figs 2a, b and 3a, b). These SNPs and the corresponding amino acid change in the coding region were described in Supporting Information, Tables S1 and S2. There was no correlation between haplotypes or allelic types relative to ssl5 or ssl8 expression. The differential expressions of ssl5 and ssl8 within a haplotype with identical upstream sequences in strains such as Newman, RN6390, and FPR3757 suggested that their expression was influenced by additional factors (Fig. 1).
Effects of staphylococcal global regulators on ssl5 and ssl8 expression
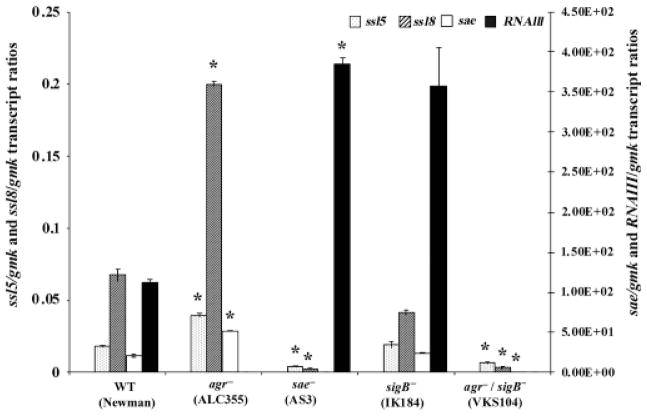
Using Newman as the model strain because of its highest expression of ssl5 and ssl8, we determined the role of known regulatory elements, Agr, Sae, and SigB, in their expression. Relative expressions of ssl5 and ssl8 were compared in the wild-type Newman strains with isogenic mutant strains of agr, sae, sigB and the agr/sigB double mutant to determine their role in ssl5 and ssl8 regulation. The mutant strains did not show any growth difference compared with the wildtype Newman strain (data not shown).
Both ssl5 and ssl8 expression showed upregulation in the agr mutant and downregulation in the sae mutant compared with the wild-type Newman strain (Fig. 4), suggesting that the Agr system is a negative regulator and Sae is a positive regulator for the expression of ssl5 and ssl8 genes. In order to clarify the role of the Agr, we also measured the RNAIII transcript level, which has been shown to regulate the expression of many exoproteins in S. aureus (Peng et al., 1988; Novick et al., 1993). In the seven strains tested, the relative RNAIII transcript levels varied and ranged from 1.5 × 10−4 to 243-folds with reference to gmk transcript levels (Fig. 1). However, no correlation between RNAIII and ssl5 or RNAIII and ssl8 expression was observed in any of the wild type reference strains tested (Fig. 1). We checked the expression of sae in all the reference strains and found that sae expression was 7–36-fold higher in the Newman strain compared with the other six strains used in this study.
Fig. 4.
Transcript ratios of ssl5/gmk, ssl8/gmk, sae/gmk, and RNAIII/gmk quantified at the early stationary phase in the Newman strain and its isogenic mutants: ALC355 (agr−), AS3 (sae−), IK184 (sigB−), and VKS104 (agr−/sigB−). The dotted and the hatched bars on the primary axis show ssl5/gmk and ssl8/gmk transcript ratios, respectively. Open and closed bars with respect to the secondary axis show sae/gmk and RNAIII/gmk transcript ratios, respectively. Data here represent the mean values of two independent measurements. An asterisk (*) shows that the values are statistically significant (i.e. P<0.05, by Student’s t-test) compared with the wild-type strain Newman.
In the sae mutant, the level of RNAIII was higher (3.5-fold), but the transcript levels of both ssl5 and ssl8 were lower by 4- and 28-fold, respectively, compared with their levels in the wild-type Newman (Fig. 4). In the agr mutant, transcript levels of sae, ssl5, and ssl8 were higher by 2.5-, 2-, and 3-fold, respectively, compared with their respective levels in the wild-type Newman. There was no change in the expression of either ssl5 or ssl8 in the Newman strain (Fig. 4) that had a sigB mutation. However, in a sigB/agr double mutant of Newman that expressed 56-fold less sae, expressions of ssl5 and ssl8 were also repressed by 3- and 20-fold, respectively, relative to the wild-type Newman strain. These data collectively suggest SaeR/S to be a major positive regulator and Agr to be a negative regulator of ssl5 and ssl8 gene expression in Newman.
Discussion
Staphylococcal extracellular virulence factors are accessory gene products that contribute significantly to S. aureus pathogenicity (Lowy, 1998; Dinges et al., 2000). Their production is often dependent on quorum sensing (Geisinger et al., 2008) and controlled by a network of global regulators including the two-component regulatory system, Agr and Sae, which act at the transcriptional level (Novick & Jiang, 2003). Sae induces the expression of several virulence factors such as coagulase (Coa), α-hemolysin (Hla), β-hemolysin (Hlb), extracellular adherence protein (Eap), extracellular matrix binding protein (Emp), protein A, and fibronectin-binding proteins (FnbA and FnbB) (Goerke et al., 2001; Harraghy et al., 2005). In contrast, the Agr inhibits the expression of coa, fnbB, and fnbA, indicating that Agr might act as an antagonist of Sae (Wolz et al., 1996). Others have reported that sae is downstream from agr in the regulatory pathway or perhaps epistatic (Giraudo et al., 2003; Novick & Jiang, 2003), suggesting that the sae transcription could be influenced by Agr in some strains, but acts independent of Agr in other strains (Ross & Novick, 2001).
In the present study, we describe the expression pattern of ssl5 and ssl8 in the early stationary phase in several S. aureus strains belonging to different clones. It appears that the regulation of ssl5 and ssl8 expression in S. aureus is strain specific as they varied even within an ST and gene haplotype (Fig. 1). Staphylococcus aureus is known to show a differential expression of genes implicated in virulence. Harraghy et al. (2005) observed marked differences in the expression of staphylococcal adhesins, eap and emp between Newman and NCTC8325 derivative strains, SH1000 (8325-4 rsbU+) and 8325-4 (rsbU−). Our data show that the ssl5 and ssl8 expression is downregulated in the sae mutant strain and upregulated in the agr mutant strain, suggesting that Sae and Agr are possible inducers and repressors, respectively, for ssl5 and ssl8 in the Newman strain (Fig. 4). Indeed, downregulation of several proteins including SSL7 and SSL11 has been observed in a Newman sae mutant strain (Rogasch et al., 2006). The Newman strain is characterized by unusually high sae levels, which have been confirmed in this study as well. The high sae expression in this strain can be attributed to a point mutation in the sensor histidine kinase of the SaeR/S two-component regulatory system (Steinhuber et al., 2003; Geiger et al., 2008). Proteomics and microarray analyses have revealed that most of the genes influenced by Sae are involved in bacterial adhesion, immune evasion, immune modulation, or toxicity (Foster, 2005; Liang et al., 2006; Rogasch et al., 2006). More importantly, it has been shown that sae is essential for virulence gene expression in vivo (Goerke et al., 2001). It was interesting to observe the suppressive effect of Agr on ssl5 and ssl8 expression, suggesting that Agr does not always act as a positive regulator for virulence gene expression in S. aureus, and inhibiting the Agr function to reduce virulence could have other consequences (Otto, 2001). Loss of Agr increases the bacterial colonization, biofilm formation, and attachment to polystyrene, suggesting that the agr mutant strain may have a greater capacity to cause chronic infections than agr-positive strains (McNamara & Bayer, 2005).
We speculated that the lack of Agr could have caused the enhanced expression of some proteins that aid in the upregulation of ssl5 and ssl8. Surprisingly, we found that the agr mutation caused increased sae transcript levels and vice versa, which indicated that the sae and agr could have an inhibitory effect on each other, and repression of ssl5 and ssl8 genes by Agr is dependent on Sae in the Newman background. This observation is in contrast to a previous report suggesting that the sae does not effect the transcription of other regulatory genes where they have observed strong signals for RNAIII in both RN6734, a Φ13 lysogen of RN6390, and its sae mutant strain, RN9808 (Novick & Jiang, 2003). However, Voyich et al. (2009) reported that ssl11 and the agr operon in a saeR/S mutant of MW2 strain is downregulated by ~16- and 2-fold, respectively, at the early stationary growth phase. In concordance with our data, Liang et al. (2006) showed, by RT-PCR analysis, that agrA mRNA levels were significantly upregulated in the saeS null mutant compared with its wild-type strain, WCUH29, a virulent clinical isolate. Taken together, these data suggest that the influence of saeR/S on the transcriptional regulation of virulence genes is probably dependent on multiple factors including the genomic background of the strain studied (Liang et al., 2006; Rogasch et al., 2006).
Interestingly, in the agr/sigB double mutant, the expressions of ssl5, ssl8, and sae was downregulated (Fig. 2). However, in the agr mutant strain, these genes were upregulated, whereas the expression of either ssl5 or ssl8 did not change in a Newman sigB mutant. This suggests that SigB probably acts synergistically with Agr, but not alone, to upregulate ssl5 and ssl8. This could very well be mediated by sae specifically in the Newman strain. An analogous phenomenon such as enhanced repression of exotoxin-encoding genes in double mutants of regulatory genes in S. aureus is not uncommon. For example, sar and agr double mutants are less virulent compared with the agr single mutant (Booth et al., 1997). Differences in the transcript levels of regulatory genes (agr, sarA, sigB, and saeR/S) have been reported between COL and Newman strains that correlate well with the expression of virulence-associated genes (Rogasch et al., 2006).
In summary, ssl5 and ssl8 expression in S. aureus clinical isolates is strain dependent and not influenced by differences in their alleles. They are positively regulated by Sae and negatively by Agr in the Newman strain. Furthermore, the ssl5 and ssl8 repression by Agr is probably achieved by the downregulation of Sae in the Newman strain. This is the first report of a negative regulation of an ssl gene by Agr. This study also highlights the potential challenges in managing infections due to S. aureus strains, which could potentially produce varying amounts of SSLs. Understanding the intricacy of global regulatory genes and their mode of regulation in different genetic backgrounds would provide an important insight into the molecular mechanisms of staphylococcal virulence. This may perhaps reveal specific targets, which would enable therapeutic intervention in S. aureus infections.
Supplementary Material
List of SNPs identified in the ssl5 coding and upstream regions in Staphylococcus aureus strains.
List of SNPs identified in the ssl8 coding and upstream regions in Staphylococcus aureus strains.
Acknowledgments
This research was funded in part by research grant RO1 AI061385 from the National Institutes of Allergy and Infectious Diseases to S.K.S. The authors thank James Burmester and Joseph Mazza, Marshfield Clinic Research Foundation, for critically reviewing the manuscript.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Al-Shangiti AM, Nair SP, Chain BM. The interaction between staphylococcal superantigen-like proteins and human dendritic cells. Clin Exp Immunol. 2005;140:461–469. doi: 10.1111/j.1365-2249.2005.02789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus VL, Langley R, Proft T, Fraser JD, Baker EN. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J Biol Chem. 2002;277:32274–32281. doi: 10.1074/jbc.M203914200. [DOI] [PubMed] [Google Scholar]
- Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker HM, Basu I, Chung MC, Caradoc-Davies T, Fraser JD, Baker EN. Crystal structures of the staphylococcal toxin SSL5 in complex with sialyl Lewis X reveal a conserved binding site that shares common features with viral and bacterial sialic acid binding proteins. J Mol Biol. 2007;374:1298–1308. doi: 10.1016/j.jmb.2007.09.091. [DOI] [PubMed] [Google Scholar]
- Bestebroer J, Poppelier MJ, Ulfman LH, Lenting PJ, Denis CV, van Kessel KP, van Strijp JA, de Haas CJ. Staphylococcal superantigen-like 5 binds PSGL-1 and inhibits P-selectin-mediated neutrophil rolling. Blood. 2007;109:2936–2943. doi: 10.1182/blood-2006-06-015461. [DOI] [PubMed] [Google Scholar]
- Booth MC, Cheung AL, Hatter KL, Jett BD, Callegan MC, Gilmore MS. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1550–1556. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MC, Wines BD, Baker H, Langley RJ, Baker EN, Fraser JD. The crystal structure of staphylococcal superantigen-like protein 11 in complex with sialyl Lewis X reveals the mechanism for cell binding and immune inhibition. Mol Microbiol. 2007;66:1342–1355. doi: 10.1111/j.1365-2958.2007.05989.x. [DOI] [PubMed] [Google Scholar]
- Diep BA, Stone GG, Basuino L, et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2008;197:1523–1530. doi: 10.1086/587907. [DOI] [PubMed] [Google Scholar]
- Dinges MM, Orwin PM, Schlievert PM. Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000;13:16–34. doi: 10.1128/cmr.13.1.16-34.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eleaume H, Jabbouri S. Comparison of two standardization methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Meth. 2004;59:363–370. doi: 10.1016/j.mimet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JR, Reid SD, Ruotsalainen E, Tripp TJ, Liu M, Cole R, Kuusela P, Schlievert PM, Jarvinen A, Musser JM. Genome diversification in Staphylococcus aureus: molecular evolution of a highly variable chromosomal region encoding the Staphylococcal exotoxin-like family of proteins. Infect Immun. 2003;71:2827–2838. doi: 10.1128/IAI.71.5.2827-2838.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–998. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190:3419–3428. doi: 10.1128/JB.01927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, George EA, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930–8938. doi: 10.1074/jbc.M710227200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Fouts DE, Archer GL, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo AT, Mansilla C, Chan A, Raspanti C, Nagel R. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr Microbiol. 2003;46:246–250. doi: 10.1007/s00284-002-3853-z. [DOI] [PubMed] [Google Scholar]
- Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol. 2001;40:1439–1447. doi: 10.1046/j.1365-2958.2001.02494.x. [DOI] [PubMed] [Google Scholar]
- Harraghy N, Kormanec J, Wolz C, Homerova D, Goerke C, Ohlsen K, Qazi S, Hill P, Herrmann M. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology. 2005;151:1789–1800. doi: 10.1099/mic.0.27902-0. [DOI] [PubMed] [Google Scholar]
- Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- Holden MT, Feil EJ, Lindsay JA, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. P Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Ohta T, Uchiyama I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- Langley RJ, Proft T, Fraser JD. Staphylococcal immune evasion toxins. In: Proft T, editor. Microbial Toxins: Current Research and Future Trends. Caister Academic Press; Wymondham, UK: 2009. pp. 147–166. [Google Scholar]
- Liang X, Yu C, Sun J, Liu H, Landwehr C, Holmes D, Ji Y. Inactivation of a two-component signal transduction system, SaeRS, eliminates adherence and attenuates virulence of Staphylococcus aureus. Infect Immun. 2006;74:4655–4665. doi: 10.1128/IAI.00322-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Fritz SA, Storch GA, Hunstad DA. Virulence gene expression in human community-acquired Staphylococcus aureus infection. J Infect Dis. 2009;199:294–301. doi: 10.1086/595982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. New Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- McNamara PJ, Bayer AS. A rot mutation restores parental virulence to an agr-null Staphylococcus aureus strain in a rabbit model of endocarditis. Infect Immun. 2005;73:3806–3809. doi: 10.1128/IAI.73.6.3806-3809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto PA, Covarrubias PC, Jedlicki E, Holmes DS, Quatrini R. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol Biol. 2009;10:63. doi: 10.1186/1471-2199-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709–2717. doi: 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides. 2001;22:1603–1608. doi: 10.1016/s0196-9781(01)00495-8. [DOI] [PubMed] [Google Scholar]
- Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;179:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogasch K, Ruhmling V, Pane-Farre J, et al. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. 2006;188:7742–7758. doi: 10.1128/JB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HF, Novick RP. sae is a key intermediary in the activation by agr of the staphylococcal virulon. In: Winans S, Bassler B, editors. Cell-cell Communication. American Society for Microbiology; Snowbird, UT: 2001. [Google Scholar]
- Singh VK, Schmidt JL, Jayaswal RK, Wilkinson BJ. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int J Antimicrob Ag. 2003;21:256–261. doi: 10.1016/s0924-8579(02)00359-x. [DOI] [PubMed] [Google Scholar]
- Smyth DS, Meaney WJ, Hartigan PJ, Smyth CJ. Occurrence of ssl genes in isolates of Staphylococcus aureus from animal infection. J Med Microbiol. 2007;56:418–425. doi: 10.1099/jmm.0.46878-0. [DOI] [PubMed] [Google Scholar]
- Steinhuber A, Goerke C, Bayer MG, Doring G, Wolz C. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J Bacteriol. 2003;185:6278–6286. doi: 10.1128/JB.185.21.6278-6286.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109–119. doi: 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol. 2001;183:7094–7101. doi: 10.1128/JB.183.24.7094-7101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Vuong C, DeWald M, et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RJ, Ward JM, Henderson B, Poole S, O’Hara BP, Wilson M, Nair SP. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect Immun. 2000;68:4407–4415. doi: 10.1128/iai.68.8.4407-4415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz C, McDevitt D, Foster TJ, Cheung AL. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect Immun. 1996;64:3142–3147. doi: 10.1128/iai.64.8.3142-3147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of SNPs identified in the ssl5 coding and upstream regions in Staphylococcus aureus strains.
List of SNPs identified in the ssl8 coding and upstream regions in Staphylococcus aureus strains.