Abstract
To improve the potency of the hypoxic cytotoxin tirapazamine (TPZ), we have constructed an analog, SN26955, with the TPZ moiety attached to an acridine chromophore to target the drug to DNA. The underlying reason for this is our previous finding that the hypoxic cytotoxicity of TPZ is a result of its ability to produce DNA double-strand breaks, whereas many of the toxicities of the drug in clinical use are likely the result of its metabolism in the cytoplasm and effects on mitochondria. We found that the DNA-targeted TPZ analog was more potent than TPZ in killing hypoxic cells by 1–2 orders of magnitude, yet it retained the hypoxic selectivity for cell killing of TPZ. We show that SN26955 is only active in producing DNA damage when it is enzymatically reduced while bound to, or in close association with, the DNA. We also show that it has a different cofactor dependence than TPZ for reduction leading to DNA double-strand breaks, suggesting the involvement of a different reductase for production of the lethal lesion than for TPZ. These results show the promise of DNA-targeting of TPZ to produce a DNA compound with greater clinical efficacy than TPZ itself.
Keywords: Tirapazamine, SN26955, Bioreductive drugs, Hypoxia, Tumor, DNA-targeting
1. Introduction
TPZ (SR 4233, 3-amino-1,2,4-benzotriazine-1,4-dioxide) is the first member of a new class of tumor-targeted drugs to enter clinical trials. Its tumor specificity rests on the fact that TPZ is a prodrug that becomes activated to its cytotoxic form only in the hypoxic microenvironment that is characteristic of solid tumors [1,2]. As hypoxic regions in tumors confer resistance to standard radiotherapy and chemotherapy [3,4], a drug selectively toxic to hypoxic cells would be both tumor specific and kill the cells resistant to conventional therapy [5,6]. TPZ has already demonstrated significant activity in Phase II and III clinical trials in combination with radiotherapy and cisplatin-based chemotherapy [7–9].
TPZ is converted by intracellular reductase(s) to a cytotoxic radical that under hypoxic conditions generates base damage, SSBs and DSBs [10,11]. However, when oxygen is present, the TPZ radical is back-oxidized to the non-toxic parent compound, thereby decreasing the cytotoxicity [12]. Studies with cells in vitro have indicated that it is the DSB that is responsible for the toxicity of TPZ under hypoxic conditions [10]. While many intracellular enzymes can convert TPZ to its cytotoxic radical [13,14], it is the nuclear reductase(s) that is responsible for the DNA damage caused by TPZ [15]. However, like the cytoplasm, the nucleus is highly compartmentalized with the nuclear matrix, a non-chromatin protein network, dictating this subnuclear organization [16]. Based on recent studies showing that TPZ can be selectively reduced by nuclear matrix-associated reductases [17], we hypothesized that the TPZ metabolized at, or close to, the nuclear matrix would affect metabolic activities associated with the nuclear matrix. Consistent with this, we have shown recently that TPZ, under hypoxic conditions, produces a marked inhibition both of DNA replication [18], one of the activities that occur at the nuclear matrix, and of the activity of topoisomerase II (topo II) [19], an enzyme that is highly enriched in the nuclear matrix and which is involved in the cytotoxicity of several chemotherapeutic agents, such as doxorubicin and etoposide [20]. Indeed, much, if not all, of the cytotoxicity of TPZ under hypoxia can be attributed to the interaction of the TPZ radical with topo II [19].
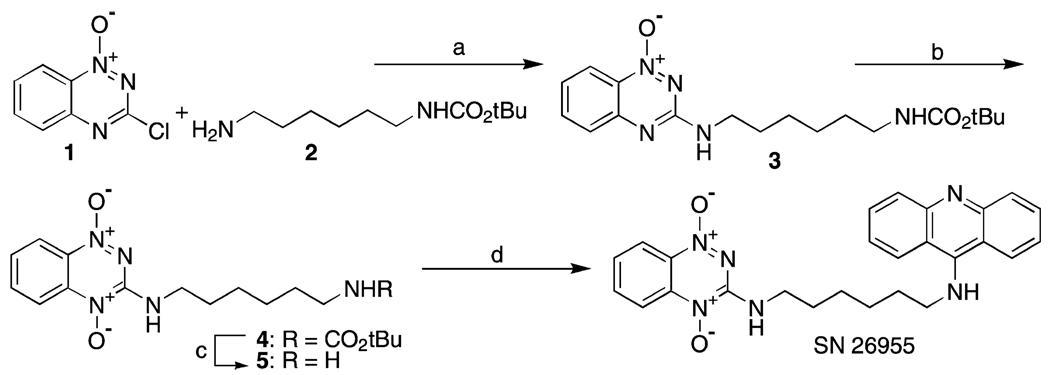
The conclusion of the above studies is that much of the metabolism of TPZ, namely that which occurs in the cytoplasm and in the nucleus at sites away from the nuclear matrix, is unproductive in that it does not contribute to damage leading to DSBs. Indeed, it is likely that such damage may be contributing to some of the toxic side-effects of TPZ including damage to mitochondria, which we have shown to be a target for TPZ-induced damage in aerobic cells [21]. In addition, any metabolism that does not directly contribute to hypoxic cytotoxicity has the potential to compromise extravascular diffusion of the drug into hypoxic zones [22,23]. A possible way in which this unproductive metabolism could be minimized would be to target TPZ directly to the DNA. To test this possibility, we have synthesized a TPZ analog attached to a DNA-intercalating acridine chromophore. We show that this compound, SN26955 (Fig. 1), is considerably more potent than TPZ in killing hypoxic cells, but retains a similar differential killing of hypoxic to aerobic cells. This drug, which establishes a proof of principle that the potency of TPZ can be increased considerably by targeting it to DNA, opens up a promising new area for further drug development of this compound.
Fig. 1.
Structure of SN26955 showing the TPZ moiety on the left attached via a linker onto a DNA-intercalating acridine ring.
2. Materials and methods
2.1. Cell lines
Cell lines were maintained in a humidified atmosphere of 95% air and 5% CO2 at 37°. HeLa cells, a cervical carcinoma cell line, were obtained from the American Type Culture Collection and maintained as monolayers in α-MEM (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum (FBS) plus 200 units/mL of penicillin and 0.2 mg/mL of streptomycin. HeLa cells were also grown in spinner flasks in RPMI medium supplemented with 10% FBS and antibiotics. HT29 cells were obtained from Dr. R.M. Sutherland (SRI International) and were cultured in McCoy’s 5A medium supplemented with 10% FBS and antibiotics as for HeLa cells. All experiments utilized cells in logarithmic-phase growth.
2.2. Drugs and chemicals
TPZ was obtained from Sanofi-Synthelabo. Dihydrorhodamine 123 and rhodamine 123 were from Molecular Probes. All other chemicals were obtained from Sigma and were of the highest analytical quality.
2.3. Synthesis of SN26955
SN26955 was prepared in five steps from 3-chloro-1,2,4-benzotriazine 1-oxide, 6-t-butyloxycarbamoylhexylamine (Aldrich) and 9-methoxyacridine (Fig. 1) as detailed below.
2.3.1. 3-[(6-t-Butyloxycarbamoylhexyl)amino]-1,2,4-benzotriazine 1-oxide (3)
A solution of 6-t-butyloxycarbamoylhexylamine 2 (12.8 g, 61.1 mmol) (Aldrich) in dichloromethane (DCM) was added to a stirred solution of 3-chloro-1,2,4-benzotriazine 1-oxide 1 (3.70 g, 20.4 mmol) [24] and Et3N (5.7 mL, 40.8 mmol) in DCM (100 mL), and the solution was stirred at 20° for 96 hr. The solvent was evaporated and the residue chromatographed, eluting with a gradient (30–100%) of EtOAc/pet. ether, to give 1-oxide 3 (4.77 g, 65%) as a yellow powder, m.p. (EtOAc/pet. ether) 154–156°; 1H NMR δ 8.26 (d, J = 8.6 Hz, 1 H, H 8), 7.70 (dd, J = 8.2, 7.2 Hz, 1 H, H 6), 7.59 (d, J = 8.5 Hz, 1 H, H 5), 7.27 (dd, J = 8.0, 7.5 Hz, 1 H,H 7), 5.34 (br s, 1 H, NH), 4.55 (br s, 1 H, OCONH), 3.51 (dd, J = 6.8, 6.6 Hz, 2 H, CH2N), 3.10–3.13 (m, 2 H, CH2N), 1.64–1.72 (m, 2 H, CH2), 1.48–1.54 (m, 2 H, CH2), 1.44 [s, 9 H, C(CH3)3], 1.38–1.43 (m, 4 H, 2 × CH2). Anal. calc. for C18H27N5O3: C, 59.8; H, 7.5; N, 19.4; found: C, 59.6; H, 7.7; N, 19.2%.
2.3.2. 3-[(6-t-Butyloxycarbamoylhexyl)amino]-1,2,4-benzotriazine 1,4-dioxide (4)
A solution of meta-chloroperbenzoic acid (MCPBA; 1.48 g, 6.02 mmol) in DCM (20 mL) was added dropwise to a stirred solution of 1-oxide 3 (1.45 g, 4.01 mmol) in DCM (100 mL) at 20°, and the solution was stirred for 4 hr. The solution was partitioned between DCM (200 mL) and saturated KHCO3 solution (200 mL). The organic fraction was dried and the solvent evaporated. The residue was chromatographed on neutral alumina, eluting with 50% EtOAc/DCM and then a gradient (0–10%) MeOH/CHCl3, to give (i) starting material 3 (0.73 g, 50%), and (ii) 1,4-dioxide 4 (0.55 g, 37%) as a yellow powder, m.p. (EtOAc/DCM) 132–134°; 1H NMR [(CD3)2SO] δ 8.30 (dd, J = 6.3, 6.1 Hz, 1 H, OCONH), 8.19 (d, J = 8.5 Hz, 1 H, H 8), 8.12 (d, J = 8.5 Hz, 1 H, H 5), 7.91–7.95 (m, 1 H, H 6), 7.53–7.57 (m, 1 H, H 7), 6.76 (br s, 1 H, NH), 3.32–3.39 (m, 2 H, CH2N), 2.87–2.92 (m, 2 H,CH2N), 1.56–1.61 (m, 2 H,CH2), 1.32–1.40 [m, 13 H, 2 × CH2, C(CH3)3], 1.25–1.31 (m, 2 H, CH2). Anal. calc. for C18H27N5O4·1/4H2O: C, 56.6; H, 7.3; N, 18.3; found: C, 56.8; H, 7.3; N, 16.8%.
2.3.3. N1-(1,4-Dioxido-1,2,4-benzotriazin-3-yl)-1,6-hexanediamine (5)
HCl gas was bubbled through a solution of carbamate 4 (204 mg, 0.54 mmol) in MeOH (20 mL) for 2 min, and the solution was stirred at 20° for 16 hr. The solvent was evaporated and the residue partitioned between CHCl3 (100 mL) and saturated KHCO3 solution (100 mL). The aqueous fraction was further extracted with CHCl3 (3 × 30 mL), the combined organic extracts were dried, and the solvent was evaporated to give amine 5 (127 mg, 85%) as a red powder, m.p. 120–122°; 1H NMR δ 8.34 (d, J = 8.5 Hz, 1 H, H 8′), 8.29 (d, J = 8.6 Hz, 1 H, H 5′), 7.87–7.90 (m, 1 H, H 6′), 7.48–7.52 (m, 1 H, H 7′), 7.13 (s, 1 H, NH), 3.60 (t, J = 7.1 Hz, 2 H, CH2N), 2.70 (t, J = 6.8 Hz, 2 H, CH2N), 1.70–1.76 (m, 2 H, CH2), 1.35–1.50 (m, 6 H, 3 × CH2). Anal. calc. for C13H19N5O2: C, 56.3; H, 6.9; N, 25.3; found: C, 56.3; H, 6.8; N, 22.2%. The compound was dissolved in MeOH, treated with HCl gas, and the solvent evaporated. The residue was crystallized to give the dihydrochloride of 5 as a red powder, m.p. (MeOH/EtOAc) 150° (dec.).
2.3.4. N1-(9-Acridinyl)-N6-(1,4-dioxido-1,2,4-benzotriazin-3-yl)-1,6-hexanediamine (SN26955)
A solution of amine 5 (64 mg, 0.23 mmol) and 9-methoxyacridine (53 mg, 0.25 mmol) in MeOH (10 mL) was stirred at reflux temperature for 10 hr. The solvent was evaporated and the residue chromatographed on neutral alumina, eluting with a gradient (0–5%) of MeOH/CHCl3, to give SN26955 (63 mg, 60%) as a red solid; 1H NMR δ 8.30 (d, J = 8.5 Hz, 1 H, H 8″), 8.29 (d, J = 8.5 Hz, 1 H, H 5″), 8.11 (d, J = 8.6 Hz, 2 H, H 1′, H 8′), 8.04 (d, J = 8.6 Hz, 2 H, H 4′, H 5′), 7.84 (ddd, J = 8.5, 7.2, 1.2 Hz, 1 H, H 6″), 7.59–7.64 (m, 2 H, H 3′, H 6′), 7.57 (ddd, J = 8.5, 7.2, 1.2 Hz, 1 H, H 7″), 7.31–7.35 (m, 2 H, H 2′, H 7′), 7.15 (br s, 1 H, NH), 3.84 (dd, J = 7.2, 7.1 Hz, 2 H, CH2N), 3.57 (dt, J = 6.7, 6.5 Hz, 2 H, CH2N), 1.78–1.85 (m, 2 H, CH2), 1.67–1.74 (m, 2 H, CH2), 1.43–1.53 (m, 4 H, 2 × CH2), NH not observed; MS (FAB+) m/z 455 (MH+, 20%), 439 (10%); HRMS (FAB+) calc. for C26H27N6O2 (MH+) m/z 455.2196, found 455.2182. The compound was dissolved in MeOH and treated with HCl gas and the solvent evaporated. The residue was crystallized from MeOH/EtOAc to give the hydrochloride of 5, m.p. (MeOH/EtOAc) 118–119°. Anal. calc. for C26H26N6O2·2HCl·1/2H2O: C, 58.2; H, 5.5; N, 15.7; found: C, 57.8; H, 5.5; N, 15.3%.
2.4. Clonogenic assay
For exposure to TPZ or SN26955, cells were seeded onto 60-mm diameter glass dishes in complete medium 2 days before the treatment such that each dish would have 1 × 106 cells at the time of drug exposure. TPZ or SN26955 was added at the appropriate concentration in medium at a final volume of 2 mL. The cells were then added to prewarmed, leak-proof aluminum jigs, placed on a shaking platform at room temperature and treated with five rapid evacuations and filling with either 95% nitrogen–5% CO2 for hypoxia or with 95% air–5% CO2 for aerobic controls. The jigs were then placed in a 37° box for 1 hr with shaking. Cells were washed with PBS, trypsinized, counted, and plated in serial dilution in complete medium for cloning. After 10–14 days, the plates were stained with crystal violet, and colonies with more than 50 cells were counted as survivors.
2.5. Radical detection with DHR123
TPZ or SN26955 radicals were detected by using DHR123 oxidation as an indicator of free radical formation. DHR123 is a non-fluorescent compound that is oxidized by the TPZ radical to form the highly fluorescent rhodamine 123 compound [25]. DHR123 (25 µM) was added to the metabolism system described above in a total volume of 2 mL. The reaction was placed in 60-mm diameter glass dishes, and hypoxia was created in the aluminum jigs with alternate flushing and filling of 95% N2, 5% CO2. After a 1-hr drug exposure at 37°, the solution was transferred to a cuvette, and the fluorescence was measured in a Perkin-Elmer LS-3 fluorescence spectrophotometer at 509-nm absorbance, 529-nm emission, with a slit-width of 10 nm. A blank containing 1× metabolism buffer and cofactors was used to zero the spectrophotometer.
2.6. Nuclei isolation
Intact nuclei were isolated from HeLa cells as previously described with minor modifications [26]. Briefly, cells were harvested in log phase, washed twice with cold PBS, and resuspended at 12.5 µL/106 cells in NI buffer (115 mM KCl, 5 mM NaCl, 1 mM KH2PO4, 20 mM HEPES, 0.3 mM MgCl2, 0.5 mM phenylmethylsulfonyl fluoride, and 0.5 mM dithiothreitol at pH 7.4). An equivalent volume of NI buffer with 1% IGEPAL CA-630 was added slowly, and the reaction was held on ice for 15 min. The permeable cells were disrupted by pipetting 25–30 times with a 1-mL pipette tip. The isolated nuclei were diluted with a 10× volume of NI buffer, centrifuged at 4°, and washed with 10 mL of NI buffer.
2.7. Alkaline comet assay
SSBs were measured using the alkaline comet assay. Isolated nuclei and whole cells were treated with TPZ or SN26955 in glass dishes under hypoxia in aluminum jigs as described above for 1 hr. Cells were treated with drug dissolved in α-medium containing 10% serum. Isolated nuclei were treated with drug in nuclei treatment buffer (100 mM KCl, 1 mM KH2PO4, 25 mM NaHCO3, 20 mM HEPES, 10 mM EDTA, 5 mM glutathione, and 0.9 mM spermine at pH 7.4). After treatment, cells and nuclei were suspended in cold PBS without Mg and CaCl2 at a concentration of 3 × 104 cells/mL. Slides for the comet assay were prepared following the protocol of Olive [27]. A 500-mL aliquot of cell or nuclei suspension was placed in a labeled 5-mL glass tube, and 1.5 mL of 1% low-gelling temperature agarose (Sigma type VII dissolved in deionized water and held in a 45° water bath) was added to the suspension. The agarose solution was pipetted onto a microscope slide and allowed to gel for 1 min at 4° on a cold metal block. The slides were then placed in an alkaline lysis solution containing 1 M sodium chloride, 0.03 M sodium hydroxide, and 0.1% N-laurylsarcosine for 1 hr at room temperature. After lysis, the slides were washed three times for 20 min in 2 mM EDTA with 0.03 M sodium hydroxide, placed in a gel box, and electrophoresed in rinse buffer at 0.6 V/cm for 20 min. The slides were then removed from the gel box and placed in water for 15 min, followed by staining in propidium iodide (2.5 µg/mL in water) for 15 min. The individual comets were scored as described by Olive [27] and the images analyzed with Loats comet assay analysis software program. The tail moment is calculated as the product of the percentage of DNA in the comet tail multiplied by the length between the means of the head and the tail distributions. Mean tail moments were determined from 150 or more individual comets for each data point.
2.8. Neutral comet assay
DSBs were measured in cells and isolated nuclei by the neutral comet assay [27]. Cells and nuclei were treated as described above, placed in agarose, pipetted onto slides, and lysed in a neutral lysis buffer according to the published protocol. The slides were rinsed in neutral buffer, electrophoresed, and visualized as described above.
2.9. DNA binding assay
DNA binding was measured by equilibrium dialysis using a Dianorm dialyzer (Diachema AG) with 20 two-cell dialysis chambers (1 mL each). SN26955 (15 µM) with calf thymus DNA (Sigma type I) (25–1000 µM) in KHE buffer (10 mM KCl, 2 mM NaHEPES, 10 µM Na2EDTA, pH 7.0) was dialyzed (2 hr at 37°) against SN26955 in KHE buffer using a Cuprophan dialysis membrane (Medicell International Ltd.) with a MW cutoff of 10,000 Da. After equilibrium conditions were reached, the contents of the dialysis chambers with KHE buffer were collected and analyzed directly by HPLC. To the samples containing DNA, 9 vol. of ice-cold ethanol was added and kept on ice for 1 hr followed by a brief spin to precipitate the DNA. Subsequently, the volume of the aqueous ethanol was reduced using a Speed-Vac concentrator and reconstituted with mobile phase followed by injection onto a high performance liquid chromatogram. Concentrations of SN26955 were calculated using an internal standard.
3. Results
3.1. SN26955 binding to DNA
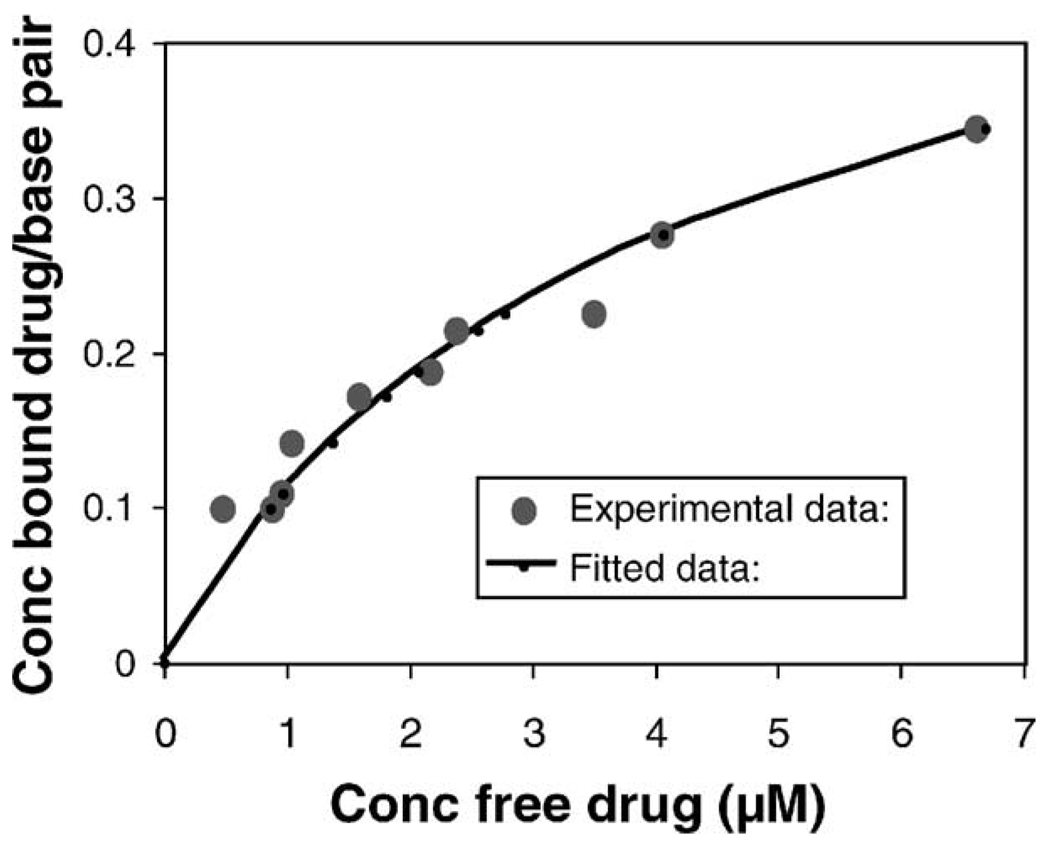
We measured the binding of SN26955 (15 µM at 37°) to calf thymus DNA by equilibrium dialysis with quantitation of the drug by HPLC. The results are shown in Fig. 2. Also shown is the fit of the binding data using the neighboring site exclusion model of McGhee and von Hippel [28]. The derived association constant [(14.3 ± 1.5) × 104 M−1] and binding site size (1.52 ± 0.09 bp) indicate that SN26955 is a moderately strong DNA binder.
Fig. 2.
Data showing the binding of SN26955 to calf thymus DNA. The line shows the fit of the binding data using the neighboring site exclusion model of McGhee and von Hippel [28].
3.2. Activity of SN26955 in vitro
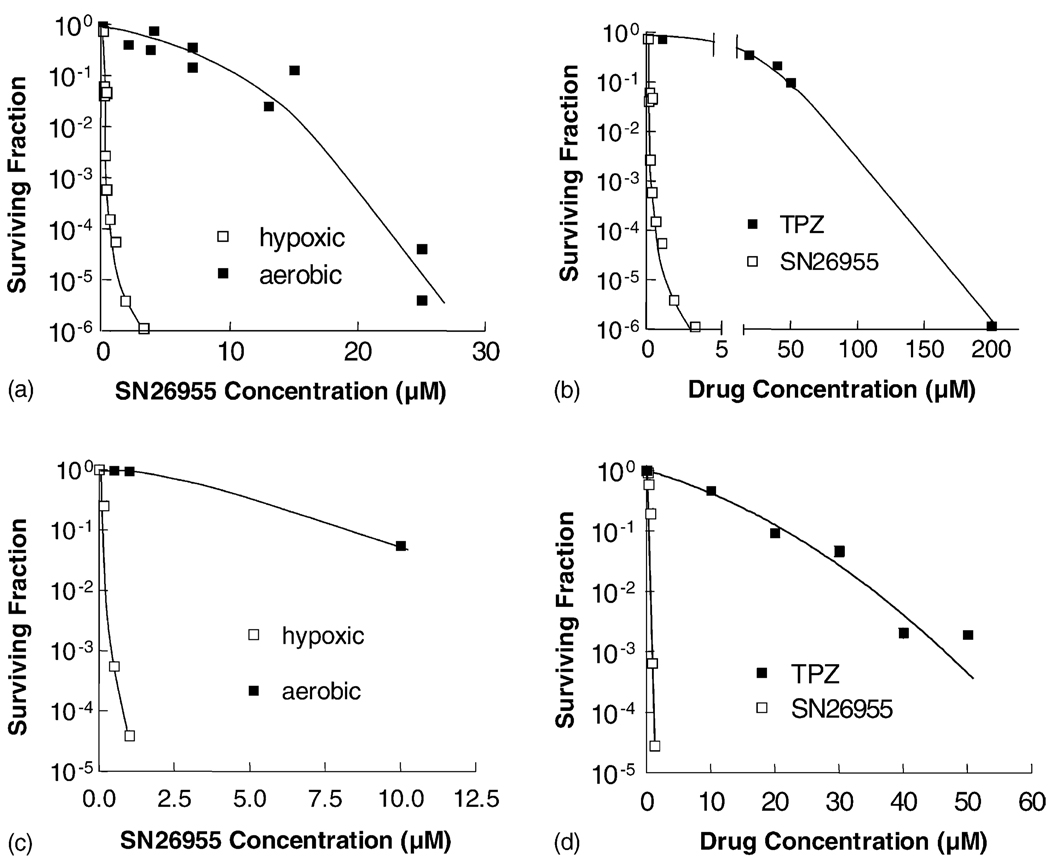
To determine whether SN26955 retained the selective hypoxic cytotoxicity of TPZ, we exposed HT29 human colon carcinoma cells and HeLa human cervix carcinoma cells to various concentrations of SN26955 under hypoxic and aerobic conditions. Panels a and c of Fig. 3 show that SN26955 was much more toxic to hypoxic cells than to aerobic cells with HCR values (defined as the ratio of concentrations under aerobic to hypoxic conditions to give 90% cell kill) of 83 and 78 for HT29 and HeLa cells, respectively. Under the same conditions the HCR for these cell lines for TPZ was 82 and 55, respectively (data not shown). Panels b and d of Fig. 3 show a comparison of the cytotoxicities of SN26955 and TPZ under hypoxic conditions and demonstrate the increased potency of the analog by a factor of 415- and 20-fold for HT29 and HeLa cells, respectively.
Fig. 3.
Sensitivity of HT29 human colon carcinoma cells (a and b) and HeLa human cervix carcinoma cells (c and d) to SN26955 under aerobic and hypoxic conditions (a and c), and in comparison to TPZ under hypoxic conditions (b and d). In all cases, the cells were exposed for 1 hr to drugs under either aerobic or hypoxic conditions, and cell viability was determined using clonogenic survival. The increased potency of both drugs under hypoxic compared to aerobic conditions was calculated by the ratios of the C10 values, as were the relative potencies of SN26955 and TPZ under hypoxic conditions. Results were pooled from two experiments for the HT29 cells and from one experiment for the HeLa cells.
To determine whether the acridine moiety in SN26955 itself was sensitizing cells to TPZ (rather than targeting the TPZ to DNA), we incubated HT29 cells under hypoxic conditions with various concentrations of TPZ with 0, 1, or 5 µM 9-(N-ethylamino)acridine, the DNA-intercalating moiety of SN26955. We found that the presence of 9-(N-ethylamino)acridine made no difference to the cytotoxicity of TPZ (data not shown). This suggests that the enhanced cytotoxicity of SN26955 compared to TPZ results from the targeting of TPZ to DNA rather than a non-specific sensitization by intercalation of the acridine into DNA.
3.3. Free radical formation by SN26955
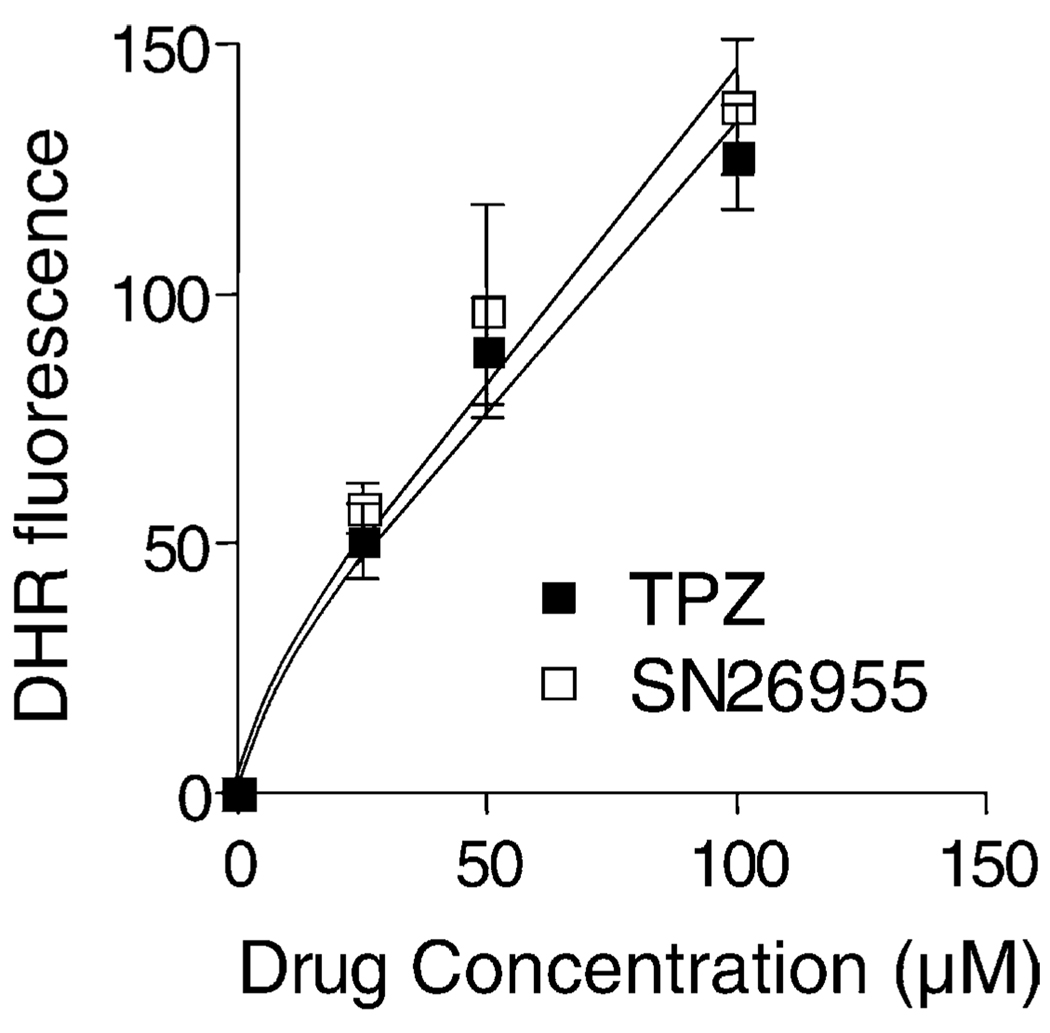
TPZ is reduced under hypoxic conditions to a free radical [29]. This can be detected by oxidation of the non-fluorescent dihydrorhodamine 123 (DHR123) to the highly fluorescent rhodamine 123 (Rh123) [17]. We used this assay to compare the extent of radical formation of TPZ and SN26955. As shown in Fig. 4, the extent of radical formation as measured by DHR fluorescence induced by enzymatic reduction by purified NADPH:cytochrome P450 reductase (0.3 µg/mL) was the same for TPZ and for SN26955. This suggests that the increased toxicity of SN26955 compared to TPZ is not the result of an improved ability to reduce SN26955 compared to TPZ at least for cytochrome P450 reductase. As an additional demonstration of the free radical mechanism for the DHR fluorescence, we added different concentrations of the radical scavenger DMSO to a solution of 50 µM SN26955 with P450 reductase (0.3 µg/mL) and found that 100 and 200 µM DMSO inhibited DHR oxidation to Rh123 by approximately 25 and 50%, respectively (data not shown).
Fig. 4.
Radical production by P450 reductase (0.3 µg/mL) in the presence of TPZ and SN26955 under hypoxic conditions in vitro. TPZ and SN26955 were incubated under hypoxia with 25 µM DHR123 for 1 hr with different concentrations of drug and saturating levels of cofactors (500 µM each of NADH and NADPH). Results pooled from three experiments with the SEM limits are shown.
3.4. Activation of SN26955 to a cytotoxic product when bound to DNA
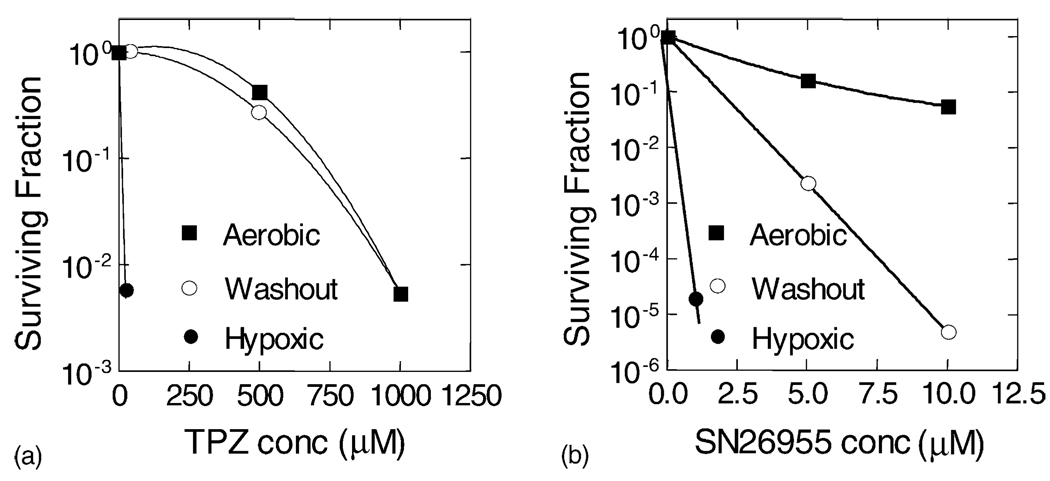
We investigated the ability of SN26955 to become activated while intercalated into DNA by incubating HeLa cells with the drug under aerobic conditions for 1 hr followed by two rinses of the cells in PBS, addition of fresh medium, and then incubation under hypoxic conditions. Fig. 5a shows that when this experiment was performed with TPZ, there was no additional cytotoxicity produced by the 1-hr exposure under hypoxic conditions in fresh medium compared to the 1-hr exposure in air alone. This demonstrates that the washing and addition of fresh medium remove essentially all TPZ from the cells and that the 1-hr exposure to hypoxia produces no cytotoxicity. However, as shown in Fig. 5b, there was increased toxicity by SN26955 produced by the 1-hr exposure under hypoxic conditions after the aerobic incubation and washing. This suggests that some of the SN26955 is retained in the cell, presumably intercalated into DNA, and is present during the hypoxic exposures in fresh medium. The fact that the cytotoxicity of SN26955 following the washout was not as great as that produced by hypoxic exposure with the drug in the medium is presumably the result of loss of some of the drug from the DNA during the washing procedure and reintroduction of fresh medium. This was confirmed by leaving the cells that had been exposed in air for 1 hr to 5 µM SN26955 for 30 min, 1, and 2 hr in fresh medium prior to the hypoxic exposure. We found that the 1- and 2-hr incubations in fresh medium exhibited no significant additional toxicity on hypoxic incubation, with the 30-min exposure being intermediate between no and 1-hr incubation in drug-free medium following the aerobic incubation with SN26955.
Fig. 5.
Washout experiments examining residual TPZ and SN26955 in cells following extensive washing after a 1-hr exposure of the cells under aerobic conditions to the respective drug. SN26955 or TPZ was added to HeLa cells for 1 hr at the indicated concentrations for the aerobic and hypoxic killing curves. In the washout experiment, cells were exposed to the drugs for 1 hr under aerobic conditions. The drug was then removed, and the cells were washed and then exposed in fresh medium under hypoxic conditions for 1 hr. Panel b shows that this hypoxic exposure produced additional killing over the 1-hr exposure of the cells to SN26955 under aerobic conditions. Pooled data from two experiments are shown.
3.5. Enzymatic requirement for DNA damage by SN26955
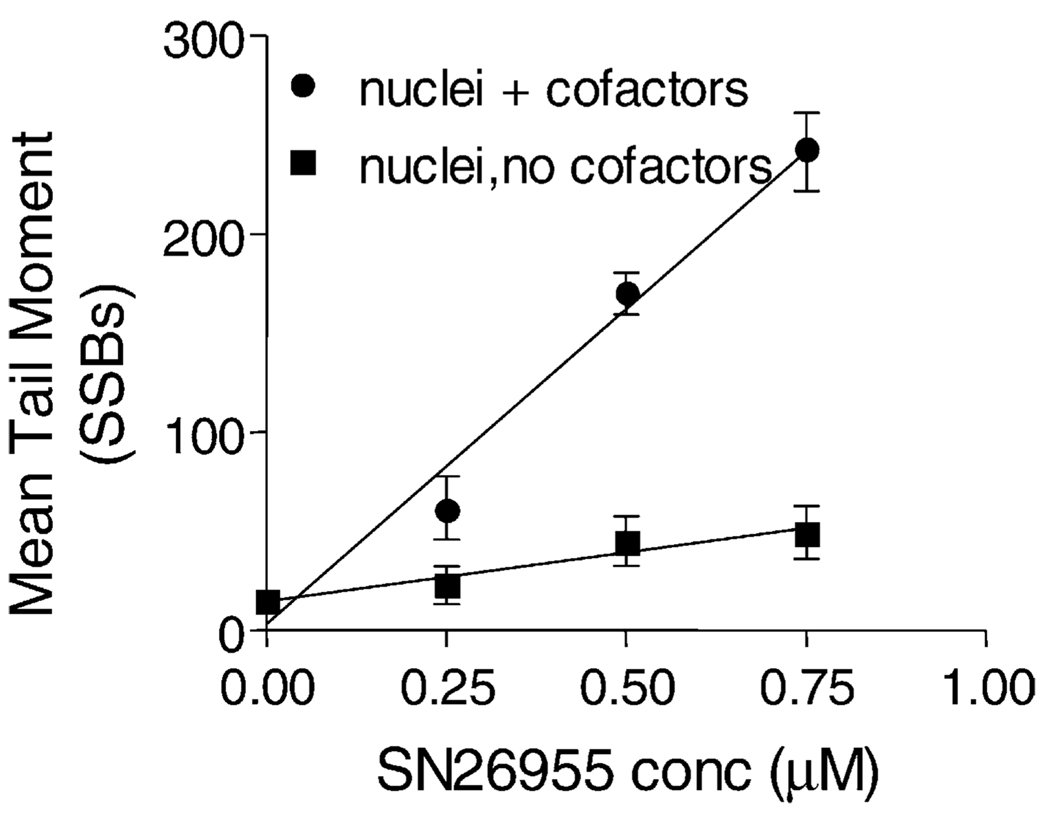
To determine whether enzymatic reduction was necessary for DNA damage produced by SN26955, we incubated isolated nuclei from HeLa cells with the drug for 1 hr with or without saturating concentrations of the two cofactors NADH and NADPH (500 µM of each). As shown in Fig. 6, SN26955 produced very few SSBs, as assayed by the alkaline comet assay, in the absence of cofactors but produced extensive DNA damage in the presence of the cofactors. This indicates that the SSBs are a result of enzymatic reduction of the drug.
Fig. 6.
SSBs produced by SN26955 in nuclei with or without cofactors (500 µM NADH and NADPH). The isolated nuclei were incubated with various concentrations of SN26955 under hypoxic conditions for 1 hr in the presence of HeLa cell nuclei with or without cofactors, and the SSBs were measured using the alkaline comet assay. A few breaks were produced in the absence of cofactors. Data from three experiments pooled with SEM limits are shown.
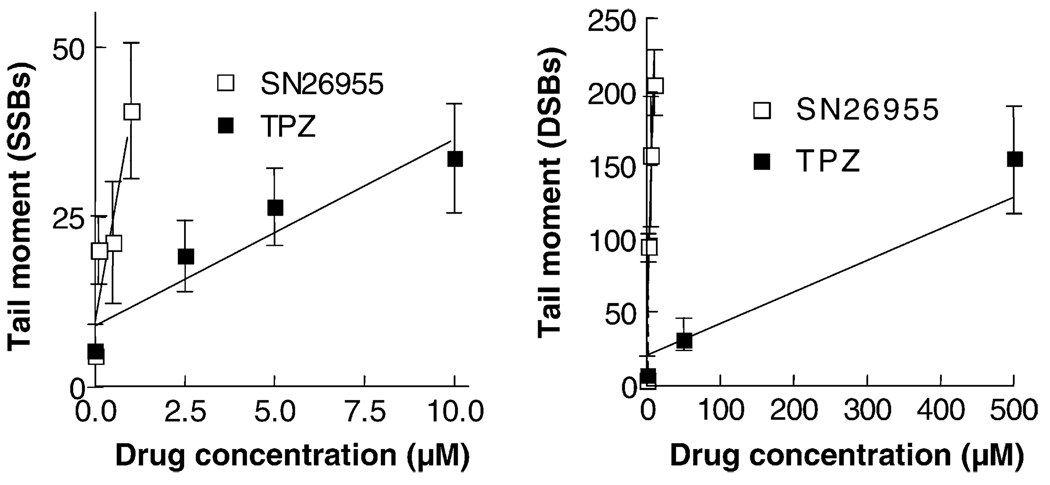
To compare the DNA damage produced by SN26955 with that of TPZ, we incubated isolated nuclei with saturating concentrations of the two cofactors, NADH and NADPH, with various concentrations of SN26955 and TPZ for 1 hr. Fig. 7 shows that SN26955 was more potent than TPZ by a factor of approximately 10 in producing SSBs and was more potent than TPZ in producing DSBs by a factor of approximately 100.
Fig. 7.
SSBs and DSBs produced in isolated nuclei by TPZ and SN26955. TPZ or SN26955 was incubated with HeLa cell nuclei under hypoxic conditions with saturating levels of cofactors (500 µM each of NADH and NADPH) for 1 hr. The SSBs and DSBs were assayed by the alkaline and neutral comet assays, respectively. SN26955 was approximately 10- and 100-fold more efficient than TPZ in producing SSBs and DSBs, respectively. Pooled data from three experiments with SEM limits are shown.
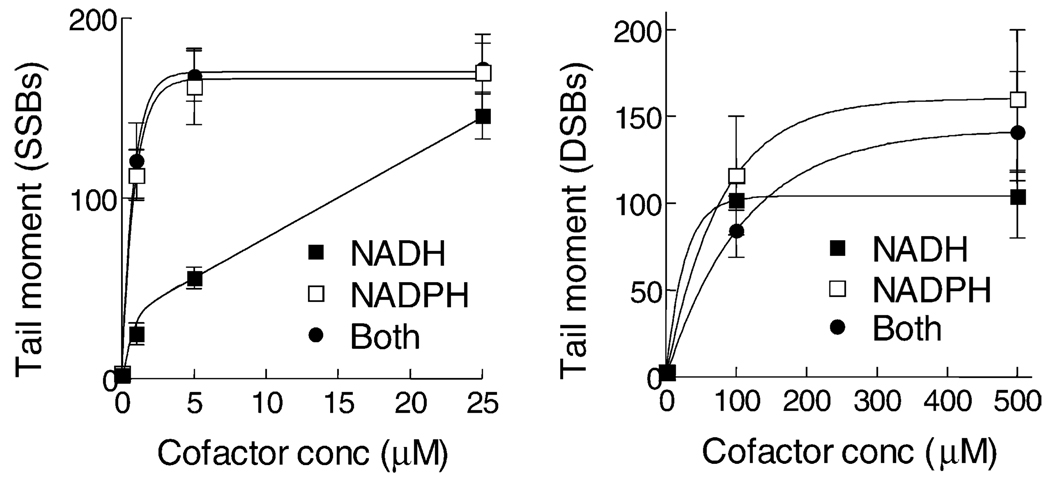
We also measured the cofactor dependence for DSBs and SSBs using isolated nuclei (Fig. 8). It can be seen that SSBs were produced by an enzyme that can use either NADH or NADPH as a cofactor although there was a strong preference for NADPH. Likewise, DSBs were produced by an enzyme that can use both NADH and NADPH as cofactors. We have performed similar experiments with TPZ previously and found a similar cofactor dependence for SSBs. However, the enzyme(s) metabolizing TPZ to produce DSBs could only use NADPH as a cofactor [17]. This different cofactor dependence for the production of DSBs between TPZ and SN26955 suggests that the DSBs are produced by different enzymes in the nucleus.
Fig. 8.
Cofactor dependencies for SSBs and DSBs produced in isolated nuclei by SN26955. HeLa cell nuclei were incubated under hypoxic conditions for 1 hr with 0.25 µM SN26955 with various concentrations of NADH or NADPH or both cofactors, and the SSBs and DSBs were measured using the alkaline and neutral comet assays, respectively. Data pooled from three experiments with the SEM limits are shown.
4. Discussion
The present series of experiments was conducted to determine whether attaching a DNA-targeting chromophore to the TPZ moiety would increase the potency of TPZ yet retain its selective toxicity towards hypoxic cells. Although a number of investigators have shown that the potency of some bioreductive drugs can be increased by targeting them to DNA [30], these examples do not guarantee the efficacy of a TPZ moiety targeted to DNA because, unlike the prior examples, the active reduction species of TPZ is a highly reactive 1-electron reduction radical. Thus, because TPZ metabolized to its active radical in the cytoplasm does not produce any DNA damage [15], it is possible that a DNA-targeted TPZ molecule would be metabolized to its reactive radical prior to entry into the nucleus and binding to DNA, thus rendering it inactive. Also, the efficacy of a TPZ tethered to DNA would depend on the accessibility of the reducing enzymes to the TPZ moiety on the DNA.
The present data, however, show that targeting of TPZ by attaching it via a linker to an acridine chromophore to create the compound SN26955 increases the potency of TPZ by 1–2 orders of magnitude, depending on the cell line, while retaining its selective toxicity towards hypoxic cells (Fig. 3). These data strongly suggest that the TPZ moiety is reduced to its reactive radical while bound by the acridine chromophore to DNA. This is consistent with our recent finding that there are TPZ-metabolizing enzymes in close association with the nuclear matrix [17], which is the organizing structure for the intranuclear DNA [16]. Further evidence for the cytotoxicity of SN26955 bound to DNA was provided by the washout experiment, in which cells incubated with the drug under aerobic conditions, washed, and then immediately exposed to hypoxia showed increased cytotoxicity compared to the aerobic exposure alone. This did not occur for TPZ itself, which is not bound to DNA and, therefore, not retained in the cell following washing and medium change (Fig. 5).
Our finding that almost all of the DNA damage assessed by SSBs in isolated nuclei produced by SN26955 depends on the presence of reduced pyridine nucleotides as cofactors (Fig. 6) demonstrates that the DNA damage, and presumably cytotoxicity, is dependent upon enzymatic reduction. We also show that the increased potency of SN26955 compared to TPZ is probably not the result of an increased rate of reduction of the compound to its oxidizing radical because the rates of radical reduction by the drug and TPZ are identical, at least for NADPH cytochrome P450 reductase (Fig. 4). Despite this similarity in the rate of formation of the oxidizing radical, we found that SN26955 in isolated nuclei produced a greater number of DNA SSBs than did TPZ (by a factor of approximately 10), and an even greater increase in the number of DSBs (by a factor of approximately 100) compared to TPZ (Fig. 7). This suggests that a larger proportion of the SSBs produced by SN26955 is converted into DSBs, presumably because of the presence of the intercalating chromophore.
When we investigated the cofactor dependence of SSBs and DSBs produced by SN26955, we found a qualitative difference between this drug and TPZ. As shown in Fig. 8, the production of both SSBs and DSBs in isolated nuclei by SN26955 required an enzyme(s) that can use either NADH or NADPH cofactor although, at least for SSBs, the NADPH cofactor is favored. Our previous work with TPZ [17] showed a similar pattern for SSBs (ability to use both cofactors but a preference for NADPH), but a different cofactor dependence with DSBs with only NADPH useable. This difference in the cofactor requirements for TPZ and SN26955 suggests that the two drugs are metabolized to the radical that eventually gives rise to DSBs by a different enzyme. As there are different TPZ-metabolizing enzymes in different locations within the nucleus [17], this suggests that the DSBs produced by TPZ and SN26955 occur in different intranuclear locations. We have shown recently that some, if not all, of the cytotoxicity of TPZ is produced by radical-stimulated damage to, or activation of, topo II [19]. This (along with the present data) is consistent with a model in which TPZ can be metabolized throughout the nucleus, but it is only the radicals produced close to topo II (located at the nuclear matrix) that induce DSBs, whereas the DSBs induced by SN26955 are produced throughout the DNA, not just in the short stretches attached to the nuclear matrix. This would imply that DSBs induced by TPZ are not randomly distributed along the DNA, but rather produce fragments consistent with the 60- to 100-kb loops of DNA between matrix attachment sites [16]. On the other hand, we would expect that DNA fragments produced by the DSBs induced by SN26955 would be more randomly distributed throughout the DNA. These predictions are currently being tested.
In summary, we have demonstrated that TPZ attached by a linker to an acridine chromophore binds to DNA and is metabolized at the DNA to produce both SSBs and DSBs. Further, this targeted construct is considerably more potent than TPZ, yet retains its selective hypoxic cytotoxicity. This increased potency and selective accumulation in the nucleus can potentially provide two important advantages in the clinical use of TPZ. First, it can greatly diminish the metabolism that occurs within mitochondria that produces loss of the transmembrane potential of mitochondria [21]. This is potentially responsible for many of the unique toxicities of TPZ in the clinic including reversible hearing loss, muscle cramps, and fatigue, all of which could be due to this effect on mitochondria in aerobic tissue. Second, by reduction of the cytoplasmic metabolism of TPZ, much of the unproductive metabolism can be eliminated, thereby allowing the drug to diffuse further in the tumor [22]. However, if the DNA binding is too great, this is likely to restrict diffusion through the tumor and reduce access to the distant hypoxic cells. Indeed, this is likely to be the case with SN26955 since it is not active in potentiating tumor cell kill by irradiation in vivo.1 Nonetheless, the present data provide proof of principle for further drug development with drugs with lower DNA-binding affinities. Such studies are in progress.
Acknowledgments
This work was funded by Grant CA 82566 from the US National Cancer Institute. We thank H.D. Sarath Liyanage for measuring the DNA binding and Dr. William R. Wilson for helpful comments.
Abbreviations
- TPZ
tirapazamine
- SSB
single-strand DNA break
- DSB
double-strand DNA break
- P450 reductase
NADPH:cytochrome P450 reductase
- HCR
hypoxic cytotoxicity ratio
- DHR123
dihydrorhodamine
Footnotes
Dorie MJ and Brown JM, unpublished data.
References
- 1.Brown JM. SR 4233 (tirapazamine): a new anticancer drug exploiting hypoxia in solid tumours. Br J Cancer. 1993;67:1163–1170. doi: 10.1038/bjc.1993.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denny WA, Wilson WR. Tirapazamine: a bioreductive anticancer drug that exploits tumour hypoxia. Expert Opin Investig Drugs. 2000;9:2889–2901. doi: 10.1517/13543784.9.12.2889. [DOI] [PubMed] [Google Scholar]
- 3.Nordsmark M, Overgaard M, Overgaard J. Pretreatment oxygenation predicts radiation response in advanced squamous cell carcinoma of the head and neck. Radiother Oncol. 1996;41:31–40. doi: 10.1016/s0167-8140(96)91811-3. [DOI] [PubMed] [Google Scholar]
- 4.Brizel DM, Sibley GS, Prosnitz LR, Scher RL, Dewhirst MW. Tumor hypoxia adversely affects the prognosis of carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 1997;38:285–289. doi: 10.1016/s0360-3016(97)00101-6. [DOI] [PubMed] [Google Scholar]
- 5.Sartorelli AC. Therapeutic attack of hypoxic cells of solid tumors: presidential address. Cancer Res. 1988;48:775–778. [PubMed] [Google Scholar]
- 6.Brown JM. The hypoxic cell: a target for selective cancer therapy—eighteenth Bruce F. Cain Memorial Award lecture. Cancer Res. 1999;59:5863–5870. [PubMed] [Google Scholar]
- 7.Craighead PS, Pearcey R, Stuart G. A phase I/II evaluation of tirapazamine administered intravenously concurrent with cisplatin and radiotherapy in women with locally advanced cervical cancer. Int J Radiat Oncol Biol Phys. 2000;48:791–795. doi: 10.1016/s0360-3016(00)00720-3. [DOI] [PubMed] [Google Scholar]
- 8.von Pawel J, von Roemeling R, Gatzemeier U, Boyer M, Elisson LO, Clark P, Talbot D, Rey A, Butler TW, Hirsh V, Olver I, Bergman B, Ayoub J, Richardson G, Dunlop D, Arcenas A, Vescio R, Viallet J, Treat J. Tirapazamine plus cisplatin versus cisplatin in advanced non-small-cell lung cancer: a report of the international CATAPULT I study group. J Clin Oncol. 2000;18:1351–1359. doi: 10.1200/JCO.2000.18.6.1351. [DOI] [PubMed] [Google Scholar]
- 9.Rischin D, Peters L, Hicks R, Hughes P, Fisher R, Hart R, Sexton M, D’Costa I, von Roemeling R. Phase I trial of concurrent tirapazamine, cisplatin, and radiotherapy in patients with advanced head and neck cancer. J Clin Oncol. 2001;19:535–542. doi: 10.1200/JCO.2001.19.2.535. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Biedermann KA, Brown JM. Repair of DNA and chromosome breaks in cells exposed to SR 4233 under hypoxia or to ionizing radiation. Cancer Res. 1992;52:4473–4477. [PubMed] [Google Scholar]
- 11.Daniels JS, Gates KS, Tronche C, Greenberg MM. Direct evidence for bimodal DNA damage induced by tirapazamine. Chem Res Toxicol. 1998;11:1254–1257. doi: 10.1021/tx980184j. [DOI] [PubMed] [Google Scholar]
- 12.Baker MA, Zeman EM, Hirst VK, Brown JM. Metabolism of SR 4233 by Chinese hamster ovary cells: basis of selective hypoxic cytotoxicity. Cancer Res. 1988;48:5947–5952. [PubMed] [Google Scholar]
- 13.Wang J, Biedermann KA, Wolf CR, Brown JM. Metabolism of the bioreductive cytotoxin SR 4233 by tumour cells: enzymatic studies. Br J Cancer. 1993;67:321–325. doi: 10.1038/bjc.1993.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson AV, Saunders MP, Chinje EC, Patterson LH, Stratford IJ. Enzymology of tirapazamine metabolism: a review. Anticancer Drug Des. 1998;13:541–573. [PubMed] [Google Scholar]
- 15.Evans JE, Yudoh K, Delahoussaye YM, Brown JM. Tirapazamine is metabolized to its DNA damaging radical by intranuclear enzymes. Cancer Res. 1998;58:2098–2101. [PubMed] [Google Scholar]
- 16.Berezney R, Wei X. The new paradigm: integrating genomic function and nuclear architecture. J Cell Biochem. 1998;30–31 Suppl:238–242. doi: 10.1002/(SICI)1097-4644(1998)72:30/31+<238::AID-JCB29>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 17.Delahoussaye YM, Evans JE, Brown JM. Metabolism of tirapazamine by multiple reductases in the nucleus. Biochem Pharmacol. 2001;62:1201–1209. doi: 10.1016/s0006-2952(01)00784-5. [DOI] [PubMed] [Google Scholar]
- 18.Peters KB, Wang H, Brown JM, Iliakis G. Inhibition of DNA replication by tirapazamine. Cancer Res. 2001;61:5425–5431. [PubMed] [Google Scholar]
- 19.Peters KB, Brown JM. Tirapazamine: a hypoxia-activated topoisomerase II poison. Cancer Res. 2002;62:5248–5253. [PubMed] [Google Scholar]
- 20.Fortune JM, Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog Nucleic Acid Res Mol Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 21.Wouters BG, Delahoussaye YM, Evans JW, Birrell GW, Dorie MJ, Wang J, MacDermed D, Chiu RK, Brown JM. Mitochondrial dysfunction after aerobic exposure to the hypoxic cytotoxin tirapazamine. Cancer Res. 2001;61:145–152. [PubMed] [Google Scholar]
- 22.Hicks KO, Fleming Y, Siim BG, Koch CJ, Wilson WR. Extravascular diffusion of tirapazamine: effect of metabolic consumption assessed using the multicellular layer model. Int J Radiat Oncol Biol Phys. 1998;42:641–649. doi: 10.1016/s0360-3016(98)00268-5. [DOI] [PubMed] [Google Scholar]
- 23.Kyle AH, Minchinton AI. Measurement of delivery and metabolism of tirapazamine to tumour tissue using the multilayered cell culture model. Cancer Chemother Pharmacol. 1999;43:213–220. doi: 10.1007/s002800050886. [DOI] [PubMed] [Google Scholar]
- 24.Robbins RF, Schofield K. Polyazabicyclic compounds. Part II. Further derivatives of benzo-1,2,4-triazine. J Chem Soc. 1957:3186–3194. [Google Scholar]
- 25.Henderson LM, Chappell JB. Dihydrorhodamine 123: a fluorescent probe for superoxide generation? Eur J Biochem. 1993;217:973–980. doi: 10.1111/j.1432-1033.1993.tb18328.x. [DOI] [PubMed] [Google Scholar]
- 26.Bump EA, Cerce BA, Al-Sarraf R, Pierce SM, Koch CJ. Radio-protection of DNA in isolated nuclei by naturally occurring thiols at intermediate oxygen tension. Radiat Res. 1992;132:94–104. [PubMed] [Google Scholar]
- 27.Olive PL. Detection of hypoxia by measurement of DNA damage in individual cells from spheroids and murine tumours exposed to bioreductive drugs. I. Tirapazamine. Br J Cancer. 1995;71:529–536. doi: 10.1038/bjc.1995.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd RV, Duling DR, Rumyantseva GV, Mason RP, Bridson PK. Microsomal reduction of 3-amino-1,2,4-benzotriazine 1,4-dioxide to a free radical. Mol Pharmacol. 1991;40:440–445. [PubMed] [Google Scholar]
- 30.Rauth AM, Melo T, Misra V. Bioreductive therapies: an overview of drugs and their mechanisms of action. Int J Radiat Oncol Biol Phys. 1998;42:755–762. doi: 10.1016/s0360-3016(98)00302-2. [DOI] [PubMed] [Google Scholar]