Abstract
Chimpanzees represent the only animal model for studies of the natural history of hepatitis C virus (HCV). To generate virus stocks of important HCV variants, we infected chimpanzees with HCV strains of genotypes 1–6 and determined the infectivity titer of acute-phase plasma pools in additional animals. The courses of first- and second-passage infections were similar, with early appearance of viremia, HCV RNA titers of >104.7 IU/mL, and development of acute hepatitis; the chronicity rate was 56%. The challenge pools had titers of 103–105 chimpanzee infectious doses/mL. Human liver–chimeric mice developed high-titer infections after inoculation with the challenge viruses of genotypes 1–6. Inoculation studies with different doses of the genotype 1b pool suggested that a relatively high virus dose is required to consistently infect chimeric mice. The challenge pools represent a unique resource for studies of HCV molecular virology and for studies of pathogenesis, protective immunity, and vaccine efficacy in vivo.
The most recent estimates suggest that 180 million people worldwide are infected with hepatitis C virus (HCV). In the United States, a total of ~3.2 million people are persistently infected with HCV, and >10,000 people die from HCV-related chronic liver disease each year. A continued increase in the number of HCV-infected patients with liver cirrhosis and hepatocellular carcinoma is expected during the next few decades, and HCV-associated end-stage liver disease is already the most common indication for liver transplantation in many Western countries. Thus, there is an urgent need for better drugs to treat this disease and for the development of a vaccine to prevent further spread. To promote such developments, it is important to define the pathogenesis of different HCV genotypes in available animal models and to develop well-characterized virus stocks that represent the various HCV variants that can be used in the development of new experimental in vitro systems and for in vivo studies of new drugs and passive and active immunization strategies.
The HCV virion contains a positive-sense, single-stranded RNA genome that has a single long open reading frame (ORF) [1, 2]. Extensive genomic sequence analysis has demonstrated that HCV strains from around the world exhibit significant genetic heterogeneity, and on the basis of phylogenetic analysis of the ORF of representative isolates, HCV has been classified into 6 major genotypes (genotypes 1–6) and a number of subtypes (subtypes a, b, and so forth) [3–5]. Important differences in the geographic distribution of these genotypes exist, and recent studies have suggested important antigenic and serologic differences [2]. Furthermore, it is well established that the current standard therapy with interferon and ribavirin has a higher sustained virologic response rate in patients infected with genotypes 2 and 3, compared with patients infected with genotypes 1 and 4 [2]. A seventh major genotype has recently been identified in 3 Canadian and Belgian patients, who were presumably infected in Central Africa [6]. However, samples from these patients have not been readily available, and experimental infection with this new genotype could therefore not be included in the present study.
The chimpanzee remains the only animal model that can be used for studies of the natural history of HCV and in challenge studies (eg, studies of immunogenicity and efficacy of HCV vaccine candidates) [7–9]. Despite the recent development of the strain JFH1 cell culture system, which permits virus propagation of a particular genotype 2a isolate in Huh7.5 cells [10], there is still no reproducible cell culture system based on the full-length sequence of other HCV genotype viruses. Thus, transmission to chimpanzees has been the only means to propagate experimentally HCV viruses of different strains.
Urokinase-type plasminogen activator (uPA)–severe combined immunodeficient (SCID) mice engrafted with primary human hepatocytes (chimeric mice) are susceptible to infection with native HCV, and they produce infectious virions with a density similar to that observed in humans and experimentally infected chimpanzees [11–13]. This small animal model has been successfully used to assess the activity of antiviral compounds [14] and to evaluate protection and passive immunization studies of HCV [15–17], but because of the lack of an immune system, this model has not been useful for studies of HCV pathogenesis. Furthermore, this model has not permitted the generation of large quantities of reagents for studies of these specific genotypes.
Our aim was to generate titrated challenge pools for the major HCV genotypes and important subtypes. Challenge viruses of genotypes 1–6 were characterized in chimpanzees and in human liver–chimeric mice.
MATERIALS AND METHODS
The housing and care of the chimpanzees were in compliance with all relevant guidelines and requirements, and the animals were housed in facilities that are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. The chimpanzees used were naive for HCV infection, because they were raised in captivity under stringent conditions of infectious disease control and housed in a facility with Animal Biosafety Level 2–3 containment, with close monitoring for HCV infection as well as for infection with other hepatitis viruses. Chimpanzees were inoculated intravenously with serum samples collected from patients infected with well-characterized HCV strains [3, 18–21] (Table 1); the HC-J4/91 (genotype 1b) inoculum was from an experimentally infected chimpanzee [22]. The 6a strain that resulted in active experimental HCV infection had not been previously characterized; this strain originated from a patient from Hong Kong with persistent HCV infection. During the acute HCV infection in the chimpanzees, plasmapheresis units were collected weekly and subsequently used to prepare a >150-mL virus pool for each genotype strain. To verify the genotype of the infecting agent, the core-E1 sequence of the recovered virus was determined as described elsewhere [23]. The HCV infectivity titer of 1.1-mL aliquots (stored at −80°C) of the plasma pools of genotypes 1–6 was determined by means of reverse titration in additional chimpanzees, as described elsewhere [24] (Table 2).
Table 1.
Characterization of Acute-Phase Infection with Hepatitis C Virus (HCV) Prototype Strains of Genotypes 1–6 in Experimentally Infected Chimpanzees
| HCV strain | Genotype | Chimpanzee ID | Appearance of viremia, weeks | Peak titer with Monitor 2.0 assay,a log10 IU/ mL (week) | First positive detection of anti-HCV, week | Peak ALT level, U/L (week) | Peak liver histology grade (week) | Outcome of HCV infection | Titer of last sample tested,a log10 IU/mL (year) |
|---|---|---|---|---|---|---|---|---|---|
| First-passage infection | |||||||||
| HC-TN | 1a | 1422 | 1–19 | 5.7 (5) | 13 | 744 (10) | 3 (10) | Cleared | Neg (week 29) |
| HC-J4/91 | 1b | 1410 | 1–>29 | 5.2 (10) | 21 | 124 (12) | 2 (11) | Persisted | 6.3 (15) |
| HC-J6 | 2a | 1417 | 1–25 | 5.0 (8) | 16 | 192 (11) | 2 (9) | Cleared | Neg (week 29) |
| HC-J8 | 2b | 1497 | 1–>28 | 4.7 (1) | 12 | 60 (5) | 1 (10) | Persisted | 3.0 (4) |
| S52 | 3a | 88A04 | 1–>27 | 5.4 (10) | 11 | 104 (14) | 1 (15) | Persisted | 4.1 (1) |
| ED43 | 4a | 1563 | 1–>28 | 5.7 (6) | 9 | 222 (11) | ND | Persisted | 6.1 (12) |
| SA13 | 5a | 1516 | 1–15 | 5.6 (8) | 14b | 320 (18) | 2 (13) | Cleared | Neg (3.5) |
| HK-6a | 6a | F15 | 2–18 | NAc | Neg | NA | NA | Cleared | Neg (week 40) |
| Second-passage infection (reverse titration of HCV plasma pools) | |||||||||
| HC-TN | 1a | 1581 | 1–>28 | 6.6 (8) | 10 | 296 (10) | 1 (10) | Persisted | 3.5 (1) |
| HC-J4/91 | 1b | 1545 | 1–>24 | 5.8 (11) | 15 | 155 (16) | 1 (20) | Persisted | 5.5 (1) |
| HC-J6 | 2a | F9 | 1–19 | 5.7 (5) | Neg | 30 (7) | 1 (6) | Cleared | Neg (1) |
| HC-J8 | 2b | 1484 | 1–18 | 4.7 (6) | Neg | 62 (7) | 2 (13) | Cleared | Neg (2.5) |
| S52 | 3a | E3 | 1–>29 | 5.4 (11) | 14 | 27 (2) | NC | Persisted | 4.1 (1.5) |
| ED43 | 4a | 1557 | 1–>30 | 6.0 (7) | 9 | 180 (11) | 3 (11) | Persisted | 4.0 (1) |
| SA13 | 5a | 1547 | 1–>24 | 5.8 (10) | 14 | 173 (13) | 2 (22) | Persisted | 7.2 (10.5) |
| HK-6a | 6a | 73 | 2–>30 | 5.0 (5) | Neg | 42 (21) | ND | Cleared | Neg (6) |
NOTE. Serum samples were collected for 24–30 weeks after inoculation and tested for HCV RNA. ALT, alanine aminotransferase; NA, not available; NC, no changes; ND, not done; neg, negative.
For a number of animals, samples were originally tested with the Monitor 1.0 (Roche), Monitor 2.0 (Roche), and/or bDNA 2.0 (Bayer) assays, which did not provide titers in international units. On the basis of these results, the sample or samples with peak titers were retested with the Monitor 2.0 assay to obtain the peak titers in international units. Titers for the last follow-up sample were obtained with the Monitor 2.0 or in-house TaqMan assay.
CH1516 was only transiently anti-HCV positive.
Not available because samples were not collected during weeks 5–13.
Table 2.
Standardized Acute-Phase Chimpanzee Pools of Hepatitis C Virus (HCV) Genotypes 1–6
| HCV strain | Genotype | Chimpanzee ID | Weeks when plasma units were collected | HCV genome titer,a log10 IU/mL | Chimpanzee infectious titer,b log10 CIDs/mL |
|---|---|---|---|---|---|
| H77Cc | 1a | 1530 | 7, 8, 9 | 4.8 | 3.5 |
| HC-TN | 1a | 1422 | 4, 5, 6 | 5.3 | 5 |
| HC-J4/91 | 1b | 1410 | 2, 3, 4, 5, 6 | 4.7 | 3 |
| HC-J6 | 2a | 1417 | 2, 3, 4, 5, 6 | 5.0 | 4 |
| HC-J8 | 2b | 1497 | 1, 2, 5 | 4.6 | 4 |
| S52 | 3a | 88A04 | 2, 3, 4, 5, 6 | 4.3 | 3 |
| ED43 | 4a | 1563 | 3, 4, 5, 6 | 5.6 | 5 |
| SA13 | 5a | 1516 | 1, 2, 3, 4, 5, 6 | 5.1 | 4 |
| HK-6a | 6a | F15 | 2, 4 | 4.9 | 3 |
Determined with the Monitor 2.0 assay (Roche).
Determined by means of reverse titration in 2 chimpanzees (for strain H77C) or in 1 chimpanzee. CID, chimpanzee infectious dose.
Monoclonal virus pool collected from CH1530 after intrahepatic transfection with RNA transcripts from clone pCV-H77C. It is included for comparison.
Serum samples were collected weekly from inoculated chimpanzees and examined for HCV antibodies (enzyme-linked immunosorbent assay 2.0; Abbott) and alanine aminotransferase (Anilytics). Initially, the infection status was determined in a highly sensitive 5′ untranslated region–based reverse transcription–nested polymerase chain reaction assay for HCV RNA detection [24], and in selected samples the HCV RNA titer was determined by means of reverse transcription–nested polymerase chain reaction on serial HCV RNA dilutions, by means of a Monitor 1.0 and/or Monitor 2.0 assay (Roche Diagnostics), by means of Versant HCV RNA branched-chain DNA 2.0 and/ or branched-chain DNA 3.0 assay (Bayer), and by means of a quantitative 5′ untranslated region–based TaqMan assay developed in house [25]. In most animals, weekly liver biopsy specimens were collected and examined for necroinflammatory changes (graded as 0 [normal], 1 [mild], 2 [mild-moderate], 3 [moderate-severe], or 4 [severe]).
To generate human liver–chimeric mice, SCID mice that were homozygous for the uPA-transgene were transplanted with cryopreserved human hepatocytes from a single HCV-uninfected female donor, as described elsewhere [13]; however, the mice inoculated with HC-J4 (genotype 1b) had been transplanted with fresh hepatocytes from other donors [13]. The animals used in the present study had a human albumin content of >2 mg/mL (a human albumin content of >1 mg/mL is generally accepted as evidence for a high-quality human liver graft). The mice were injected intraperitoneally with defined doses of the HCV challenge pools (Table 3). The study protocol was approved by the animal ethics committee of the Ghent University Faculty of Medicine and Health Sciences. Collected mouse plasma samples were analyzed for human albumin levels with a human albumin enzyme-linked immunosorbent assay quantitation kit (Bethyl Laboratories) and for HCV RNA titers with the Monitor 2.0 assay or the TaqMan48 assay (Roche Diagnostics).
Table 3.
Infection of Human Liver–Chimeric Mice with Hepatitis C Virus (HCV) Genotypes 1–6 Strains from Titrated Challenge Pools
| HCV genotype pool inocula | Plasma HCV RNA titer by week, log10 IU/mL | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| uPA-SCID mouse ID | Plasma human albumin level at week 0, mg/mL | Genotype | Strain | Dose,a log10 CID | Dose,a log10 IU | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Test used |
| K202 | 5.2 | 1a | H77C | 2.6 | 3.9 | NA | 7.7 | NA | NA | NA | NA | NA | NA | T |
| K230 | 4.0 | 1a | H77C | 2.6 | 3.9 | NA | 7.7 | NA | NA | NA | NA | NA | NA | T |
| K292 | 2.2 | 1a | H77C | 2.6 | 3.9 | NA | 7.8 | NA | NA | NA | NA | NA | NA | T |
| K292R | 2.7 | 1a | H77C | 2.6 | 3.9 | NA | 7.6 | NA | NA | NA | NA | NA | NA | T |
| K16R | 2.1 | 1b | HC-J4 | 1.3 | 3.0 | NA | 6.4 | NA | 7.9 | NA | 7.5 | NA | NA | M |
| K21L | 7.4 | 1b | HC-J4 | 1.7 | 3.4 | NA | >2.7 | NA | NA | NA | NA | NA | NA | M |
| K7 | 2.8 | 1b | HC-J4 | 2.0 | 3.7 | NA | >2.7 | NA | NA | NA | 7.9 | NA | NA | M |
| K7L | 3.0 | 1b | HC-J4 | 2.0 | 3.7 | NA | >2.7 | NA | NA | NA | 7.9 | NA | 7.2 | M |
| K179-L | 4.7 | 2a | HC-J6 | 3.0 | 4.0 | NA | NA | 7.4 | NA | 6.5 | NA | 5.9 | 4.9 | M |
| K637 | 2.1 | 2b | HC-J8 | 3.0 | 3.6 | <2.9 | +b | 4.1 | 5.3 | 5.6 | 5.3 | 5.1 | NA | T |
| K595 | 6.4 | 3a | S52 | 2.0 | 3.3 | 4.6 | 6.0 | 6.6 | 6.5 | 6.6 | 6.2 | 5.9 | NA | T |
| K328-R | 6.2 | 4a | ED43 | 3.6 | 4.2 | NA | 7.4 | NA | + | + | NA | NA | NA | T |
| K698 | 2.6 | 5a | SA13 | 3.0 | 4.1 | +b | 4.5 | 4.8 | 5.4 | NA | NA | NA | NA | T |
| K686 | 4.6 | 6a | HK-6a | 2.0 | 3.9 | NA | 4.0 | 6.2 | 7.1 | 7.2 | NA | NA | NA | T |
NOTE. Plus signs indicate unquantified positive detections of HCV RNA. Uninfected mice are not shown (see text). CID, chimpanzee infectious dose; M, Roche HCV Monitor 2.0 assay; NA, not available; T, Roche HCV TaqMan48 assay; uPA-SCID, urokinase-type plasminogen activator–severe combined immunodeficient.
The titer in CIDs is the titer listed in Table 2, corrected to reflect the relative amount of the inoculum.
HCV RNA level, <1500 IU/mL.
RESULTS
In 1992, in the absence of culture systems to propagate and titrate HCV, we initiated a study to generate challenge pools of the different genotypes of HCV by inoculating naive chimpanzees with serum containing well-characterized HCV isolates. We have achieved robust infection of chimpanzees with strains HC-TN (1a), HC-J4 (1b), HC-J6 (2a), HC-J8 (2b), S52 (3a), ED43 (4a), SA13 (5a), and HK-6a (6a). We previously reported on the genotype 1a and 5a infections [24, 26]. The first inoculation with HC-J8 (2b) resulted in suboptimal HCV titers (data not shown), and therefore the infection reported below resulted from inoculation of a second animal; both animals developed a persistent infection. In all other cases, the reported infection was the result of the first inoculation performed. However, it should be noted that a number of inoculations with other HCV strains (S83 [2c], Z4 [4b], Z7 [4c], DK13 [4d], SA1 [5a], SA7 [5a], and HK2 [6a]) [3] did not result in infections, most likely because of the presence of neutralizing antibodies in the chronic-phase source samples or because of suboptimal storage conditions. To our knowledge, this study permitted for the first time a comparative study of replication fitness and pathogenicity of the different genotypes of HCV in chimpanzees.
Experimental infection of chimpanzees with prototype strains of genotypes 1–6
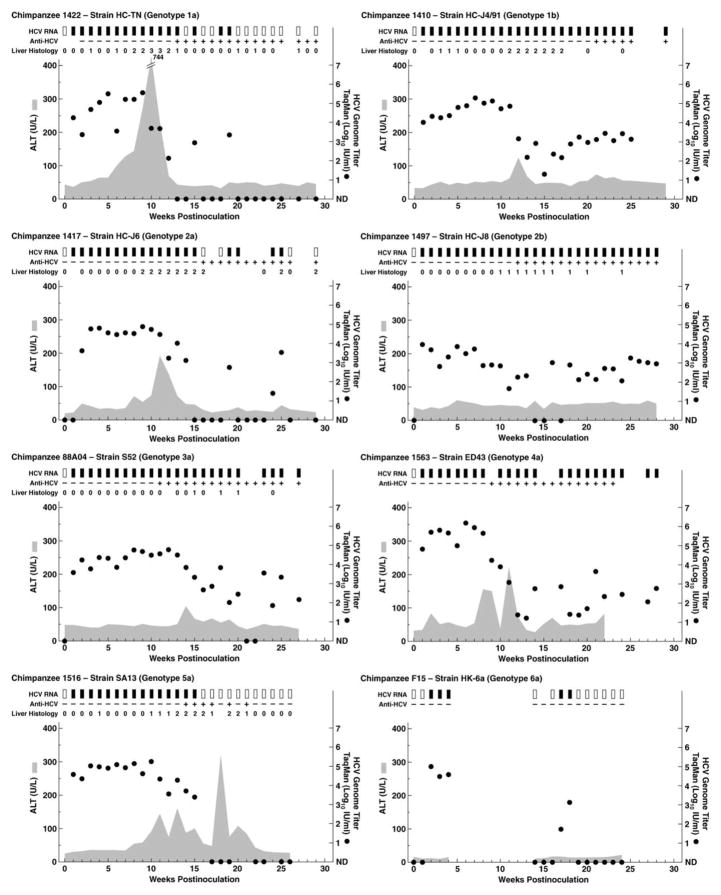
The 8 chimpanzees inoculated intravenously with serum originating from patients infected with strains of genotypes 1–6 became viremic at week 1 or 2, and the peak HCV RNA titers were 104.7–105.7 IU/mL (Table 1 and Figure 1). In the animals infected with genotype 1a, 2a, 5a, or 6a, viral clearance was observed at week 20, 26, and 15 and after week 30, respectively. In the animals infected with genotype 1b, 2b, 3a, or 4a, the virus persisted for >1 year of follow-up (Table 1). Thus, the HCV carrier rate in the first-passage animals was 50%. HCV antibody seroconversion was observed between weeks 9 and 21; the animal infected with genotype 6a did not become anti-HCV positive. Finally, all animals that were evaluated had evidence of acute hepatitis with elevated liver enzymes (peak serum alanine aminotransferase levels, 60–744 U/L) and necroinflammatory changes in liver biopsy specimens (Table 1 and Figure 1); biopsy specimens were not available from animals CH1563 and CH-F15. It is noteworthy that in the animals that developed a persistent infection, the HCV RNA titers were relatively low late in the acute infection (Figure 1), which suggests some viral control. However, in the 2 animals (CH1410 and CH1563) for which we had samples that were collected >10 years later, the HCV RNA titers had increased by 2–3 logs and were similar to or higher than the peak titers observed during the acute infection (Table 1).
Figure 1.
Course of first-passage infection with hepatitis C virus (HCV) genotypes 1–6 in chimpanzees. Serum samples collected weekly during 24–29 weeks of follow-up were tested for HCV RNA by means of in-house reverse transcription–nested polymerase chain reaction (PCR) with 5′ untranslated region primers and/or by means of quantitative tests (see Materials and Methods) (black rectangles, positive by reverse transcription–nested PCR and/or quantitative test; white rectangles, negative by reverse transcription–nested PCR). The estimated HCV RNA titers (black circles), determined by means of an in-house TaqMan assay, were plotted against time; samples below the detection limit of ~10 IU/mL are shown as not detected (ND). Anti-HCV antibodies were detected in the second-generation enzyme-linked immunosorbent assay (plus signs, positive detections; minus signs, negative detections). The shaded area represents serum alanine aminotransferase (ALT) level. Weekly liver biopsy specimens were collected and examined for necroinflammatory changes (graded as 0 [normal], 1 [mild], 2 [mild-moderate], 3 [moderate-severe], or 4 [severe]).
Preparation and titration of challenge pools of HCV genotypes 1–6
For each genotype, a pool was made from plasmapheresis units obtained from infected chimpanzees during the first 6 weeks of follow-up prior to anti-HCV seroconversion (Table 2). Plasma units were thawed and pooled on ice and divided into 1.1-mL aliquots; at least 100 aliquots were prepared from each pool. For each pool, the genotype strain was initially confirmed by means of sequence analysis of core-E1 (data not shown). In other studies, the complete consensus ORF sequence was determined for viruses recovered from the pools of genotypes 1a, 1b, 2a, and 5a (GenBank accession no. AF177036, AF054247, EF621489, and AF064490, respectively) [24, 26–28], and the core-NS2 sequence was determined for viruses recovered from the pools of genotypes 2b, 3a, 4a, and 6a (GenBank accession no. FJ230882, EU204645, EU363760, and FJ230883, respectively) [29–32].
The HCV genome titers of the different pools were 104.3–105.6 IU/mL [25] (Table 2). Most importantly, the infectious titer of each genotype pool was determined by means of reverse titration in a naive chimpanzee. For each dilution tested, we used a new pool aliquot and made dilutions in preinoculation plasma from the chimpanzee. Typically, 2 or 3 higher dilutions were tested before the animal became infected. The infectious titer determined for the pools of genotypes 1–6 were 103–105 chimpanzee infectious doses (CIDs)/mL (Table 2). However, it should be noted that because this titer was determined by means of reverse titration with 10-fold dilutions in a single animal, the titer might be ± 1log10.
The 8 chimpanzees used for reverse titration of the individual genotype pools became viremic at week 1 or 2, and peak HCV RNA titers were 104.7–106.6 IU/mL (Table 1). Viral persistence for >1 year of follow-up was observed in 5 (63%) of the 8 chimpanzees with second-passage infections (Table 1); these animals seroconverted between weeks 9 and 15. However, the 3 animals with acute resolving infection did not become anti-HCV positive. Finally, 6 of the 8 animals had evidence of acute hepatitis with elevated liver enzymes and/or necroinflammatory changes in liver biopsy specimens (Table 1); biopsy specimens were not available from animal CH73.
Susceptibility of human liver–uPA-SCID mice to HCV of genotypes 1–6
We inoculated chimeric mice with 102.0–103.6 CIDs of the plasma pools of genotypes 1–6 (Table 3); strain H77C was used for genotype 1a inoculations. The results of the infections with genotypes 1a and 1b were reported elsewhere [13, 16]. For the mice inoculated with genotype 1a, 1b, 2a, 4a, or 6a, we obtained HCV genome titers of >107 IU/mL within the first 4 weeks of follow-up. In the mice inoculated with genotype 2b, 3a, or 5a, the HCV genome titers reached levels of >105.4 IU/mL within the first 5 weeks of follow-up (Table 3). Thus, human liver–chimeric mice can be used for studies with all the different genotype pool strains that have been developed in the present study.
Additional studies were performed with the genotype 1b pool to compare the chimpanzee infectivity in the chimpanzee with that in the human liver–chimeric mouse. In the 1b pool, the genome titer was 1.7log10 higher than the infectivity titer determined by means of reverse titration in a single chimpanzee, which suggests that the determined infectivity titer of this pool was not overestimated (Table 2). First, we challenged 3 chimeric mice with 20 CIDs, and only 1 mouse became infected (Table 3). Subsequently, we challenged 2 mice with 50 CIDs, and 1 mouse became infected (Table 3). However, as described above, challenge with 100 CIDs resulted in robust HCV infection in both animals tested. All the mice used in this titration study had human albumin levels of >2 mg/mL, which indicates that the negative results obtained after challenge with 50 CIDs (1 mouse) and 20 CIDs (2 mice) were not due to suboptimal human liver grafts. In fact, 2 of the 3 animals that remained HCV negative after challenge had human albumin levels that were equal to or higher than those of the mice challenged with 100 CIDs. It is noteworthy that the mouse that became infected after inoculation with only 20 CIDs had a very robust HCV infection with peak genome titers at week 4 of 107.9 IU/mL. Overall, this titration study suggests that the human liver–uPA-SCID mouse model is less susceptible to HCV infection, compared with chimpanzees (1 mouse infectious dose is equal to ~50 CIDs), but if infection occurs, the viremia course is apparently not dependent on the virus dose that was used.
Because we used different liver grafts for the studies of human liver–chimeric uPA-SCID mice, it was of interest to determine whether different grafts have similar susceptibility to HCV infection. We previously found that mice engrafted with primary hepatocytes (cryopreserved) from the donor, which were used for most inoculations in the present study, had a 100% infection rate when inoculated with 104 IU of a mouse-passaged pool of chimpanzee-derived H77C [16] (Table 3). Here we found that 5 mice engrafted with hepatocytes (cryo-preserved) from a new donor (human albumin level, ≥3) had robust HCV infection with titers of ~106 IU (at week 2 or 3) after inoculation with 104 IU of the same mouse-passaged pool of chimpanzee-derived H77C. Thus, these data suggested that the HCV susceptibility of this new liver donor is at least as good as that observed for the old donor.
DISCUSSION
We have demonstrated and/or confirmed that chimpanzees are susceptible to all 6 published HCV genotypes and important subtypes. For many of the genotypes, this is the first time the course of infection in chimpanzees has been described, to our knowledge. Overall, we found that the course of acute infection is similar in animals infected with the different HCV genotypes, with early appearance of viremia, relatively high peak genome titers, and development of acute hepatitis. It should be noted, however, that infection in 2 animals does not necessarily represent the typical course for a particular genotype or, for that matter, that particular strain. Also, we cannot rule out the possibility that the infection observed in animals after reverse titration could be influenced by exposure to apparently non-infectious virus challenges. However, we observed robust infections in both first- and second-passage animals, with no apparent difference. This is in agreement with data previously published by our group, in which it was found that chimpanzees inoculated with high doses (1,000,000 infectious doses of the H77 strain) of HCV had infections that were similar to those observed after inoculation with lower doses, only with a shorter incubation period [33].
The chronicity rate of HCV in experimentally infected chimpanzees has varied in different studies, from as low as 33% to as high as 67% [9]. In the present study, we found a chronicity rate of 56% among 16 experimentally infected chimpanzees. It is noteworthy that in the available samples taken from the same animals during the late chronic-phase infection (CH1410, CH1563, and CH1547), there was a clear indication of an increase in viral titers over time, compared with samples taken from the same animals during the late acute-phase infection (Table 1 and Figure 1), which suggests a relative loss of the host’s control of viremia [34]. We observed that 5 of 7 animals with acute resolving infection did not seroconvert (4 animals) or had transient seroconversion (1 animal), whereas all 9 animals that developed a persistent infection seroconverted during the acute infection. Although it must be acknowledged that the humoral immune response of chimpanzees might differ from that of humans [35, 36], these data are in agreement with data from long-term follow-up studies of patients with transfusion-associated HCV infection [37].
The virus pools characterized in the present study have been useful for a number of studies with chimpanzees. The chimpanzee is the only animal model in which to study protective immunity following resolved experimental infections or immunization with vaccine candidates. However, titrated challenge viruses previously were available only for genotype 1a and 1b strains. The current virus panel has made it possible to perform more extensive heterologous rechallenges. In a study by Prince et al [38], in which chimpanzees with resolved genotype 1 infections were rechallenged with 100 CIDs of these respective genotype pools, it was found that heterologous challenge would lead to frequent viral persistence. In a recent study, we found that an animal with apparent sterilizing immunity against challenge with the homologous genotype 1a was not protected against acute infection with genotype 1b or 2a after challenge with 100 CIDs of the respective virus pools [39]. In the testing of vaccine candidates, it is important to challenge with a virus dose that consistently infects the animal. Titrated virus pools of genotype 1a have been important for a number of vaccine studies with chimpanzees [40]. The 1b pool presented here has been used in other vaccine studies [41, 42]. The virus-containing plasma pools could also be useful in the development of diagnostic assays, such as quantitative diagnostic assays or genotyping assays, and they have already been distributed to a number of investigators for these purposes.
Advances in HCV research have been hampered by a lack of readily available in vitro systems. The development of infectious HCV clones of strain H77 in 1997 paved the way for the development of such systems [43, 44]; genotype 1a (pHC-TN), 1b (pCV-J4L6S), and 2a (pJ6CF) infectious clones have since been developed from the virus pools characterized in the present study [26–28]. A subgenomic replicon system for strain Con1 (genotype 1b) was developed in 1999 [45]; however, this system currently is available only for genotype 1a, 1b, and 2a HCV strains. A retroviral pseudoparticle system bearing the HCV glycoproteins was developed in 2003 [46], and the strains of our genotype pools have contributed to the development of this system for genotypes 1b, 2a, 3a, 4a, 5a, and 6a [30, 47]. In 2005, the JFH1 (genotype 2a)–based cell culture system was developed; this system provides robust HCV infection in Huh7.5 cells [10]. The genotype pools presented here have served as the virus source for the development of JFH1-based intragenotypic and intergenotypic recombinants of genotypes 1b, 2a, 2b, 3a, 4a, 5a, and 6a, producing viruses with the genotype-specific core, E1, E2, p7, and NS2 in Huh7.5 cells [29, 31, 32, 48, 49]. Thus, the virus genotype pools have contributed to the development of new experimental systems for HCV. The use of prototype strains in the development of such systems provides the added advantage of permitting the use of identical HCV strains in different experimental systems. For example, antibodies identified in vitro in the HCV pseudoparticle systems or in the JFH1-based cell culture systems could be used for passive immunoprophylaxis studies with chimpanzees in which a matched challenge virus from the genotype pools is used.
In the human liver–uPA-SCID mouse model, we observed robust infection with HCV strains that represent HCV genotypes 1–6, in most cases with peak HCV RNA titers that were 1log10–3log10 higher than those observed for the same strains in chimpanzees. Robust infection was seen in mice with different human liver grafts, but titration data indicated that HCV infectious doses that are 1–2 logs higher than those required to infect chimpanzees are required to infect human liver–chimeric mice. Plasma from infected mice with relatively high HCV RNA titers (often of >107 IU/mL; Table 3) can be used to make additional antibody-free challenge pools of these HCV strains, as we reported for the H77C strain [16]. The susceptibility of the human liver–chimeric mice to well-defined challenge viruses will permit more detailed studies of the cross-neutralization potential of antibody preparations. In addition, new drugs can be tested for therapeutic potential against the different genotypes [14–16, 50].
In summary, we have developed titrated challenge pools of genotypes 1–6 of HCV. These reference viruses are available to the scientific community and will contribute to studies of active and passive immunity against HCV. In addition, these challenge pools of genotypes 1–6 will permit controlled studies in vitro and in vivo, because different experimental systems have already been developed for these strains: for the HC-J6 (genotype 2a) strain, a consensus molecular clone that is infectious for chimpanzees, a retroviral pseudoparticle system that permits entry into Huh7 cells, and a JFH1-based intragenotypic recombinant that is infectious for Huh7 cells have been developed [28, 30, 47, 49]. We have demonstrated that infection of chimpanzees with all HCV genotypes resulted in relatively high HCV titers and liver inflammation, and that viruses from the challenge genotype pools readily infect human liver–chimeric mice, which will permit further studies with both animal models of HCV infection.
Acknowledgments
We thank Doris Wong, Carl Apgar, Alicia Brockington, Kathleen Mihalik, Lieven Verhoye, and Thomas Vanwolleghem for technical assistance. We thank Christina Eder, Max Shapiro, and Charlene Shaver, as well as the animal technical staff at the Michale E. Keeling Center for Comparative Medicine and Research, for animal care. We thank Shunji Mishiro, Hiroaki Okamoto, and Makoto Mayumi for providing the HC-J4, HC-J6, and HC-J8 inocula, Patrizia Farci for providing the TN and S52 inocula, Peter Simmonds for providing the ED43 inoculum, Michael Kew for providing the SA13 inoculum, Anna Lok for providing the HK-6a inoculum, and Christopher Walker for providing chronic-phase serum samples from animals CH1410, CH1547, and CH1563. We also thank Alfred Prince and Michael Houghton for their support of these studies.
Financial support: Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health (contract NO1-AO-62713); University of Copenhagen (professorship to J.B.); Lundbeck Foundation (external funding to J.B.); Ghent University (Concerted Action Grant 01G00507); the Belgian State via the Interuniversity Attraction Poles Program (grant P6/36 HEPRO); European Union (6th Framework–HEPACIVAC); The Research Foundation Flanders (FWO-Vlaanderen; postdoctoral fellowship to P.M.).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Gottwein JM, Bukh J. Cutting the gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 2.Lemon SM, Walker CM, Alter MJ, Yi MK. Hepatitis C virus. In: Knipe D, Howley P, Griffin D, et al., editors. Fields Virology. 5. Philadelphia, PA: Wolters Kluwer, Lippincott Williams & Wilkins; 2008. pp. 1253–1304. [Google Scholar]
- 3.Bukh J, Purcell RH, Miller RH. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci U S A. 1993;90:8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds P, Holmes EC, Cha TA, et al. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 5.Simmonds P, Bukh J, Combet C, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 6.Murphy D, Chamberland J, Dandavino R, Sablon E. A new genotype of hepatitis C virus originating from central Africa. Hepatology. 2007;46:623A. doi: 10.1128/JCM.02831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukh J, Forns X, Emerson SU, Purcell RH. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Inter-virology. 2001;44:132–142. doi: 10.1159/000050040. [DOI] [PubMed] [Google Scholar]
- 8.Lanford RE, Bigger C, Bassett S, Klimpel G. The chimpanzee model of hepatitis C virus infections. ILAR J. 2001;42:117–126. doi: 10.1093/ilar.42.2.117. [DOI] [PubMed] [Google Scholar]
- 9.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 10.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindenbach BD, Meuleman P, Ploss A, et al. Cell culture–grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mercer DF, Schiller DE, Elliott JF, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 13.Meuleman P, Libbrecht L, De VR, et al. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- 14.Meuleman P, Leroux-Roels G. The human liver–uPA-SCID mouse: a model for the evaluation of antiviral compounds against HBV and HCV. Antiviral Res. 2008;80:231–238. doi: 10.1016/j.antiviral.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 15.Law M, Maruyama T, Lewis J, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 16.Vanwolleghem T, Bukh J, Meuleman P, et al. Polyclonal immunoglobulins from a chronic hepatitis C virus patient protect human liver–chimeric mice from infection with a homologous hepatitis C virus strain. Hepatology. 2008;47:1846–1855. doi: 10.1002/hep.22244. [DOI] [PubMed] [Google Scholar]
- 17.Meuleman P, Hesselgesser J, Paulson M, et al. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48:1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 18.Chamberlain RW, Adams N, Saeed AA, Simmonds P, Elliott RM. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78:1341–1347. doi: 10.1099/0022-1317-78-6-1341. [DOI] [PubMed] [Google Scholar]
- 19.Farci P, Munoz SJ, Shimoda A, et al. Experimental transmission of hepatitis C virus–associated fulminant hepatitis to a chimpanzee. J Infect Dis. 1999;179:1007–1011. doi: 10.1086/314653. [DOI] [PubMed] [Google Scholar]
- 20.Okamoto H, Okada S, Sugiyama Y, et al. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto H, Kurai K, Okada S, et al. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto H, Kojima M, Okada S, et al. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology. 1992;190:894–899. doi: 10.1016/0042-6822(92)90933-g. [DOI] [PubMed] [Google Scholar]
- 23.Corbet S, Bukh J, Heinsen A, Fomsgaard A. Hepatitis C virus subtyping by a core-envelope 1–based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J Clin Microbiol. 2003;41:1091–1100. doi: 10.1128/JCM.41.3.1091-1100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bukh J, Apgar CL, Engle R, et al. Experimental infection of chimpanzees with hepatitis C virus of genotype 5a: genetic analysis of the virus and generation of a standardized challenge pool. J Infect Dis. 1998;178:1193–1197. doi: 10.1086/515683. [DOI] [PubMed] [Google Scholar]
- 25.Engle RE, Russell RS, Purcell RH, Bukh J. Development of a TaqMan assay for the six major genotypes of hepatitis C virus: comparison with commercial assays. J Med Virol. 2008;80:72–79. doi: 10.1002/jmv.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai A, Takikawa S, Thimme R, et al. In vivo study of the HC-TN strain of hepatitis C virus recovered from a patient with fulminant hepatitis: RNA transcripts of a molecular clone (pHC-TN) are infectious in chimpanzees but not in Huh7.5 cells. J Virol. 2007;81:7208–7219. doi: 10.1128/JVI.01774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagi M, St Claire M, Shapiro M, Emerson SU, Purcell RH, Bukh J. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology. 1998;244:161–172. doi: 10.1006/viro.1998.9092. [DOI] [PubMed] [Google Scholar]
- 28.Yanagi M, Purcell RH, Emerson SU, Bukh J. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology. 1999;262:250–263. doi: 10.1006/viro.1999.9889. [DOI] [PubMed] [Google Scholar]
- 29.Gottwein JM, Scheel TK, Hoegh AM, et al. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology. 2007;133:1614–1626. doi: 10.1053/j.gastro.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Meunier JC, Engle RE, Faulk K, et al. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc Natl Acad Sci U S A. 2005;102:4560–4565. doi: 10.1073/pnas.0501275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheel TK, Gottwein JM, Jensen TB, et al. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A. 2008;105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottwein JM, Scheel TK, Jensen TB, et al. Development and characterization of hepatitis C virus genotype 1–7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu YK, Weiner AJ, Rosenblatt J, et al. Early events in hepatitis C virus infection of chimpanzees. Proc Natl Acad Sci U S A. 1990;87:6441–6444. doi: 10.1073/pnas.87.16.6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez J, Taylor D, Morhardt DR, et al. Long-term persistence of infection in chimpanzees inoculated with an infectious hepatitis C virus clone is associated with a decrease in the viral amino acid substitution rate and low levels of heterogeneity. J Virol. 2004;78:9782–9789. doi: 10.1128/JVI.78.18.9782-9789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassett SE, Brasky KM, Lanford RE. Analysis of hepatitis C virus–inoculated chimpanzees reveals unexpected clinical profiles. J Virol. 1998;72:2589–2599. doi: 10.1128/jvi.72.4.2589-2599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bassett SE, Thomas DL, Brasky KM, Lanford RE. Viral persistence, antibody to E1 and E2, and hypervariable region 1 sequence stability in hepatitis C virus–inoculated chimpanzees. J Virol. 1999;73:1118–1126. doi: 10.1128/jvi.73.2.1118-1126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeff LB. Natural history of chronic hepatitis C. Hepatology. 2002;36:S35–S46. doi: 10.1053/jhep.2002.36806. [DOI] [PubMed] [Google Scholar]
- 38.Prince AM, Brotman B, Lee DH, et al. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J Infect Dis. 2005;192:1701–1709. doi: 10.1086/496889. [DOI] [PubMed] [Google Scholar]
- 39.Bukh J, Thimme R, Meunier JC, et al. Previously infected chimpanzees are not consistently protected from reinfection or persistent infection following reexposure to the identical hepatitis C virus strain. J Virol. 2008;82:8183–8195. doi: 10.1128/JVI.00142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mikkelsen M, Bukh J. Current status of a hepatitis C vaccine: encouraging results but significant challenges ahead. Curr Infect Dis Rep. 2007;9:94–101. doi: 10.1007/s11908-007-0003-6. [DOI] [PubMed] [Google Scholar]
- 41.Rollier C, Depla E, Drexhage JA, et al. Control of heterologous hepatitis C virus infection in chimpanzees is associated with the quality of vaccine-induced peripheral T-helper immune response. J Virol. 2004;78:187–196. doi: 10.1128/JVI.78.1.187-196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rollier CS, Paranhos-Baccala G, Verschoor EJ, et al. Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology. 2007;45:602–613. doi: 10.1002/hep.21573. [DOI] [PubMed] [Google Scholar]
- 43.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 44.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 46.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003;197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logvinoff C, Major ME, Oldach D, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci U S A. 2004;101:10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jensen TB, Gottwein JM, Scheel TK, Hoegh AM, Eugen-Olsen J, Bukh J. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J Infect Dis. 2008;198:1756–1765. doi: 10.1086/593021. [DOI] [PubMed] [Google Scholar]
- 49.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 50.Inoue K, Umehara T, Ruegg UT, et al. Evaluation of a cyclophilin inhibitor in hepatitis C virus–infected chimeric mice in vivo. Hepatology. 2007;45:921–928. doi: 10.1002/hep.21587. [DOI] [PubMed] [Google Scholar]