Abstract
Objective:
The objective of this study was to provide insight into the molecular mechanisms of acute ischemic cerebrovascular syndrome (AICS) through gene expression profiling and pathway analysis.
Methods:
Peripheral whole blood samples were collected from 39 MRI-diagnosed patients with AICS and 25 nonstroke control subjects ≥18 years of age. Total RNA was extracted from whole blood stabilized in Paxgene RNA tubes, amplified, and hybridized to Illumina HumanRef-8v2 bead chips. Gene expression was compared in a univariate manner between stroke patients and control subjects using t test in GeneSpring. The significant genes were tested in a logistic regression model controlling for age, hypertension, and dyslipidemia. Inflation of type 1 error was corrected by Bonferroni and Ingenuity Systems Pathway analysis was performed. Validation was performed by QRT-PCR using Taqman gene expression assays.
Results:
A 9-gene profile was identified in the whole blood of ischemic stroke patients using gene expression profiling. Five of these 9 genes were identified in a previously published expression profiling study of stroke and are therefore likely biomarkers of stroke. Pathway analysis revealed toll-like receptor signaling as a highly significant canonical pathway present in the peripheral whole blood of patients with AICS.
Conclusions:
Our study highlights the relevance of the innate immune system through toll-like receptor signaling as a mediator of response to ischemic stroke and supports the claim that gene expression profiling can be used to identify biomarkers of ischemic stroke. Further studies are needed to validate and refine these biomarkers for their diagnostic potential.
GLOSSARY
- AICS
= acute ischemic cerebrovascular syndrome;
- BBB
= blood–brain barrier;
- IPA
= Ingenuity Systems Pathway analysis;
- PBMC
= peripheral blood mononuclear cell;
- rtPA
= recombinant tissue plasminogen activator;
- TLR
= toll-like receptor.
Stroke is the third leading cause of death in the United States1 and one of the most common causes of disability in industrialized countries.2 Despite significant advances in neuroimaging and clinical management resulting in increased survival,3 morbidity remains high secondary to complications and a lack of alternative treatments.
The development of microarray systems for gene expression profiling permits screening of large numbers of genes for involvement in biologic processes. Gene expression profiles can be used to reflect and predict pathologic processes. Thus, gene expression profiling has been utilized for the characterization of several neurologic and immune disorders4–6 and has facilitated the refinement of tumor subtypes,7 distinction between good-prognosis and poor-prognosis tumors,8 and the prediction of response to treatment.9 The method has the potential to lead to a better understanding of the molecular mechanisms underlying disease, identify risk for secondary complications, and aid in the development of novel therapeutics.
In the context of ischemic stroke, there are 2 reports of peripheral blood mononuclear cells (PBMCs)10,11 and 1 of peripheral whole blood12 examining gene expression in peripheral blood. These studies provide a novel and sophisticated approach to identify biomarkers of stroke. However, the study designs are not complementary; nor are the findings. Thus, the purpose of our study was to determine the gene expression profile of peripheral whole blood following acute ischemic stroke in a larger cohort of subjects, adjusting for common stroke risk factors to provide insight into the molecular mechanisms involved in ischemic stroke and attempt to replicate previous findings.
METHODS
A retrospective case-control study utilizing prospectively collected data from 2 different study sources was undertaken. Recruitment of stroke patients occurred from June 2007 through September 2008 when the following inclusion criteria were met: age ≥18 years; MRI diagnosed definite acute ischemic cerebrovascular syndrome (AICS13); and blood draw within 24 hours from symptom onset. Patients with probable/possible AICS and hemorrhage were excluded from this study. Time of onset was determined as the time the patient was last known to be free of the acute stroke symptoms. Recombinant tissue plasminogen activator (rtPA) was given to patients with disabling symptoms within 3 hours from onset. Premorbid deficits were determined by the modified Rankin Scale score for status prior to stroke and severity of injury was determined by the NIH Stroke Scale score at the time of blood draw after stroke. Control subjects were recruited as a consecutive convenience sample under a separate NIA/NIH protocol if they were neurologically normal per neurologist assessment at the time of enrollment. Peripheral whole blood was collected into Paxgene™ blood RNA tubes (PreAnalytiX, Qiagen) after consent. Demographic data were collected from the patient or significant other by trained neurologists.
Standard protocol approvals, registrations, and consents.
This study received approval for human subject research from the institutional review boards of the National Institute of Neurological Disorders and Stroke and National Institute on Aging at NIH and Suburban Hospital, Bethesda, MD. Written informed consent was obtained from all subjects or their authorized representatives prior to performing any study procedures.
RNA extraction and amplification.
Paxgene RNA tubes were inverted 8–10 times and placed in a −80°C freezer until RNA extraction. Tubes were thawed on a rotating bed at room temperature for 24 hours prior to RNA isolation. RNA was extracted per Paxgene Blood RNA Extraction Kit (PreAnalytiX, Qiagen). Globin reduction was not conducted on any sample in this study since it has been shown to have little impact on probe detection when using the Illumina platform.14
Biotinylated, amplified RNA was generated from the Illumina TotalPrep RNA amplification kit (Applied Biosystems). RNA quantity was determined by the Nanodrop and RNA quality was determined by A260/A280 ratio and the presence of 2 distinct ribosomal bands on gel electrophoresis.
Array hybridization.
Samples were randomly hybridized to Illumina HumanRef-8 v2 expression bead chips, capable of analyzing >22,000 genes and alternative splice variants. Bead arrays were scanned by the Illumina BeadStation 500× and raw intensity values were saved in Illumina's Bead Studio program manager. Sample labeling, hybridization, and scanning were conducted using standard Illumina protocols.
Statistical analysis.
Baseline demographic statistics were conducted in SPSS (version 15, SPSS, Inc., Chicago, IL). Comparisons were made using χ2 analysis for gender, race, comorbidities (hypertension, diabetes, and hyperlipidemia), and medication history. Student t test was used to analyze the significance of age among the groups. The level of significance was established at 0.05 for 2-sided hypothesis testing.
Probe level analysis.
Probe expression was filtered in GeneSpring GX v10 (Agilent Technologies) resulting in a 24,424 final probe set. Robust multiarray analysis normalization collated the probe data in the following order: 1) background correction—perfect match probe information; 2) quantile normalization—probe level normalization; and 3) summarization—expression measure summary in log base 2 scale with median to fit a linear model. Unsupervised clustering was performed to determine phylogenetic distances to detect outliers.
Gene expression level analysis.
Gene expression analysis was conducted in Illumina BeadStudio Gene Expression (GX) Module (version 1, Illumina®, San Diego, CA) and verified in GeneSpring GX v10 (Agilent Technologies). Genes with at least a 2-fold difference in expression were compared in a univariate manner between stroke patients and control subjects through the use of Illumina's custom model (modified t test) in BeadStudio and t test comparisons in GeneSpring. The influence of multiple testing was evaluated using the Bonferroni family-wise error.
Pathway analysis.
Ingenuity Systems Pathway analysis (IPA) (Ingenuity® Systems, www.ingenuity.com) was used to assess genes with a 1.5-fold difference or greater expression. Gene identifiers and signal intensities were compared to the Canonical Pathways in the Ingenuity Systems knowledge base; significant pathways (p < 0.05) were identified and a set of networks (35 genes/gene products) was generated. A score, derived from a p value, was generated for each network according to the fit of the set of significant genes. Scores of 2 or higher have at least a 99% confidence of not being generated by chance alone. Biologic functions were calculated and assigned to each network. The significance of the association was measured by 1) the number of molecules in a pathway that met the 1.5-fold cutoff divided by the total number of molecules of that pathway; and 2) a right-tailed Fisher exact test to calculate a p value.
Logistic regression for identification of off-target effects.
Given the significant difference of age by group, a post hoc logistic regression was performed. The normalized intensities for each gene were entered separately with age and then hypertension and dyslipidemia as the covariates of interest. A Bonferroni-corrected p of ≤0.005 (0.05/9) was significant.
PCR validation.
cDNA was generated per Invitrogen SuperScript III first strand synthesis kit. QRT-PCR reactions were performed using Taqman® gene expression probes (Applied Biosystems) for ARG1, CCR7, LY96, and MMP9 by the 7900HT QRT-PCR system. β-Actin normalized the relative expression of chosen genes. Fold change differences were calculated by the Δ CT method.15,16 Validation was confirmed if t test revealed significance (p ≥ 0.05) and QRT-PCR results correlated with microarray signal intensity (Pearson r ≥ 0.5 and p ≥ 0.05).
Sample size estimation.
Sample size estimation was conducted using PASS Power Analysis and Sample Size system and JMP. Twenty-two patients and 22 control subjects achieves 90.68% power for each gene to detect a difference in expression with at least a 1.5-fold change and a SD of 1.5 with a false discovery rate of 0.05 using a 2-sided 1-sample t test.
RESULTS
Clinical characteristics.
Ninety-two subjects (67 stroke patients and 25 control subjects) were recruited. Of the 67 stroke patients enrolled, 39 were definite AICS.13 Based on the Trial of Org 10172 in Acute Stroke Treatment, 43.6% (n = 17) of the cases were cardioembolic stroke, 28.2% (n = 11) were undetermined, 12.8% (n = 5) were large artery embolus/thrombosis, and the remaining 15.4% (n = 6) were small vessel or other cause.
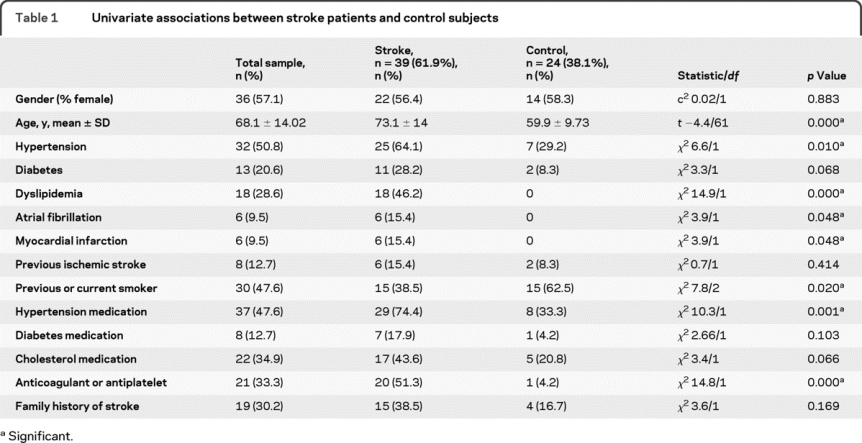
The mean time from symptom onset to blood draw was 10:06 hours ± 6:31. Nine (23.1%) patients received rtPA; only one patient had blood drawn before rtPA administration. Median prestroke modified Rankin Scale score was zero and the severity of stroke was mild with a median baseline NIH Stroke Scale score of 3 (range 0–23). There was no difference by race or gender between the groups. However, stroke patients were significantly older (t = −4.03; p = 0.000) and, as expected, vascular risk factors were more common in the stroke group (table 1).
Table 1 Univariate associations between stroke patients and control subjects
Array quality control.
RNA A260/A280 was 1.9–2.2 and yield was >1–2 μg from 2.5 mL of whole blood. Negative control, background, and noise signals were low (<200) across all arrays and housekeeping and biotin signals were consistently high (>20,000). The average signal for internal controls was similar across the arrays. Control plots were consistent with high-quality data.
Nine-gene profile for stroke.
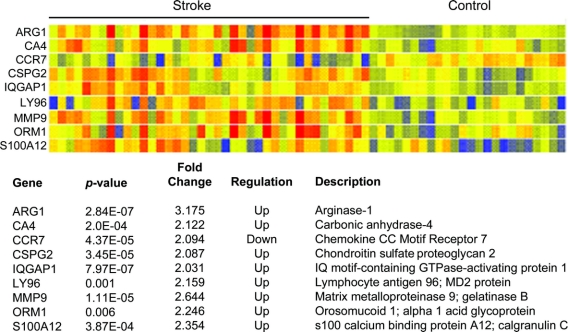
BeadStudio identified 19 genes with a 2-fold difference in expression (table e-1 on the Neurology® Web site at www.neurology.org). GeneSpring identified 16 genes with a 2-fold difference in expression (table e-1). After comparison, there were 9 genes significantly different between stroke patients and control subjects with at least a 2-fold difference in expression and corrected p < 0.05 (figure 1). Five of these 9 genes were significant in the first whole blood gene expression profiling study of stroke (ARG1, CA4, LY96, MMP9, S100A12).12
Figure 1 Stroke gene profile
After comparison between both statistical packages, there were 9 genes differentially expressed with at least a 2-fold difference in expression and Bonferroni-corrected p < 0.05 between stroke patients and control subjects (ARG1, CA4, CCR7, CSPG2, IQGAP1, LY96, MMP9, ORM1, S100A12).
Logistic regression for identification of off-target effects.
s100A12 was the only gene that fell out of the model after correction (p = 0.014). ARG1 (p = 0.002), CA4 (p = 0.002), CCR7 (p = 0.005), CSPG2 (p = 0.003), IQGAP1 (p = 0.003), and MMP9 (p = 0.002) remained significantly associated with stroke after controlling for age, history of hypertension, and dyslipidemia.
Pathway analysis.
A total of 355 genes were eligible for pathway analysis (≥1.5-fold difference and corrected p < 0.05). The 5 most significant pathways in the peripheral whole blood of stroke patients were CD28 signaling (p = 4.03E00), nuclear factor of activated T cells in regulation of the immune response (p = 4.03E00), dendritic cell maturation (p = 3.4E00), toll-like receptor (TLR) signaling (p = 3.33E00), and calcium-induced T-lymphocyte apoptosis (p = 2.92E00) (figure e-1). There were more genes expressed in the TLR signaling pathway than in any other pathway, implying it is the most significant for this dataset (figure e-2).
Taqman gene expression assay validation.
QRT-PCR validated significant mRNA changes of 4 genes in all samples. Secondary to the availability of RNA, the entire gene profile could not be validated. ARG1, LY96, and MMP9 were chosen because they were significant in previous work and CCR7 was the only downregulated gene in our study (figure e-3).
DISCUSSION
Peripheral whole blood gene expression profiling identified 9 genes with ≥2-fold difference in expression following ischemic stroke: Arginase 1 (ARG1), carbonic anhydrase 4 (CA4), chondroitin sulfate proteoglycan 2 (CSPG2), IG motif-containing GTPase activation protein 1 (IQGAP1), lymphocyte antigen 96 (LY96), matrix metalloproteinase 9 (MMP9), orosomucoid 1 (ORM1), and s100 calcium binding protein A12 (s100A12), and one downregulated gene, chemokine receptor 7 (CCR7).
A limitation of our study was a younger control group; however, post hoc analyses controlling for age and stroke risk factors supported the primary analysis findings. The fact that 5 of the 9 genes identified in this study were also found to be differently expressed in a previous study12 using a different microarray platform, independent cohorts of patients and controls, and other methodologic aspects provides validation for both studies and confirms the strength of this method to unveil molecular mechanisms involved in ischemic stroke.
The majority of genes induced 2–24 hours following stroke occurs in neutrophils and monocytes.17 Therefore, peripheral whole blood gene expression may be the most useful for identifying biomarkers of ischemic stroke in humans. The inconsistency between the first 3 peripheral blood gene expression profiling studies of stroke is most attributable to the different sources of RNA under study. Some have also interpreted this to treatment of patients with rtPA11 and variable time between symptom onset and blood draw. In this study, there was one gene coincident with the first PBMC study (CSPG2)11 and 5 genes overlapping with the previous whole blood study (ARG1, CA4, LY96, MMP9, S100A12).12 Nine patients (23%) received rtPA, a similar proportion in the PBMC study, and the mean time from symptom onset to blood draw was 10:06 hours, more than the 3 hours previously suggested useful.12 It is remarkable that besides these differences, 50% of the genes identified in the previous studies have been replicated in our study, and all of the genes shared between our study and the previous whole blood study were expressed at all time points (<3 hours, 5 hours, 24 hours). This indicates that the differences between the first expression studies were in fact due to differences in cell populations under study and that coincident findings with the previous whole blood study are not reflecting changes secondary to administration of rtPA as previously considered, but rather changes associated with ischemic stroke. These results suggest that the relative expression of ARG1, CA4, LY96, MMP9, and S100A12 taken together have strong evidence for diagnostic capability in acute ischemic stroke.
Interestingly, MMP9 and various isoforms of S100 at the protein level have been implicated as predictors of stroke outcome. One of our recent publications suggests baseline serum MMP9 may help predict the occurrence of blood–brain barrier (BBB) disruption18 and high level of S100 serum protein is associated with worse clinical outcome,19 implying MMP9 and S100 may also be useful as prognostic markers in ischemic stroke.
In addition to the common genes identified by previous studies, we have also identified novel biomarkers of ischemic stroke: CCR7, IQGAP1, and ORM1.
Chemokine receptor 7 (CCR7).
CCR7 is a G-coupled chemokine receptor and the data on CCR7 function in humans are rather sparse. However, increased CCR7+ T cells have been reported in peripheral blood leukocytes of patients with mild to moderate ischemic stroke 1 week following stroke.20 We found a downregulation of CCR7 in the peripheral blood in the acute phase of ischemic stroke. Differences in the direction of this regulation could be explained as differences in tissue/cell-specific immune response following stroke and/or blood draw time. Dramatic decreases of CCR7 expression in the acute phase of ischemic stroke could be followed by an increase in expression with stroke progression. Further studies of CCR7 and stroke should investigate this issue.
IQ Motif-containing GTPase activating protein 1 (IQGAP1).
IQGAP1 is an evolutionarily conserved scaffold protein that plays a fundamental role in cell polarity. Rho-family GTPases are small signaling G proteins that require IQGAP1 to regulate actin cytoskeleton to produce a gradient of signaling molecules.21 Experimental evidence suggests the expression of RhoA increases in aortic and basilar arteries with age,22 therefore RhoA, and indirectly IQGAP1, may play a role in altered vascular responses associated with aging.22 Studies in vitro have suggested that leukocyte transmigration and changes in endothelial permeability are facilitated by RhoA.23 The upregulation of IQGAP1 expression in the context of ischemic stroke suggests an increase in cellular signaling and transcription in the acute phase of ischemic stroke leading to increased permeability of the BBB. IQGAP1 may mediate the disruption of the BBB as a means by which signals from the brain enter the periphery to augment cellular recruitment.
Oromucosid 1 (ORM1).
Finally, ORM1, also known as α-1 acid glycoprotein, is an acute phase protein that suppresses lymphocyte response to lipopolysaccharides (thereby preventing ongoing tissue damage by neutrophil proteases), decreases platelet aggregation (and further platelet recruitment), and enhances cytokine secretion.24 It exhibits anti-inflammatory effects, playing a significant role in immunomodulation of the acute phase response.25 The upregulation of ORM1 in the context of ischemic stroke suggests an acute phase response in ischemic stroke similar to trauma, infection, and systemic tissue injury.
IPA analysis identified TLR signaling as the most significant pathway in the peripheral blood of ischemic stroke patients. Activation of innate immunity, through TLRs, is a primary component of proinflammatory cytokine generation following ischemic brain injury.26 Products of protein degradation damaged DNA, fibrinogen, and heat shock proteins activate the TLR pathway through a mechanism known as damage associated molecular pattern recognition.27,28
Activation of TLRs has been implicated as a negative moderator of the innate immune response.29 Downregulation of TLR4 results in diminished infarct volume and better outcome in mouse middle cerebral artery occlusion. Human studies have identified that increased activation of TLR4 following ischemic stroke corresponds to worse clinical outcome.30,31
The predominance of the innate and inflammatory immune pathways in our study reinforces their importance in ischemic stroke. A better understanding of how these pathways react and respond to ischemic stroke may lead to the emergence of new avenues for therapeutic intervention. The TLR pathway is a promising and worthy target to be considered and studied with more detail.
The results of this study provide insight into the molecular mechanisms involved in ischemic stroke. A 9-gene profile for acute ischemic stroke has been identified and replicates previous work,12 therefore supporting the use of gene expression profiling of peripheral whole blood to identify biomarkers of ischemic stroke. Future studies including a stroke mimic cohort and a TIA group will help determine the differential diagnostic capability and clinical utility of these results.
AUTHOR CONTRIBUTIONS
Statistical analysis was conducted by Dr. J. Ding.
ACKNOWLEDGMENT
The authors thank Mark Cookson, PhD, and Marcel P. Van der Brug, PhD, for expert review and guidance.
DISCLOSURE
Dr. Barr has a patent pending re: diagnosis of ischemic stroke using gene expression profiling and receives research support from the Division of Intramural Research of the NIH/NINDS/NIA and a predoctoral training fellowship from NINR. Dr. Conley serves on the editorial board of Biological Research for Nursing; has a patent pending re: panel of markers to diagnose stroke and receives royalties (Optherion, Inc.) for a patent re: susceptibility genes for age-related maculopathy; receives research support from the NIH (NINR R01NR008424–01 [coinvestigator], NINDS P01NS030318 [coinvestigator], NEI R01EY09859 [coinvestigator], NICHD R01HD048162 [subcontract PI], NINR R01NR004339 [sponsor], NINR F31NR011379 [codirector], NINR T32NR009759 [training faculty], and NICHD T32HD040686 [PI]), the US Department of Defense (W81XWH-07–0701 [coinvestigator]), and the Oncology Nursing Society. Dr. Ding and A. Dillman report no disclosures. Dr. Warach serves on the editorial boards of the Journal of Cerebral Blood Flow and Metabolism, Stroke, The Lancet Neurology, the International Journal of Stroke, and Cerebrovascular Diseases and receives research support from the NIH/NINDS, Division of Intramural Research. Dr. Singleton serves on the editorial boards of Annals of Neurology, The Lancet Neurology, Neurogenetics, and Neurodegenerative Diseases; has a patent pending re: panel of markers to diagnose stroke; and receives NIH Intramural funding (Department of Defense W81XWH-09–2-0128 [PI]). Dr. Matarin reports no disclosures.
Supplementary Material
Address correspondence and reprint requests to Dr. Taura L. Barr, National Institute of Nursing Research, Tissue Injury Unit Building, 10 Hatfield Clinical Research Center, Room 2-1339, Bethesda, MD 20812 barrt@mail.nih.gov
Supplemental data at www.neurology.org
Study funding: Supported by the Division of Intramural Research of the NIH/NINDS/NIA (Z01 AG000957-05) and by a predoctoral Intramural Research Training Award via the Graduate Partnerships Program through the National Institute of Nursing Research, NIH (to T.L.B.).
Disclosure: Author disclosures are provided at the end of the article.
Received February 12, 2010. Accepted in final form May 20, 2010.
REFERENCES
- 1.Thom T, Haase N, Rosamond W, et al. Heart disease and stroke statistics: 2006 update: a report from the American Heart Association statistics committee and stroke statistics subcommittee. Circulation 2006;113:e85–151. [DOI] [PubMed] [Google Scholar]
- 2.McKay J, Mensah GA, ebrary Inc. The Atlas of Heart Disease and Stroke. Geneva: World Health Organization; 2005. [Google Scholar]
- 3.Kim D, Liebeskind DS. Neuroimaging advances and the transformation of acute stroke care. Semin Neurol 2005;25:345–361. [DOI] [PubMed] [Google Scholar]
- 4.Tajouri L, Fernandez F, Griffiths LR. Gene expression studies in multiple sclerosis. Curr Genomics 2007;8:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherzer CR, Eklund AC, Morse LJ, et al. Molecular markers of early Parkinson's disease based on gene expression in blood. Proc Natl Acad Sci USA 2007;104:955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maes OC, Xu S, Yu B, Chertkow HM, Wang E, Schipper HM. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol Aging 2007;28:1795–1809. [DOI] [PubMed] [Google Scholar]
- 7.Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature 2000;406:536–540. [DOI] [PubMed] [Google Scholar]
- 8.Jones MH, Virtanen C, Honjoh D, et al. Two prognostically significant subtypes of high-grade lung neuroendocrine tumours independent of small-cell and large-cell neuroendocrine carcinomas identified by gene expression profiles. Lancet 2004;363:775–781. [DOI] [PubMed] [Google Scholar]
- 9.Chang JC, Wooten EC, Tsimelzon A, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 2003;362:362–369. [DOI] [PubMed] [Google Scholar]
- 10.Grond-Ginsbach C, Hummel M, Wiest T, et al. Gene expression in human peripheral blood mononuclear cells upon acute ischemic stroke. J Neurol 2008;255:723–731. [DOI] [PubMed] [Google Scholar]
- 11.Moore DF, Li H, Jeffries N, et al. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation 2005;111:212–221. [DOI] [PubMed] [Google Scholar]
- 12.Tang Y, Xu H, Du X, et al. Gene expression in blood changes rapidly in neutrophils and monocytes after ischemic stroke in humans: a microarray study. J Cereb Blood Flow Metab 2006;26:1089–1102. [DOI] [PubMed] [Google Scholar]
- 13.Kidwell CS, Warach S. Acute ischemic cerebrovascular syndrome: diagnostic criteria. Stroke 2003;34:2995–2998. [DOI] [PubMed] [Google Scholar]
- 14.Expression profiling of whole blood specimens on Illumina Beadchips: expression analysis tech note: October 2007. Available at: www.Expressionanalysis.com.
- 15.Amplification efficiency of TaqMan gene expression assays, stock# 127ap05–03. Available at: www3.Appliedbiosystems.com.
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y, Gilbert DL, Glauser TA, Hershey AD, Sharp FR. Blood gene expression profiling of neurologic diseases: a pilot microarray study. Arch Neurol 2005;62:210–215. [DOI] [PubMed] [Google Scholar]
- 18.Barr TL, Latour LL, Lee KY, et al. Blood-brain barrier disruption in humans is independently associated with increased matrix metalloproteinase-9. Stroke 2009;41:e123–e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brea D, Sobrino T, Blanco M, et al. Temporal profile and clinical significance of serum neuron-specific enolase and s100 in ischemic and hemorrhagic stroke. Clin Chem Lab Med 2009;47:1513–1518. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Greer JM, Etherington K, et al. Immune activation in the peripheral blood of patients with acute ischemic stroke. J Neuroimmunol 2009;206:112–117. [DOI] [PubMed] [Google Scholar]
- 21.Fukata M, Nakagawa M, Kaibuchi K. Roles of rho-family gtpases in cell polarisation and directional migration. Curr Opin Cell Biol 2003;15:590–597. [DOI] [PubMed] [Google Scholar]
- 22.Miao L, Calvert JW, Tang J, Parent AD, Zhang JH. Age-related rhoa expression in blood vessels of rats. Mech Ageing Dev 2001;122:1757–1770. [DOI] [PubMed] [Google Scholar]
- 23.Wojciak-Stothard B, Ridley AJ. Rho gtpases and the regulation of endothelial permeability. Vascul Pharmacol 2002;39:187–199. [DOI] [PubMed] [Google Scholar]
- 24.Hochepied T, Berger FG, Baumann H, Libert C. Alpha(1)-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev 2003;14:25–34. [DOI] [PubMed] [Google Scholar]
- 25.Fournier T, Medjoubi NN, Porquet D. Alpha-1-acid glycoprotein. Biochim Biophys Acta 2000;1482:157–171. [DOI] [PubMed] [Google Scholar]
- 26.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol 2001;1:135–145. [DOI] [PubMed] [Google Scholar]
- 27.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol 2007;81:1–5. [DOI] [PubMed] [Google Scholar]
- 28.Rubartelli A, Lotze MT. Inside, outside, upside down: damage-associated molecular-pattern molecules (DAMPs) and redox. Trends Immunol 2007;28:429–436. [DOI] [PubMed] [Google Scholar]
- 29.Kilic U, Kilic E, Matter CM, Bassetti CL, Hermann DM. Tlr-4 deficiency protects against focal cerebral ischemia and axotomy-induced neurodegeneration. Neurobiol Dis 2008;31:33–40. [DOI] [PubMed] [Google Scholar]
- 30.Yang QW, Li JC, Lu FL, et al. Upregulated expression of toll-like receptor 4 in monocytes correlates with severity of acute cerebral infarction. J Cereb Blood Flow Metab 2008;28:1588–1596. [DOI] [PubMed] [Google Scholar]
- 31.Urra X, Cervera A, Obach V, Climent N, Planas AM, Chamorro A. Monocytes are major players in the prognosis and risk of infection after acute stroke. Stroke 2009;40:1262–1268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.