Abstract
Reduction of various biological processes is a hallmark of the parasitic lifestyle. Generally, the more intimate the association between parasites and hosts the stronger the parasite relies on its host's physiology for survival and reproduction. However, some systems have been held to be indispensable, for example, the core pathways of carbon metabolism that produce energy from sugars. Even the most hardened anaerobes that lack oxidative phosphorylation and the tricarboxylic acid cycle have retained glycolysis and some downstream means to generate ATP. Here we describe the deep-coverage genome resequencing of the pathogenic microsporidiian, Enterocytozoon bieneusi, which shows that this parasite has crossed this line and abandoned complete pathways for the most basic carbon metabolism. Comparing two genome sequence surveys of E. bieneusi to genomic data from four other microsporidia reveals a normal complement of 353 genes representing 30 functional pathways in E. bieneusi, except that only 2 out of 21 genes collectively involved in glycolysis, pentose phosphate, and trehalose metabolism are present. Similarly, no genes encoding proteins involved in the processing of spliceosomal introns were found. Altogether, E. bieneusi appears to have no fully functional pathway to generate ATP from glucose. Therefore, this intracellular parasite relies on transporters to import ATP from its host.
Keywords: microsporidia, parasite, glycolysis, carbon metabolism, reduction, evolution
Introduction
Microsporidia are notoriously reduced and derived intracellular parasites, some with genomes smaller than those of many bacteria and a reduced set of about 2,000 genes (Katinka et al. 2001; Cornman et al. 2009; Corradi et al. 2009). Even so, the complete genome of the model pathogenic microsporidian Encephalitozoon cuniculi has a fully functional set of genes for several core carbon metabolic pathways, as well as some lipid metabolism (Katinka et al. 2001). Three other microsporidian genomes that have been surveyed at depth, Nosema ceranae, Antonospora locustae, and Octosporea bayeri (this species is in the process of being renamed, but the genome survey was under the name O. bayeri), contain more or less the same complement of genes for core functions (Cornman et al. 2009; Corradi et al. 2009). However, all three of these genomes also contain several genes that are not present in E. cuniculi, raising questions about whether this pool of genes adequately represents the potential metabolic diversity of microsporidia.
This is particularly striking in the case of energy metabolism. Glycolysis, the pentose phosphate pathway, and trehalose metabolism have been taken to represent the backbone of microsporidian energy metabolism because their mitochondria are massively reduced and all species investigated to date lack the tricarboxylic acid cycle and oxidative metabolism (Williams and Keeling 2003; van der Giezen et al. 2005), with the exception of the alternative oxidase (AOX) in some species (Williams et al. 2010). However, a recent 3× coverage genome sequence survey of a human isolate of Enterocytozoon bieneusi revealed that genes relating to energy metabolism were underrepresented in general (Akiyoshi et al. 2009). Whether this survey accurately represents the content of the E. bieneusi genome is a challenging question because of its very low coverage (Milinkovitch et al. 2010). Low coverage is particularly problematic when it concerns the potential absence of genes because any given gene may be missing due to sampling error and only the functional relationship of these genes allowed the suggestion that they may actually be absent. In addition, the parasite is not presently cultivable and may only be isolated directly from infected host animals and is therefore not amenable to biochemical analysis to prove the absence of enzymatic activity.
To test this question, we have used clone-independent, 10× coverage 454 genome sequencing to compare an independent survey of the E. bieneusi genome with the previous survey, as well as with the genomes of four distantly related microsporidians. If the absence of genes in the previous 3× coverage was due to sampling error resulting from low coverage, we would expect a resampling (at even higher coverage) to produce a different but overlapping set of genes. If, on the other hand, the genes identified in the 3× coverage did adequately represent the content of the genome, then we would expect to find very few additional genes in a second survey, and in particular, no additional genes for energy metabolism would be expected to be found (supporting their absence from the genome). Here we report an almost perfect correlation between the gene content of the original genome survey and that of the present, 10× coverage 454 survey. Specifically, most functional pathways present in other microsporidia are well represented in both E. bieneusi surveys, with the exception of all pathways relating to energy generation. From these pathways, only the same two genes were found in both surveys, a result not consistent with sampling error. Interestingly, we found that no genes were present relating to the removal of spliceosomal introns and that genes for fatty acid metabolism are also reduced, suggesting that these functions are also reduced or lost. Overall, these results show that E. bieneusi has lost not only introns and the spliceosome (which have otherwise only been recorded to be lost in a single nucleus-derived organelle: Lane et al. 2007) but also more interestingly lost the ability to generate energy from sugars, a level of host dependence that has never been observed in any other parasite.
Materials and Methods
The genome of E. bieneusi human isolate H348, karyotype H1, was sequenced using Roche GS-FLX (454) with library construction and sequencing performed essentially as described (Margulies et al. 2005). Briefly, 2 μg of high molecular weight DNA (Akiyoshi et al. 2009) was sheared by nebulization and size selected to generate 300–800 bp fragments. DNA fragment ends were repaired and phosphorylated using T4 DNA polymerase and T4 polynucleotide kinase. Adaptor oligonucleotides “A” and “B” supplied with the 454 Life Sciences shotgun sequencing reagent kit were ligated to the DNA fragments using T4 DNA ligase. Purified DNA fragments were hybridized to DNA capture beads and clonally amplified by emulsion polymerase chain reaction. DNA capture beads containing amplified DNA were deposited onto the GS-FLX PicoTiterPlate for sequencing. The equivalent of a full FLX run was done but as two half plates on two separate dates. The sequencing produced 300,865 reads, totaling 71.3 Mb of sequence data. The sequence data were assembled using Roche's Newbler assembler, version 1.1.03.24, and open reading frames were identified using GLIMMER3 (Delcher et al. 1999) and the ‘getorf’ module of the EMBOSS package (Rice, Longden, and Bleasby 2000). New sequences were both assembled alone and with the original Sanger survey data. The assemblies were similar suggesting that the 10× coverage was not significantly different than the original survey. Additional homologues to yeast and E. cuniculi genes were identified using Blast (TBlastN, TBlastX, and BlastX; Altschul et al. 1997) to search the assembly. Gene identification was generally based on Blast searches with a cutoff of 10−10. Those genes involved in energy metabolism in other microsporidia that were not identified were specifically sought using TBlastN with a lower cutoff. Even at 10−3 and with a close inspection of the resulting alignments, no additional candidates for energy metabolism were identified. The complement of genes identified in the original sequence survey and the 454 sequence survey were compared by reciprocal Blast. All new sequences have been deposited in GenBank under accession SRX019563 and in MicrosporidiaDB (www.microsporidiadb.org) at EuPathdB.
Results
The genome of E. bieneusi was resampled to an estimated 10× coverage using 454 pyrosequencing and compared with the original survey (Akiyoshi et al. 2009). The pool of genes identified in the two surveys were found to correlate remarkably well (table 1, supplementary table S1, Supplementary Material online). Indeed, out of 353 genes representing 30 functional pathways common to the other microsporidia, only two genes were identified in the present survey that were not identified previously (table 1, supplementary table S1, Supplementary Material online). Finding the same pool of identifiable genes in two surveys suggests that both surveys have sampled the array of E. bieneusi genes to near completion. At face value, this is not easily reconcilable with the estimated genome size of 6 Mb for E. bieneusi, which suggested that only 60% of the genome had been covered in the previous survey (Akiyoshi et al. 2009). However, partial genome duplications have been shown in other microsporidia (Nassonova et al. 2005), and the presence of such duplications in E. bieneusi would lead to the observed inconsistency.
Table 1.
Numbers of Genes Corresponding to Core Carbon Metabolic Pathways and Other Representative Pathways in Five Diverse Microsporidian Genomes
| Biochemical Pathways | Encephalitozoon cuniculi | Antonospora locustae | Octosporea bayeri | Nosema ceranae | Enterocytozoon bieneusi | |
| Genomic data available | Sanger (complete) | Sanger | Solexa | 454 | Sanger | 454 |
| Estimated genome size (Mb) | 2.9 | 5.6 | ≤24.2 | ≤10 | 6 | |
| (A) Pathways with strong differences among species | ||||||
| Glycolysis | 12 | 12 | 12 | 12 | 1 | 1 |
| Pentose phosphate pathway | 5 | 5 | 5 | 5 | 1 | 1 |
| Trehalose metabolism | 4 | 4 | 4 | 4 | 0 | 0 |
| Fatty acids biosynthesis | 20 | 19 | 20 | 19 | 5 | 6 |
| Number of ORFs | 41 | 40 | 41 | 40 | 7 | 8 |
| (B) Pathways present in all species | ||||||
| Transcriptional control | 44 | 41 | 44 | 34 | 39 | 39 |
| 60S ribosomal proteins | 46 | 39 | 39 | 39 | 45 | 43 |
| 40S ribosomal proteins | 31 | 31 | 29 | 25 | 26 | 27 |
| tRNA synthetases | 21 | 21 | 21 | 21 | 21 | 21 |
| rRNA processing | 20 | 20 | 18 | 19 | 18 | 18 |
| Recombination and DNA repair | 22 | 20 | 21 | 20 | 19 | 19 |
| Meiosis | 18 | 18 | 15 | 18 | 14 | 13 |
| Protein kinases | 19 | 18 | 14 | 17 | 18 | 19 |
| Transcription initiation factors | 18 | 18 | 18 | 15 | 17 | 17 |
| Cell growth and cell polarity | 11 | 8 | 9 | 10 | 10 | 9 |
| Deoxyribonucleotide metabolism | 11 | 6 | 7 | 4 | 11 | 10 |
| C-compound and carbohydrate metabolism | 9 | 9 | 9 | 8 | 7 | 7 |
| DNA-directed DNA polymerases | 8 | 8 | 8 | 8 | 8 | 8 |
| DNA replication factors A and C | 8 | 7 | 6 | 5 | 8 | 8 |
| DNA replication licensing factors, MCM family | 8 | 8 | 8 | 8 | 8 | 8 |
| tRNA modification | 7 | 7 | 7 | 6 | 7 | 7 |
| tRNA synthesis | 7 | 7 | 7 | 6 | 6 | 6 |
| DNA-dependent helicases, ligases, telomerases | 7 | 7 | 7 | 7 | 7 | 7 |
| Chromosome segregation proteins, SMC family | 5 | 5 | 5 | 5 | 5 | 5 |
| Phosphate metabolism | 5 | 3 | 5 | 5 | 5 | 5 |
| Amino acid metabolism | 4 | 4 | 3 | 4 | 3 | 3 |
| Transcription elongation factors | 3 | 3 | 2 | 3 | 3 | 3 |
| DNA topoisomerases | 3 | 3 | 3 | 2 | 3 | 3 |
| Nitrogen and sulfur metabolism | 3 | 3 | 3 | 2 | 2 | 2 |
| Polynucleotide degradation | 2 | 2 | 2 | 2 | 2 | 2 |
| tRNA processing | 2 | 2 | 2 | 1 | 2 | 2 |
| Origin recognition complex | 2 | 2 | 2 | 1 | 2 | 2 |
| Glycerol phosphate shuttle | 2 | 2 | 2 | 2 | 1 | 1 |
| Sugar transporters | 3 | 2 | 2 | 3 | 3 | 3 |
| ADP/ATP transporters | 4 | 3 | 1 | 4 | 4 | 4 |
| Number of ORFs | 353 | 327 | 319 | 304 | 324 | 321 |
| Total ORFs | 394 | 367 | 360 | 344 | 331 | 329 |
MCM, minichromosome maintenance; ORF, open reading frame; rRNA, ribosomal RNA; SMC, structural maintenance of chromosomes; tRNA, transfer RNA.
Moreover, with very few exceptions (see below), the genes identified in E. bieneusi surveys are the same genes found in other phylogenetically diverse microsporidian genomes (table 1, supplementary table S1, Supplementary Material online). For example, the E. cuniculi genome encodes 46 identifiable large subunit ribosomal proteins, and we identified 45 of these, and no additional large subunit ribosomal proteins, in E. bieneusi (table 1, supplementary table S1, Supplementary Material online). The same holds true for most of the 353 genes and 30 functional pathways analyzed, of which more than 91% were also identified in E. bieneusi (table 1, supplementary table S1, Supplementary Material online). Altogether, this pattern suggests that the majority of E. bieneusi genes have been sampled and identified.
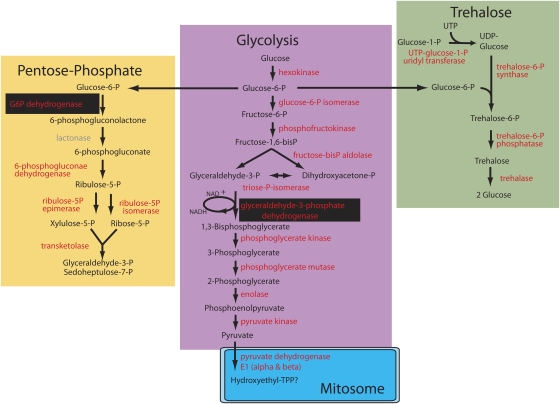
Although most functional pathways were similarly represented in E. bieneusi and other microsporidia, several exceptions stand out. The most important of these are genes relating to several core carbon metabolic pathways. All other microsporidian genomes contain complete pathways for glycolysis, including two subunits of the downstream pyruvate dehydrogenase enzyme (12 genes in total) and trehalose metabolism (four genes). The pentose phosphate pathway is also complete (five genes), with the exception of lactonase. The E. bieneusi genome, on the other hand, encodes genes for only one glycolytic enzyme (glycerol-3-phosphate dehydrogenase), one enzyme in the pentose phosphate pathway (glucose-6-phosphate dehydrogenase), and no enzymes whatsoever for trehalose metabolism (fig. 1, table 1, supplementary table S1, Supplementary Material online). Importantly, these two enzymes and only these two were found in both E. bieneusi surveys. Similarly, only 6 of the 20 genes involved in lipid metabolism in other microsporidia were found in E. bieneusi, and only 1 of 2 genes for the glycerol-3-phosphate shuttle system was found (table 1, supplementary table S1, Supplementary Material online). The overall pattern, therefore, demonstrates a clearly non-random pattern of gene content as the majority of functional categories represented in the E. bieneusi genome matches closely that found in other microsporidia, but genes related to energy generation and carbon metabolism are severely reduced in the E. bieneusi genome (fig. 1, table 1). This leads us to conclude that these genes and their corresponding activities have been lost.
FIG. 1.—
Core carbon metabolic pathways believed to form the backbone of energy metabolism in microsporidia. Enzymes are in red and metabolites in black. All the enzymes shown are present in the genomes of Encephalitozoon cuniculi, Antonospora locustae, Nosema ceranae, and Octosporea bayeri, with the single exception of lactonase (in gray) which has not been identified. In contrast, only two enzymes in any of these pathways are present in Enterocytozoon bieneusi (shown in red on black).
The only functional class of proteins outside of core carbon metabolism found to be similarly affected is the spliceosome: no genes relating to the removal of spliceosomal introns were identified, which is consistent with the original genome survey where no evidence of introns was found (Akiyoshi et al. 2009).
Discussion
The implications of our observations are striking: although there seems to be some residual capacity to modify small carbohydrates and lipids in E. bieneusi, complete pathways to produce energy from sugar are entirely absent (fig. 1). No other pathways are found in the genome that could perform this function, so this intracellular parasitic eukaryote lacks any apparent mechanism to make ATP from glucose. This in turn suggests that E. bieneusi must be directly dependent on its host for both sugars (which it cannot make) and ATP. Intriguingly, the E. cuniculi genome encodes several ATP transporters possibly derived by horizontal gene transfer (Richards et al. 2003; Tsaousis et al. 2008). A subset of these have been shown to recruit ATP from the host cytosol in E. cuniculi, whereas others recruit ATP from the cytosol to the relict mitochondrion (Tsaousis et al. 2008; Williams et al. 2008), and the same family of transporters is seen in E. bieneusi and other microsporidian genomes that have been analyzed (supplementary table S1, Supplementary Material online).
The existence of such transporters may have laid the foundations for the eventual loss of sugar metabolism altogether in E. bieneusi, but it is also possible that this trait is more widespread in microsporidia than we now imagine. This is because glycolysis generates not only ATP but also nicotinamide adenine dinucleotide hydrogenase (NADH). Without some mechanism to eliminate the accumulating reducing potential, this system is unsustainable. Trypanosomes solve this problem by shunting reducing potential to the mitochondrion using the glycerol-3-phosphate shuttle where it is metabolized by the mitochondrial AOX. Recently, a mitochondrial AOX has been identified in several microsporidia (Corradi et al. 2009; Williams et al. 2010). The AOX gene is not found in either genome survey of E. bieneusi, as expected, but it is also absent from the complete genome of E. cuniculi. Moreover, E. cuniculi has also been shown to have a disrupted glycerol-3-phosphate shuttle because it is lacking enzymes normally used to process the mitochondrial partner in the shuttle (Buri et al. 2006), and localization of this protein suggests that it is now cytosolic (Williams et al. 2008). It is difficult to see how glycolysis could be sustained to generate energy in E. cuniculi without either the glycerol-3-phosphate shuttle or a terminal oxidase such as AOX, leading to the intriguing possibility that E. cuniculi also does not use its glycolysis pathway for energy generation (Williams et al. 2010). An interesting alternative speculation is that glycolysis is primarily retained for the production of NADH in E. cuniculi (with ATP being a useful by-product).
Regardless of what the core function of glycolysis is in other microsporidia, it is clear from genomic data that E. bieneusi has not only lost this function but also been metabolically reduced beyond that observed for any other eukaryote to date, relying on host processes for even the most basic of metabolic activities. This extreme reliance on its host and corresponding lack of intrinsic biochemical pathways might also go some way to explain why this parasite is so refractory to cultivation.
Supplementary Material
Supplementary table S1 is available at Genome Biology and Evolution Online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by grants from the Canadian Institutes for Health Research (MOP-84265), the National Institutes of Health (NIH AI31788, R21 AI52792, and R21 AI064118), and the National Science Foundation (MCB-0135272). N.C. is a Scholar of the Canadian Institute for Advanced Research and is supported by a fellowship from the Swiss National Science Foundation (NSF) (PA00P3-124166). D.E. is supported by the Swiss NSF. P.J.K. is a Fellow of the Canadian Institute for Advanced Research and a Senior Scholar of the Michael Smith Foundation for Health Research. We acknowledge the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution for the use of data included in the Antonospora locustae Genome Project funded by NSF award number 0135272.
References
- Akiyoshi DE, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog. 2009;5:e1000261. doi: 10.1371/journal.ppat.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buri L, Williams BAP, Bursac D, Lithgow T, Keeling PJ. Progressive reduction of complexity in the mitosomal targeting machinery of microsporidian parasites. Proc Natl Acad Sci USA. 2006;103:15916–15920. doi: 10.1073/pnas.0604109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, et al. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Haag KL, Pombert JF, Ebert D, Keeling PJ. Draft genome sequence of the Daphnia pathogen Octosporea bayeri: insights into the gene content of a large microsporidian genome and a model for host-parasite interactions. Genome Biol. 2009;10:R106. doi: 10.1186/gb-2009-10-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci USA. 2007;104:19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milinkovitch MC, Helaers R, Depiereux E, Tzika AC, Gabaldon T. 2X genomes—depth does matter. Genome Biol. 2010;11:R16. doi: 10.1186/gb-2010-11-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassonova E, et al. Chromosomal composition of the genome in the monomorphic diplokaryotic microsporidium Paranosema grylli: analysis by two-dimensional pulsed-field gel electrophoresis. Folia Parasitol (Praha) 2005;52:145–157. doi: 10.14411/fp.2005.019. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Richards TA, Hirt RP, Williams BA, Embley TM. Horizontal gene transfer and the evolution of parasitic protozoa. Protist. 2003;154:17–32. doi: 10.1078/143446103764928468. [DOI] [PubMed] [Google Scholar]
- Tsaousis AD, et al. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Tovar J, Clark CG. Mitochondrion-derived organelles in protists and fungi. Int Rev Cytol. 2005;244:175–225. doi: 10.1016/S0074-7696(05)44005-X. [DOI] [PubMed] [Google Scholar]
- Williams BA, Cali A, Takvorian PM, Keeling PJ. Distinct localization patterns of two putative mitochondrial proteins in the microsporidian Encephalitozoon cuniculi. J Eukaryot Microbiol. 2008;55:131–133. doi: 10.1111/j.1550-7408.2008.00315.x. [DOI] [PubMed] [Google Scholar]
- Williams BA, et al. A broad distribution of the alternative oxidase in microsporidian parasites. PLoS Pathog. 2010;6:e1000761. doi: 10.1371/journal.ppat.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BAP, Keeling PJ. Cryptic organelles in parasitic protists and fungi. Adv Parasitol. 2003;54:9–67. doi: 10.1016/s0065-308x(03)54001-5. [DOI] [PubMed] [Google Scholar]