Abstract
A genome's ability to produce two separate sexually dimorphic phenotypes is an intriguing biological mystery. Microarray-based studies of a handful of model systems suggest that much of the mystery can be explained by sex-biased gene expression evolved in response to sexually antagonistic selection. We present the first whole-genome study of sex-biased expression in the red flour beetle, Tribolium castaneum. Tribolium is a model for the largest eukaryotic order, Coleoptera, and we show that in whole-body adults, ∼20% of the transcriptome is differentially regulated between the sexes. Among T. castaneum, Drosophila melanogaster, and Anopheles gambiae, we identify 416 1:1:1 orthologs with conserved sex-biased expression. Overrepresented functional categories among sex-biased genes are primarily those involved in gamete production and development. The genomic distribution of sex-biased genes in T. castaneum is distinctly nonrandom, with the strongest deficit of male-biased genes on the X chromosome (9 of 793) of any species studied to date. Tribolium also shows a significant enrichment of X-linked female-biased genes (408 of 793). Our analyses suggest that the extensive female bias of Tribolium X chromosome gene expression is due to hyperexpression of X-linked genes in both males and females. We propose that the overexpression of X chromosomes in females is an evolutionary side effect of the need to dosage compensate in males and that mechanisms to reduce female X chromosome gene expression to autosomal levels are sufficient but imperfect.
Keywords: dosage compensation, sex chromosomes, microarrays, Tribolium
Introduction
The genomes of organisms with separate sexes constantly experience selection to maintain two distinct, and potentially antagonistic, gender-specific phenotypes. In some cases, a beneficial phenotype for one sex will pose a fitness cost to the other sex. Because the difference in gene composition between sexes in species with chromosomal sex determination is typically limited to a few genes on the Y (or W) chromosome, mitigation of these trade-offs is accomplished primarily via sex-specific gene regulation (Rinn and Snyder 2005; Ellegren and Parsch 2007). Genes with differential expression between sexes are termed “sex-biased genes.” The relatively few species surveyed for sex-biased genes show that the proportion of the genome that is more highly expressed in males (male biased) or females (female biased) is often extensive but can vary widely among closely related species (Sturgill et al. 2007; Zhang et al. 2007) and tissues within a species (Yang et al. 2006).
In addition to demonstrating that much of the genome is regulated differently between sexes, genome-wide surveys show that the distribution of sex-biased genes is not consistent across the genome. In particular, the proportion of sex-biased genes on the shared sex chromosome (X or Z) often differs significantly from what is expected based on the chromosome size and genome-wide proportion of sex-biased genes. For instance, the X chromosome of fruit flies and nematodes has fewer than expected male-biased genes (Parisi et al. 2003; Ranz et al. 2003; Reinke et al. 2004), whereas the X chromosome of human (Lercher et al. 2003) and mouse (Wang et al. 2001; Yang et al. 2006) appears to be enriched for male-biased genes (but see Khil et al. 2004). In the ZW sex determination systems of chicken (Kaiser and Ellegren 2006; Storchova and Divina 2006) and silkworm (Arunkumar et al. 2009), the deficit of female-biased genes found on the Z chromosome is similar to the pattern in fruit flies and nematodes—a deficit of heterogametic sex-biased genes on the shared sex chromosome—but the underlying causes are different.
Several hypotheses have been put forward to explain the peculiar genomic distribution of sex-biased genes. First, Rice (1984) proposed that hemizygosity could explain inequitable distributions of sex-biased genes under a model of sexually antagonistic selection. In XY systems, recessive mutations that benefit males are able to increase in frequency on the X chromosome until balanced by the fitness cost of homozygosity in females. Subsequent modifiers that restrict expression to the appropriate sex may result in fixation of the male beneficial mutation. Assuming optimization in this scenario is achieved by higher expression of the original recessive mutation, theory predicts a net excess of male-biased genes (and/or deficit of female-biased genes) on the X. Alternatively, X-linked mutations that are dominant are governed by their increased residence time in females, such that 2/3 of the alleles are available to selection in females. In this case, selection for expression modifiers is ultimately expected to yield excess female-biased genes (and/or deficit of male-biased genes) on the X. Although there is evidence for sexual antagonism (Rice 1987), it remains unclear whether this is a general explanation for the distribution of sex-biased genes among chromosomes. Unique, a priori predictions for the distribution of X-linked sex-biased genes are difficult based on this hypothesis because the expectations depend on the dominance of new mutations, something that is rarely known.
A second potential explanation for observed deficits of X-linked male-biased genes is avoidance of meiotic sex chromosome inactivation (MSCI) during spermatogenesis (Hense et al. 2007; Potrzebowski et al. 2008). Under this hypothesis, translocation of testes-expressed genes from the X to an autosome may be favored by selection if it is beneficial for the gene to be expressed during a time when it would otherwise be inactivated during male meiosis. The hypothesis predicts a deficit of male-biased X-linked genes expressed during meiotic and postmeiotic stages of spermatogenesis, a pattern consistent with observations in mouse (Khil et al. 2004) and Drosophila melanogaster (Vibranovski, Lopes, et al. 2009). Also consistent with the MSCI hypothesis is the bias of X to autosome retrotranspositions that often maintain (or acquire) testes expression (Betran et al. 2002; Emerson et al. 2004), and in some mammals, genes retrotransposed from the X to autosomes may functionally compensate for their X-linked parental genes (Potrzebowski et al. 2008). The generality of the MSCI hypothesis is challenged by the paucity of X-linked male-biased genes in tissues that do not experience X inactivation (Parisi et al. 2003; Sturgill et al. 2007), and the pattern of X to autosome movement for duplicative events other than retrotransposition is unclear (cf. Vibranovski, Zhang, and Long 2009; Meisel et al. 2010). Furthermore, prior to the initiation of meiosis, the mouse X is enriched for genes expressed in spermatogonia, a pattern initially ascribed to the sexual antagonism hypothesis summarized above (Wang et al. 2001; Khil et al. 2004).
A third hypothesis proposes that the observed paucity of male-biased genes on the X chromosome is a limitation imposed by dosage compensation (Vicoso and Charlesworth 2009). When dosage compensation is attained by hypertranscription of the X chromosome, all else being equal, it may be more difficult to increase transcription of an X-linked gene in males. A unique prediction of this hypothesis is that the magnitude of expression of male-biased genes should be negatively correlated with the frequency of occurrence on the X chromosome (i.e., fewer highly expressed male-biased genes). The available data for sex-biased genes in Drosophila fit the predictions. However, counter to the expectation, nonbiased genes with high expression are actually overrepresented on the fly X chromosome and mammals do not show the same paucity of X-linked male bias in spite of X hypertranscription (Vicoso and Charlesworth 2009).
Finally, overrepresentation of female-biased genes on the X or male-biased genes on the Z could simply reflect incomplete compensation for the 2:1 ratio of gene dose (XX female vs. XY male or ZZ male vs. ZW female). This does not seem to be the case in most of the well-studied XY systems because although they solve the problem differently, Drosophila, Caenorhabditis elegans, and mammals (except platypus; Deakin et al. 2008), all dosage compensate such that the average expression level of X and autosomes is balanced in males and females (X/A = 1), and average X chromosome expression is the same between sexes (Xfemale/Xmale = 1) (Straub and Becker 2007). Additionally, the recent discovery of incomplete dosage compensation in a fish with XY sex determination may be due to limited divergence between the nascent sex chromosomes (Leder et al. 2010). Unlike XY systems, the dramatic overrepresentation of male-biased genes on chicken, zebra finch, and silk moth Z chromosomes suggests that a lack of chromosome-wide dosage compensation is common among ZW sex determination systems (Ellegren et al. 2007; Itoh et al. 2007; Zha et al. 2009). Results from the ZW taxa challenge the decades old paradigm that differentiation of the sex chromosomes into X and Y (or Z and W) must be accompanied by a mechanism to modify gene expression levels to compensate for the gene dosage imbalance between sexes (Charlesworth 1978). Interestingly, this paradigm has been so widely accepted that initial descriptions of sex bias in chicken concluded that overrepresentation of male-biased Z-linked genes was due to sexually antagonistic selection rather than lack of global dosage compensation (Kaiser and Ellegren 2006; Storchova and Divina 2006). Because not all Z-linked genes are sex biased, it has also been argued that either 1) dosage compensation occurs locally (i.e., on a gene-by-gene basis) (Mank and Ellegren 2009) or 2) the Z is dosage compensated to the extent necessary for maintenance of critical biochemical pathways and the disproportionate number of male-biased genes is a product of stronger sexual selection on males, male-biased mutation, and increased residence of Z chromosomes in males (Naurin et al. 2010).
Following, we report the first genome-wide survey of transcriptional differences between males and females of Tribolium castaneum, the red flour beetle. Tribolium castaneum is a world-wide pest of stored grains and serves as a model organism for the most speciose eukaryotic order, Coleoptera. We hybridize samples from whole, virgin, adult males and females between 48 and 168 h old to custom 385 k NimbleGen microarrays containing probes for >98% of known and predicted expressed sequences. Comparing male and female expression, we describe the proportions of sex-biased genes (including splice variants) and their functional representation, genomic distribution, and conservation among T. castaneum, D. melanogaster, and Anopheles gambiae. Our analyses reveal a significant overrepresentation of female-biased genes on the X chromosome, a result of chromosome-wide hyperexpression of the X in females. We propose that the gene expression imbalance in females represents a novel resolution to the antagonistic dosage compensation requirements of males and females.
Materials and Methods
Microarray and Experimental Design
In collaboration with Roche/NimbleGen Inc. we designed a custom 385 k microarray to target each of the genes in the consensus set identified by the Tribolium genome sequencing consortium (Richards et al. 2008). In total, 80% (56,919/71,259) of T. castaneum exons are present on the array, and multiple (if not all) exons are present for 98% (16,130/16,434) of the genes. An additional 304 regions (custom ranges) not annotated as coding sequences are also represented on the array based on evidence for expression from tiling array data generated in conjunction with the genome project (Richards et al. 2008). Some of these custom ranges are likely to be unannotated exons, whereas other may be noncoding RNAs. Ninety-six percent of exons and custom ranges are queried by three, 60-mer probes with the remainder having only 1 or 2 probes per sequence for a total of 167,538 unique probes. Each probe is printed on the array in duplicate.
Ga-2 strain T. castaneum (genome reference strain) were originally provided by Dr Richard W. Beeman, United States Department of Agriculture–Agricultural Research Service. Beetles were reared on standard medium (95% organic whole-wheat flour with 5% Brewers yeast) at 29 °C. Each sex was replicated by four samples of 20 adult 2- to 7-day-old virgin beetles. One replicate of each sex was split and hybridized to separate arrays to provide an estimate of technical reproducibility. Groups of adults for each sample were placed directly into lysis solution and homogenized in microcentrifuge tubes by pestle and syringe. RNA was extracted from resulting homogenate using the Qiagen Rneasy Kit following the manufacturer’s instructions. All extractions were performed on the same day.
RNA samples were initially checked for quality with a Nanodrop ND 1000 Spectrophotometer. Total RNA samples were converted to double-stranded cDNA using the Invitrogen Superscript Double-Stranded cDNA Synthesis Kit. Quality of cDNA was assessed by Nanodrop and gel electrophoresis. We submitted >2.5 μg of cDNA for each sample to Roche/Nimblegen (NimbleGen Systems), who subsequently performed additional quality control analyses, Cy3-labeling, array hybridizations, data acquisition, and normalization.
MIAME-compliant data sets are provided in the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/) and can be accessed through GEO series accession number GSE18087.
Computational Methods
Average normalized log2 expression intensity values were exported for each exon or custom range with ArrayStar 3.0 (DNASTAR Inc). For gene-based analyses, we averaged the normalized expression values across exons. Significant female- and male-biased genes were identified at false discovery rate (FDR) ≤ 0.01 (α = 0.01721235) using the optimal discovery procedure (Storey et al. 2007) as implemented in EDGE (Leek et al. 2006). We used Blast2GO (Conesa et al. 2005) to obtain gene ontology (GO) terms and implement Fisher’s exact test with multiple testing correction to assess overrepresentation of sex-biased genes relative to all annotated genes in the Tcas_3 reference set.
To identify instances of alternative splicing between sexes, we tested for a sex-by-exon interaction with the program R (http://www.R-project.org), using the following linear model: Yijk = μ + ai + bj + abij + ϵijk, where Yijk is the expression intensity for sex i, exon j, and replicate k. μ is the overall mean of expression intensity for that gene, a is the effect of sex, b is the effect of exon, ab is the effect of the interaction between sex and exon, and ϵ is the error. We used the program Q-value (Storey and Tibshirani 2003) to evaluate P values with FDR ≤ 0.01. Expected values for the number of alternatively spliced genes per chromosome were calculated as the product of the genome-wide proportion of alternatively spliced genes and the number of genes on a particular chromosome.
Similarly, expected values for the number of sex-biased genes on each chromosome were calculated as the product of the number of genes on a chromosome and the genome-wide proportion of genes that were significantly sex biased at FDR ≤ 0.01. Expected values for the number of female- or male-biased genes on each chromosome were calculated as the product of the number of genes on a chromosome and the genome-wide proportion of female- or male-biased genes at FDR ≤ 0.01.
To assess universal dosage compensation of the X chromosome in males, we compared expression of ribosomal proteins in males and females. Fifty-two autosomal and four X-linked ribosomal proteins were retrieved from GenBank annotations. Two of the autosomal genes had expression patterns that were clearly outliers. These proteins seem unlikely to be involved in the ribosomal complex and were excluded from the analysis.
To identify homologous genes with a similar pattern of sex-biased expression between the beetle, fruit fly, and mosquito, we used the D. melanogaster composite data set from the Sex Bias Database (Gnad and Parsch 2006) and the A. gambiae data set from Marinotti et al. (2006) as downloaded from VectorBase (Lawson et al. 2007). All data sets were screened for sex-biased expression at FDR ≤ 0.01, and peptide sequences were assigned using Tcas_3 from BeetleBase (Wang et al. 2007), Dmel_r5.21 from FlyBase (Tweedie et al. 2009), and AgamP3.5 from VectorBase (Lawson et al. 2007). FASTA version 35 (Pearson and Lipman 1988) was used to search for homology at an E value cutoff of 0.0001.
Data Deposition
GEO series accession number GSE18087.
Results
Analysis of four hybridizations for each sex and one technical replicate for each sex indicates good reproducibility among experiments. The correlation among duplicate probes within an array is 0.99. The correlation between technical replicates is 0.99 for males and 0.98 for females. The average correlation among biological replicates among male samples is 0.95 and 0.87 among females. Due to the high reproducibility of expression estimates within sexes, we are able to detect significant expression differences at fold changes as low as 1.08. The mean and median fold change among significantly sex-biased genes were 2.53 and 1.74, respectively.
Overall Sex Bias
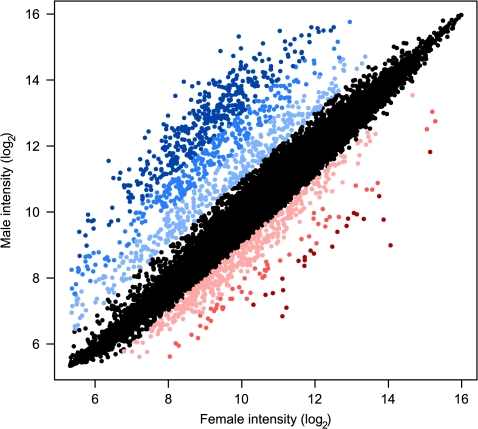
We identified 3,209 (∼20%) differentially transcribed genes between adult male and female T. castaneum (FDR ≤ 0.01). Of these, a slight majority (58%; 1,870/3,209) are more highly expressed in females. However, at a threshold of 2-fold difference, 75% (1,023/1,359) are male-biased, and as the degree of expression bias increases, the proportion of male-biased genes increases rapidly, such that at 4-fold difference, 90% (715/795) are male-biased and 95% (418/442) at 8-fold (fig. 1). Reassuringly, the most significant sex-biased GO terms include gene families clearly involved in sex-specific reproductive functions (e.g., oogenesis and flagellar motililty). A complete list of sex-biased genes with GO annotations is given in supplementary table S1 (Supplementary Material online).
FIG. 1.—
Genome-wide sex-biased expression. Colors indicate higher expression in females (red) or males (blue). Light shades indicate ≥2-fold, medium shades indicate ≥4-fold, and darkest shades indicate ≥8-fold difference between sexes. Black indicates <2-fold difference in expression.
In addition to whole genes that are sex biased, we identified 265 genes that have significant sex-by-exon interactions, indicating that they are differentially spliced between the sexes (FDR ≤ 0.01). These genes have a variety of functions, such as protein binding, ion binding, nucleic acid binding, and transcription factor activity (supplementary table S2, Supplementary Material online). Examining the chromosomal distribution of these genes shows more genes with sex-specific expression of alternative transcripts on the X chromosome than expected (23 observed, 13 expected, chi-square = 7.91, P < 0.005); no significant deviations are observed on autosomes. These 265 genes represent 2% of the genes with multiple exons in T. castaneum (265/11,382) compared with 85% (9,694/11,382) of all genes with multiple exons that have significantly different expression among exons but no significant sex-by-exon interaction (FDR ≤ 0.01).
Evolutionary Conservation of Expression Bias
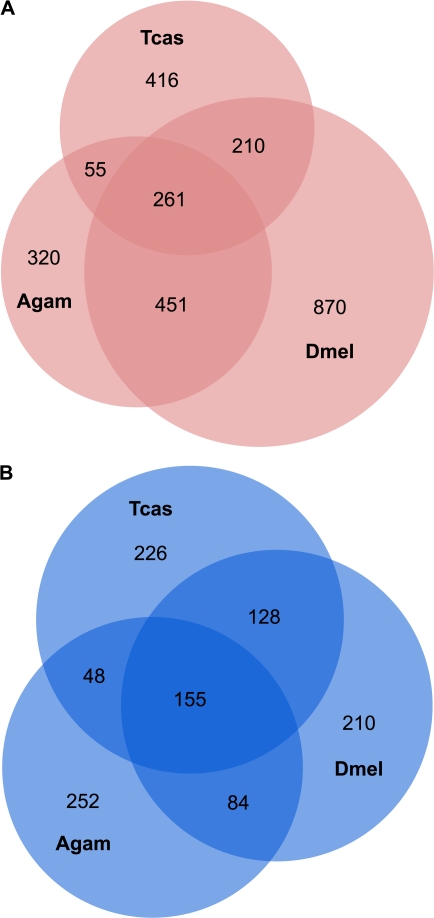
Among T. castaneum, A. gambiae, and D. melanogaster, we identified 2,583 orthologs with female-biased expression and 1,103 with male-biased expression in at least one species. There are 977 female-biased and 415 male-biased genes with conserved sex-biased expression between at least two of the taxa compared (fig. 2). Except for female-biased genes shared between A. gambiae and D. melanogaster, the largest categories for both male- and female-biased genes are those that have unique expression in each species. However, there are still many genes (261 female-biased genes and 155 male-biased genes) that have conserved sex-biased expression in all three species. Analysis of GO categories reveals that while some of the genes in this core set are involved in spermatogenesis or oogenesis, the overrepresented GO terms indicate a role in basic cellular and metabolic functions. The complete list of orthologous sex-biased genes and associated GO terms are provided in supplementary table S3 (Supplementary Material online).
FIG. 2.—
Conservation of sex-biased genes across Tribolium castaneum, Drosophila melanogaster, and Anopheles gambiae. Orthologous genes with conserved female-biased (A) or male-biased (B) expression. Numbers in regions of overlap reflect 1:1 or 1:1:1 orthologs. Genes shown to be unique to a species are homologous to a nonbiased gene in at least one of the other two species. The number of sex-biased genes in each species where no homology was found in the other taxa: female-biased Tcas = 431, Dmel = 248, Agam = 104; male-biased: Tcas = 467, Dmel = 632, Agam = 93.
Chromosomal Distribution
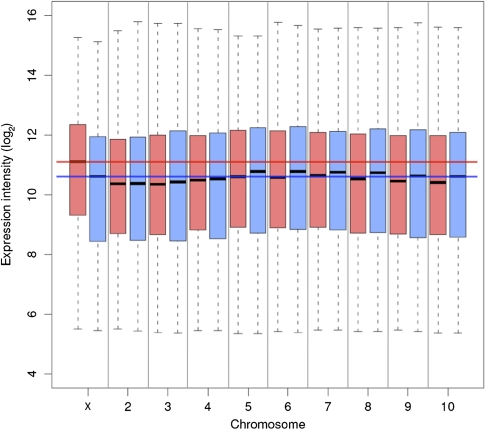
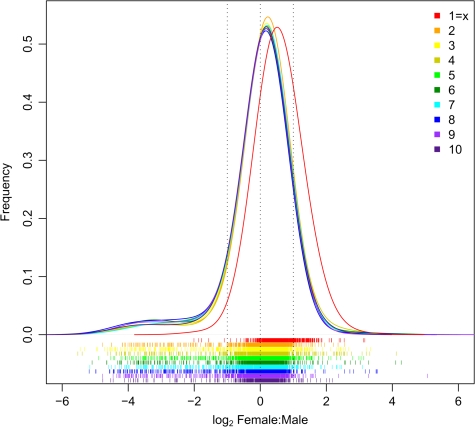
Comparison of average expression levels between sexes and across chromosomes indicates that the most transcriptionally active chromosome is the X in females (fig. 3). Unlike any of the previously studied XY systems, where chromosome-wide dosage compensation results in Xmale/Xfemale = Amale/Afemale = 1; in beetle, there is a clear shift toward higher expression of the X in females such that while Amale/Afemale ≈ 1, Xmale/Xfemale = 0.79 (fig. 4). The imbalance of X chromosome expression is further born out by the X/autosome ratios within each sex. On average, males express X-linked genes at approximately the same level as autosomal genes (median Xmale/Amale = 1.0; mean = 0.83), whereas females express X-linked genes at a higher level than autosomal genes (median Xfemale/Afemale = 1.53; mean X/A = 1.12).
FIG. 3.—
Average expression of genes on each chromosome in females (pink) and males (blue). Median (black lines), ±25th–75th quartile (box), and range of chromosome-wide hybridization intensities. Red (female) and blue (male) lines highlight the median expression values of the X chromosome in each sex relative to the autosomes.
FIG. 4.—
The female/male gene expression ratio for each chromosome illustrating the female-biased shift in X chromosome expression relative to autosomes. Higher expression in females is indicated by positive values on the x axis. Horizontal bars below the histogram mark female/male expression ratios for each gene.
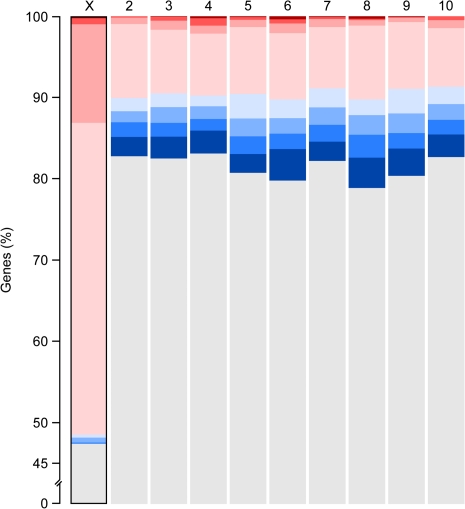
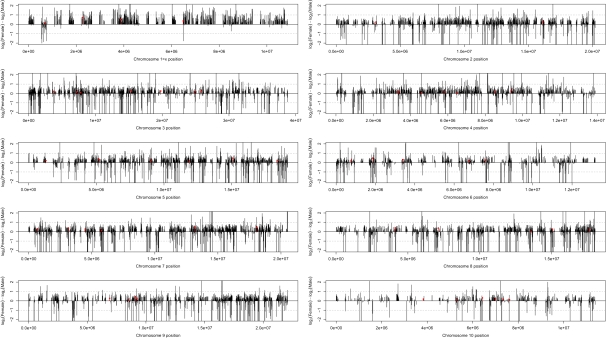
Overall, 53% (417/793) of genes on the X chromosome are significantly sex biased (FDR < 0.01; fig. 5). This is a 2.6-fold excess over the expectation from the genome-wide proportion of sex-biased genes and the total number of genes on the X chromosome (417 observed, 162 expected, chi-square = 401.61, P < 0.0001). Among the X-linked sex-biased genes, there is a significant excess of female-biased genes (408 observed, 243 expected, chi-square = 112.66, P < 0.0001) and a significant paucity of male-biased genes (9 observed, 174 expected, chi-square = 156.81, P < 0.0001). Furthermore, the male-biased genes are nonrandomly distributed, with 8 of the 9 genes residing within a 240-kb window (fig. 6). The remarkable bias and spatial distribution found on the X chromosome stands in stark contrast to the autosomes which each contain ∼20% sex-biased genes (range 17–21%) that are split roughly equally between male and female bias (figs. 5 and 6).
FIG. 5.—
Proportion of genes with nonbiased (gray), female-biased (red), and male-biased (blue) expression on each chromosome. Sex-biased expression with the lightest color is significant at FDR < 0.01 but <2-fold enrichment, medium is ≥2-fold, darker shading is ≥4-fold, and darkest is ≥8-fold. The X chromosome (outlined) is the only chromosome that differs significantly from the expected distribution of sex-biased genes.
FIG. 6.—
Spatial distribution of sex-biased gene expression across the X chromosome and autosomes. Each bar represents a single gene. Expression bias is evaluated as log2(female expression) − log2(male expression) such that bars above and below the centerline are female or male biased, respectively. Red bars indicate locations of the ribosomal protein genes used in regression analysis (see text). Dotted lines represent 2-fold expression difference.
Discussion
The overall proportion of genes exhibiting sex-biased expression in T. castaneum (∼20%) is consistent with what has been seen in comparable studies of other taxa. A study of whole-body adults for seven Drosophila species shows the overall proportion of sex-biased genes to be between 12% and 32% (Zhang et al. 2007). Unlike Tribolium, most Drosophila species tested (5 of 7) have more male-biased genes at FDR ≤ 0.01. The difference is due in part to the unique bias of the Tribolium X discussed below but because the pattern of increased proportion of male-biased genes with increasing fold difference is the same in the two species, statistical differences in the power to detect sex bias at low fold change may also contribute. It is worth noting here that even comparisons across other whole-body expression studies are complicated by the inability to separate the number of sex-biased genes from the statistical power to detect them. For example, using very high replication, Ayroles et al. (2009) found that 88% of D. melanogaster genes showed statistically significant sex-biased expression; a much higher proportion than previous reports for whole flies.
That 20% of genes in whole-body adult Tribolium are sex biased should be viewed as a conservative estimate for other reasons as well. First, Yang et al. (2006) showed that as the number of tissues that are individually analyzed increases, the number of sex-biased genes increases. Because we investigated whole-body adults, our study will miss differences among tissues or in the number of tissues where a gene is expressed in each sex. Second, sex-biased expression at other life stages and or under different physiological conditions may involve other genes. For example, like Tribolium, A. gambiae shows excess female-biased genes at low fold difference (Hahn and Lanzaro 2005; Marinotti et al. 2006); but in contrast to Tribolium and Drosophila, the proportion of female-biased genes increases with fold difference. The reason for overall female bias in the A. gambiae genome may be explained by their unique biology wherein only the females take blood meals in preparation to reproduce. Marinotti et al. (2005) compared male A. gambiae with nonblood-fed females and blood-fed females and found that blood feeding caused an increase in the number of sex-biased genes as females switched their metabolism and began egg production. Additionally, in Daphnia pulex, mature females have a higher proportion of female-biased genes than juvenile females (Eads et al. 2007), and D. melanogaster reared under better nutritional conditions show exaggerated expression bias (Wyman et al. 2010).
Evolutionary Conservation of Expression Bias
In spite of the statistical and sampling considerations above, we were able to identify a substantial number of genes with conserved sex-biased expression across D. melanogaster, A. gambiae, and T. castaneum. Genes with sex-biased expression often evolve more rapidly than nonbiased genes, both in sequence and expression (reviewed in Ellegren and Parsch 2007; Zhang et al. 2007). In particular, several studies have shown that male-biased genes are among the most rapidly diverging proteins in the genome (e.g., Zhang et al. 2004; Baines et al. 2008). It is unclear whether the overall paucity of male-biased orthologs relative to female-biased orthologs across our data set (compare fig. 2A vs 2B) is due to protein divergence or gene loss, but the absence is particularly interesting in Drosophila and Tribolium where the total number of male-biased genes in each genome is similar to, or greater than, female-biased genes. Rapid turnover of male-biased genes would be consistent with interspecific Drosophila comparisons where male-biased genes have higher rates of turnover and sequence divergence than female-biased genes or nonbiased genes (Zhang et al. 2007; but see Jiang and Machado 2009). In any case, since there are many more male-biased genes in each species that lack orthologs (and consequently cannot have conserved expression), limiting our analyses to orthologous genes artificially inflates the proportion identified as having conserved expression and makes the proportions in each sex appear the same (38%) when in fact conservation is lower in males.
Female Bias of the Tribolium X: Chromosome-Wide Process or Gene-By-Gene Selection?
Among all X-linked genes, 51% (408/793) are significantly female biased, whereas only 1% (9/793) are male biased (fig. 5). Though the deficit of male-biased genes on the X chromosome is in the same direction as observed in several Drosophila species (Zhang et al. 2007), C. elegans (Reinke et al. 2004), and meiotic mouse testes (Khil et al. 2004), Tribolium is far more extreme than any of these reports. Several lines of evidence suggest that extensive feminization of the Tribolium X chromosome is predominantly due to imperfect resolution of the chromosome-wide antagonistic dosage compensation requirements of males and females.
First, the distribution of sex-biased genes across the X chromosome is difficult to explain by hypotheses that require evolution in response to gene-by-gene selection pressures (e.g., sexually antagonistic selection or escape of meiotic X inactivation). All but one of the nine X-linked male-biased genes reside in a 240-kb region, and with few exceptions, the rest of the chromosome exhibits varying degrees of female bias (fig. 6). Although gene-by-gene processes probably contribute to unique sex bias in mammal, fly, and worm X chromosomes (Reinke et al. 2000; Wang et al. 2001; Parisi et al. 2003; Khil et al. 2004; Hense et al. 2007; Sturgill et al. 2007), the end result is a more heterogeneous spatial distribution than the male-biased window we observe in Tribolium.
The odd distribution of sex bias on the Tribolium X is puzzling. One possible explanation is that the region is pseudoautosomal (i.e., also present on the Y), and the rest of the X lacks complete dosage compensation. Alternatively, the region may simply experience regional dosage compensation that is more finely tuned than the rest of the X. None of the annotated genes in the window have conspicuously male-related functions (i.e., expressed in gonads or reproduction) which would suggest a history of strong selection to maintain male-biased expression (supplementary table S2, Supplementary Material online), but additional studies are required to distinguish between these alternative hypotheses.
The average expression level of X and autosomes also suggest that feminization of the X is due to broad scale dosage compensation inequities rather than gene-by-gene mechanisms (figs. 3 and 4). Although the overall pattern in Tribolium is consistent with recently reported ZW systems that lack chromosome-wide dosage compensation (Ellegren et al. 2007; Itoh et al. 2007; Zha et al. 2009), the underlying mechanism appears to be different. In the bird and moth examples, extensive Z-linked male bias arises because the hemizygous sex (female) fails to increase transcription of the Z to autosomal levels. However, in Tribolium, the hemizygous sex (male) does appear to increase X expression to autosomal levels, and the expression imbalance arises because the homogametic sex (female) also increases X expression.
Given that the X chromosome in male Tribolium appears to be dosage compensated, we further explored the Vicoso and Charlesworth (2009) hypothesis that dosage compensation may limit male-bias expression on the X (although it would still not explain excess female-biased genes). We examined the distribution of sex-biased genes on the X chromosome at low, medium, and high expression to see if the degree of male bias differs according to the absolute magnitude of gene expression. If hypertranscription of the X is limiting males’ ability to further increase expression, we expect male-biased genes to be underrepresented in the high expression category; however, the nine X-linked genes with significant male-biased expression are evenly distributed across high, medium, and low expression intensity. With only nine genes having significant male-biased expression on the X chromosome, firm conclusions await additional tissue-specific experiments. So, although our present results suggest that dosage compensation plays a role in the extreme deficit of X-linked male-biased genes in Tribolium, it is not for the reason proposed by Vicoso and Charlesworth (2009).
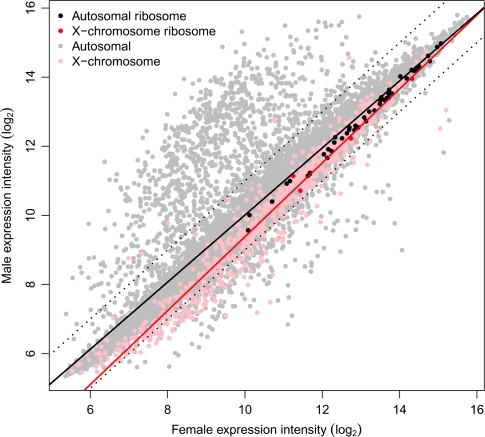
To further explore whether the observed deficit of male-biased genes and enrichment of female-biased genes on the X chromosome is due to a chromosome-wide effect, we followed the approach of Parisi et al. (2003) who examined the expression of genes that encode ribosomal proteins. Ribosomal proteins provide a good test set because they are present on both the X and autosomes, and they are expected to maintain 1:1 stoichiometry. If there is a chromosome-wide regulatory effect in either sex, we expect the relationship between X-linked ribosomal proteins to differ from that of the autosomal proteins because of the imbalance in gene dose between sexes. We identified 54 ribosomal protein genes in the current Tribolium annotation (see supplementary table S4, Supplementary Material online); 50 of them are on autosomes and 4 are on the X chromosome. The small sample of X-linked genes limits the strength of conclusions based on this analysis, but the expression of X and autosomal ribosomal proteins in males and females are similar (fig. 7). This suggests that X-linked and autosomal gene expression are balanced for at least some genes. However, we believe this compensation is localized, or gene specific, rather than chromosome-wide because: 1) one of the ribosomal protein genes is in the male-biased region, 2) a second one is adjacent to the outlying male-biased gene, and 3) the remaining two are among the most female biased of the ribosomal proteins (fig. 6). Furthermore, if we consider expression of all genes in a similar way to the ribosomal protein subset, X-linked genes are clearly skewed toward high expression in females compared with autosomes (fig. 7).
FIG. 7.—
Regression of X-linked and autosomal genes highlighting ribosomal protein gene expression. The solid lines represent the regression of all autosomal (black) and all X-linked (red) expression between sexes. Dotted lines represent 2-fold expression difference.
Do Female Beetles Lack “Counter–Compensation” for Evolution of X Hypertranscription in Males?
Although dosage alteration of one or a few genes can be buffered through biochemical pathways with no or limited consequences to an organism’s phenotype; simultaneously altering expression of many genes (e.g., large segmental duplications or deletions or aneuploidy) is typically lethal (Oliver 2007). So how do Tribolium females survive with what appears to be functional aneuploidy of the X chromosome? Although we cannot answer this question conclusively with our current data, evidence from other XY systems is instructive. In humans, mice, Drosophila, and C. elegans males increase expression of their single X chromosome to the same level as autosomes. This hypertranscription of the X in males poses the same potential “overcompensation” problem for females in each of these species that we see in Tribolium; if the dosage compensation machinery is not sex specific upregulating the X in females results in expression ratios of Xfemale/Afemale > 1. The other XY systems each resolve this antagonism differently. In mammals, females inactivate one X, whereas leaving the other hyperexpressed (Lin et al. 2007). In C. elegans, hermaphrodites suppress expression of both X chromosomes to bring expression in line with autosomes (Gupta et al. 2006). In Drosophila, the dosage compensation complex (DCC) is not assembled in females, so they avoid the necessity of counter–compensation. However, Zhang and Oliver (2010) recently demonstrated that the chromatin structure of the D. melanogaster X is different from autosomes in both males and females. Combined with their earlier reports of slightly elevated female X chromosome expression (Gupta et al. 2006; Sturgill et al. 2007), these recent results suggest that the evolution of male X chromosome hypertranscription may collaterally increase female transcription even in the absence of the DCC.
Our results suggest that Tribolium has followed a similar trajectory in the evolution of dosage compensation to that of other XY systems. However, female beetles somehow avoid the necessity of complete counter–compensation for X chromosome hypertranscription. Without more information about the molecular mechanisms responsible for upregulation of the X chromosome, it is impossible to know whether the current status in Tribolium is an intermediate stage in the evolution of complete dosage compensation in both sexes or a stable alternative (i.e., females do compensate to the degree that it is physiologically necessary). Given that the sex chromosomes in Tribolium are probably quite old, perhaps predating the origin of the superfamily Tenebrionoidea at >100 MYA (Sokoloff 1972; Hunt et al. 2007), the later alternative seems more probable to us.
Because so little is known about dosage compensation in Tribolium, we used TBlastN (Altschul et al. 1997) to search Beetlebase (Wang et al. 2007) for homologs of the dosage compensation systems of other well-studied systems (Drosophila, C. elegans, and mammals). We only found clear homologs for three genes of the Drosophila DCC (mle, msl3, and mof). One of these, msl3, has two copies in Tribolium (male-specific lethal like 1 on LG10 = TC011005 and male-specific lethal like 1 on LG6 = TC015251). Homologs of the mle, msl3, and mof genes are responsible for histone acetyltransferase activity in a wide range of higher eukaryotes, including humans (Smith et al. 2005). Their role in male dosage compensation is so far unique to Drosophila and involves male-specific translation of msl2 to localize the DCC to the male X chromosome. As far as we can tell, msl2 is absent in Tribolium, and our expression data show that each of the DCC homologs present in Tribolium are expressed in both sexes. Consequently, although our data provide evidence for male dosage compensation in Tribolium, the precise genetic mechanism is unclear and will require additional experiments to elucidate.
Conclusions
In addition to being the first description of sex-biased expression in the largest eukaryotic order and identifying conservation of sex-biased genes across three insect species, our study provides the first example of an XY sex determination system where there is an imbalance between X and autosome gene expression that is not due to gene dose. Tribolium males dosage compensate their X chromosomes so that X:Autosome gene expression ratios are near 1:1; but the X is also upregulated in females, making them functionally XXX(X):AA. We propose that the overexpression of X chromosomes in females is an evolutionary side effect of the need to dosage compensate in males and that this may reflect a stable equilibrium wherein females have struck a sufficient, though imperfect, balance between X and autosomal gene expression. If our hypothesis is correct, it may offer further evidence that males are more sensitive to dosage imbalance than females; an idea proposed by Mank (2009) in response to the dichotomy between the presence or absence of dosage compensation between XY and ZW systems.
Supplementary Material
Supplementary tables S1–S4 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
We thank Ester Betran, Michael Wade, Pawel Michalak, and anonymous reviewers for helpful insights and comments on the manuscript and analyses. Terence Murphey and Hee Shin Kim provided information to help resolved assembly, annotation, and version inconsistencies between National Center for Biotechnology Information and Beetlebase. Sue Olsen and David Guffey (Roche/NimbleGen) provided technical support for the array design and quality control. This work was supported by National Institutes of Health (grant number 2R01GM065414-05A1) and The University of Texas at Arlington startup funding to J.P.D.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunkumar KP, Mita K, Nagaraju J. The silkworm Z chromosome is enriched in testis-specific genes. Genetics. 2009;182:493–501. doi: 10.1534/genetics.108.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betran E, Thornton K, Long M. Retroposed new genes out of the X in Drosophila. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y-chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Graves JAM. The status of dosage compensation in the multiple X chromosomes of the Platypus. PLoS Genet. 2008;4:e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads BD, Colbourne JK, Bohuski E, Andrews J. Profiling sex-biased gene expression during parthenogenetic reproduction in Daphnia pulex. BMC Genomics. 2007;8:464. doi: 10.1186/1471-2164-8-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betran E, Long M. Extensive gene traffic on the mammalian X chromosome. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:2577–2579. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Gupta V, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Lanzaro GC. Female-biased gene expression in the malaria mosquito Anopheles gambiae. Curr Biol. 2005;15:R192–193. doi: 10.1016/j.cub.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Machado CA. Evolution of sex-dependent gene expression in three recently diverged species of Drosophila. Genetics. 2009;183:1175–1185. doi: 10.1534/genetics.109.105775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Ellegren H. Nonrandom distribution of genes with sex-biased expression in the chicken genome. Evolution. 2006;60:1945–1951. [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- Lawson D, et al. VectorBase: a home for invertebrate vectors of human pathogens. Nucleic Acids Res. 2007;35:D503–D505. doi: 10.1093/nar/gkl960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder E, et al. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq031. doi:10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Evidence that the human X chromosome is enriched for male-specific but not female-specific genes. Mol Biol Evol. 2003;20:1113–1116. doi: 10.1093/molbev/msg131. [DOI] [PubMed] [Google Scholar]
- Lin H, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. The W, X, Y and Z of sex-chromosome dosage compensation. Trends Genet. 2009;25:226–233. doi: 10.1016/j.tig.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity. 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Marinotti O, et al. Genome-wide analysis of gene expression in adult Anopheles gambiae. Insect Mol Biol. 2006;15:1–12. doi: 10.1111/j.1365-2583.2006.00610.x. [DOI] [PubMed] [Google Scholar]
- Marinotti O, Nguyen QK, Calvo E, James AA, Ribeiro JM. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–373. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- Meisel RP, Han MV, Hahn MW. A complex suite of forces drives gene traffic from Drosophila X chromosomes. Genome Biol Evol. 2010;2009:176–188. doi: 10.1093/gbe/evp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Bensch S, Hasselquist D. Why does dosage compensation differ between XY and ZW taxa? Trends Genet. 2010;26:15–20. doi: 10.1016/j.tig.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Oliver B. Sex, dose, and equality. PLoS Biol. 2007;5:e340. doi: 10.1371/journal.pbio.0050340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, et al. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Reinke V, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Richards S, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Smith ER, et al. A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol Cell Biol. 2005;25:9175–9188. doi: 10.1128/MCB.25.21.9175-9188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokoloff A. The biology of tribolium vol. 1. London: Oxford University Press; 1972. [Google Scholar]
- Storchova R, Divina P. Nonrandom representation of sex-biased genes on chicken Z chromosome. J Mol Evol. 2006;63:676–681. doi: 10.1007/s00239-006-0022-1. [DOI] [PubMed] [Google Scholar]
- Storey JD, Dai JY, Leek JT. The optimal discovery procedure for large-scale significance testing, with applications to comparative microarray experiments. Biostatistics. 2007;8:414–432. doi: 10.1093/biostatistics/kxl019. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Becker PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Lopes HF, Karr TL, Long M. Stage-specific expression profiling of Drosophila spermatogenesis suggests that meiotic sex chromosome inactivation drives genomic relocation of testis-expressed genes. PLoS Genet. 2009;5:e1000731. doi: 10.1371/journal.pgen.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibranovski MD, Zhang Y, Long M. General gene movement off the X chromosome in the Drosophila genus. Genome Res. 2009;19:897–903. doi: 10.1101/gr.088609.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–583. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li Y, Paradesi MS, Brown SJ. BeetleBase: the model organism database for Tribolium castaneum. Nucleic Acids Res. 2007;35:D476–D479. doi: 10.1093/nar/gkl776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Wyman MJ, Agrawal AF, Rowe L. Condition-dependence of the sexually dimorphic transcriptome in Drosophila melanogaster. Evolution. 2010 doi: 10.1111/j.1558-5646.2009.00938.x. doi:10.1111/j.1558-5646.2009.00938.x. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha XF, et al. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem Mol Biol. 2009;39:315–321. doi: 10.1016/j.ibmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Oliver B. An evolutionary consequence of dosage compensation on Drosophila melanogaster female X-chromatin structure? BMC Genomics. 2010;11:6. doi: 10.1186/1471-2164-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus Drosophila. Nature. 2007;450:233–237. doi: 10.1038/nature06323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]