Abstract
Bitter taste perception likely evolved as a protective mechanism against the ingestion of harmful compounds in food. The evolution of the taste receptor type 2 (TAS2R) gene family, which encodes the chemoreceptors that are directly responsible for the detection of bitter compounds, has therefore been of considerable interest. Though TAS2R repertoires have been characterized for a number of species, to date the complement of TAS2Rs from just one bird, the chicken, which had a notably small number of TAS2Rs, has been established. Here, we used targeted mapping and genomic sequencing in the white-throated sparrow (Zonotrichia albicollis) and sample sequencing in other closely related birds to reconstruct the history of a TAS2R gene cluster physically linked to the break points of an evolutionary chromosomal rearrangement. In the white-throated sparrow, this TAS2R cluster encodes up to 18 functional bitter taste receptors and likely underwent a large expansion that predates and/or coincides with the radiation of the Emberizinae subfamily into the New World. In addition to signatures of gene birth-and-death evolution within this cluster, estimates of Ka/Ks for the songbird TAS2Rs were similar to those previously observed in mammals, including humans. Finally, comparison of the complete genomic sequence of the cluster from two common haplotypes in the white-throated sparrow revealed a number of nonsynonymous variants and differences in functional gene content within this species. These results suggest that interspecies and intraspecies genetic variability does exist in avian TAS2Rs and that these differences could contribute to variation in bitter taste perception in birds.
Keywords: bitter taste receptors, molecular evolution, inversion, duplication
Introduction
Taste perception is necessary to identify nutritious and potentially harmful substances in food (Bachmanov and Beauchamp 2007). The perception of bitter taste is thought to have evolved as a protective mechanism for the avoidance of toxic substances, especially those produced by plants (Glendinning 1994). Bitter taste perception is mediated through bitter taste receptors (TAS2R), which are G protein–coupled receptors expressed on the microvilli of taste receptor cells located in the taste buds on the tongue (Lindemann 2001). Although compounds that activate specific bitter taste receptors have been identified for some rodent and human TAS2Rs (Chandrashekar et al. 2000; Bufe et al. 2002, 2005; Behrens et al. 2004, 2009; Pronin et al. 2004; Maehashi et al. 2008; Meyerhof et al. 2010), little is known about their natural ligands (Hofmann 2009). Nonetheless, genetic variation within some mouse, human, and chimpanzee TAS2Rs have been linked to differences in sensitivity to bitter compounds (Chandrashekar et al. 2000; Kim et al. 2003; Sandell and Breslin 2006; Wooding et al. 2006). Therefore, differences in bitter taste perception between individuals and between species can be attributed to genetic variation and the interspecies divergence of TAS2Rs.
TAS2Rs are expressed in discrete locations outside of taste buds as well, suggesting that they function as defensive chemoreceptors in contexts other than bitter taste perception in the oral cavity (Finger et al. 2003). Specifically, in mice and rats, TAS2Rs expressed in the nasal epithelium on solitary chemosensory cells can detect airborne irritants and trigger a protective respiratory response (Finger et al. 2003). TAS2Rs are also expressed on the motile cilia of human airway epithelial cells and, when activated by bitter compounds in vitro, increase the ciliary beat frequency, thereby potentially providing a defensive response that aids in the elimination of inhaled noxious compounds (Shah et al. 2009). Finally, signaling through TAS2Rs expressed in the gut enteroendocrine cells is hypothesized to participate in the molecular pathway that invokes protective responses, such as vomiting, after ingesting harmful compounds (Sternini et al. 2008). Thus, intraspecific variation and interspecific divergence in the TAS2Rs is likely to have significant functional consequences beyond bitter taste perception.
Because the ability to perceive harmful airborne or food-borne compounds can directly affect an individual’s survival, genetic variation within the TAS2Rs is an obvious target for natural selection. The molecular evolution of these genes has therefore been of considerable interest and has led to a number of studies focusing on the pattern in which TAS2Rs evolve. For example, signatures of balancing selection, which could provide heterozygotes with a fitness advantage by providing a broader sense of bitter taste perception, and positive selection, which could result from selection for individuals with increased sensitivity to harmful toxins in new food sources, have been associated with specific human TAS2Rs (Wooding et al. 2004; Soranzo et al. 2005). Furthermore, in mammals, there is a tendency for rapidly evolving sites in the TAS2R proteins to be enriched in the extracellular domains, which presumably have a central role in determining ligand-binding specificity (Shi et al. 2003; Pronin et al. 2004; Fischer et al. 2005; Go et al. 2005). TAS2R genes are often found in clusters that have undergone dynamic patterns of gene gain and loss during the course of vertebrate evolution (Go et al. 2005; Go 2006; Dong et al. 2009). For example, in a small but diverse set of mammals, there have been an estimated 87 independent gains and 71 losses of TAS2Rs since the most recent common ancestor (MRCA) of those species. As a consequence, the functional repertoire of TAS2Rs in sequenced mammalian genomes varies from as few as four genes in the platypus to as many as 36 in rat (Dong et al. 2009). Though the gain and loss of TAS2Rs in different lineages may simply be a neutral byproduct of the inherent instability of gene clusters (Nei et al. 2008), the extant differences in functional TAS2R repertoires between species are intriguing candidates for dietary adaptations and interspecies differences in taste perception.
The morphological variation in the size and shape of beaks in Darwin’s finches is a classic example of adaptive change associated with differences in diet within populations and between species (Lack 1983). However, despite the precedent for dietary adaptations in birds, little is known about avian bitter taste perception and the molecular evolution of TAS2Rs in this vertebrate lineage. In particular, although we know that birds do perceive bitter taste and bitter taste perception can vary within- and between-species (Kare and Mason 1986), to date there have been no complementary biochemical or genetic studies characterizing TAS2Rs in birds. Indeed, the only bird for which TAS2R sequences have been described is the chicken and that genome encodes just three intact TAS2Rs (Hillier et al. 2004). Here, we report the sequencing of a TAS2R cluster linked to an evolutionary inversion break point in the white-throated sparrow (Zonotrichia albicollis), a member of the Emberizinae subfamily, whose >800 members (Sibley and Monroe 1990) are the product of a radiation which began with a dispersal into North America and initial diversification ∼16–20 Ma and continued with a more recent radiation into South America (Burns 1997; Yuri and Mindell 2002; Barker et al. 2004). In addition, we report the reconstruction of the evolutionary history and patterns of evolution for the avian TAS2R gene cluster as well as the linked inversion.
Materials and Methods
BAC Library Construction
A white-throated sparrow bacterial artificial chromosome (BAC) library was constructed from frozen kidney tissue from a single white female (ID #822) following the methods described in Osoegawa et al. (1998). The library (CHORI-264) consists of 196,354 recombinant clones with an average insert size of 144 kb and has been arrayed into 528 microtiter dishes (384-well) and gridded onto eleven 22 × 22-cm nylon high-density filters for screening by probe hybridization (see http://bacpac.chori.org/library.php?id=469). Note that the individual from which the BAC library was constructed was verified to be a female (ZW) by polymerase chain reaction (PCR) (Griffiths et al. 1998) and by visual inspection of the gonads, and confirmed to have the expected genotype for a ZAL2/2m individual by direct sequencing of 58 loci linked to the polymorphism (LY Huynh and JW Thomas, unpublished data).
Physical Mapping of the Evolutionary Inversion Break Point Intervals
The white-throated sparrow BAC library was screened with overgo hybridization probes designed from zebra finch sequences conserved with chicken within the regions likely to contain the inversion break points using the Custom option on the Uprobe Web site (Sullivan et al. 2008). Pools of overgo probes were then hybridized to the white-throated sparrow library, and probe content and restriction enzyme fingerprint maps of each targeted region were constructed using previously described methods (Thomas et al. 2002).
BAC End Sequencing and Comparative Mapping
Quality trimmed and repeat-masked BAC end sequences (BESs) generated at the BC Cancer Agency Genome Sciences Centre were mapped to the zebra finch genome assembly (taeGut1, Warren et al. 2010) using MEGABlast (-t 16, -N 2, -W 11, -e 1 × 10−30) (Zhang et al. 2000). Individual white-throated sparrow BESs were initially classified as either mapping to 0, 1, or >1 location in the zebra finch genome. In cases where both of the mate-pair reads from a single BAC mapped to one location in the zebra finch genome, the orientation and distance between the mate-pair alignments were used to classify clones as “concordant” (orientation: + and – strand; distance: 90–200 kb) or “discordant” (orientation: + and + strand or – and – strand; distance: <90 kb or >200 kb). BESs that displayed no matches to the zebra finch genome assembly at the initial e value cutoff were reanalyzed and classified using a less stringent e value cutoff (1 × 10−10 or 1 × 10−20). To define the most likely position of BESs that mapped to more than one location in the zebra finch genome, when applicable, the location of a uniquely mapped mate pair was used to search the alternative genomic locations for one that would meet the criteria set for concordant clones. All BESs have been deposited in GenBank (for the mapping information and GenBank accession numbers, see supplementary tables S1–S3, Supplementary Material online).
Selection of Haplotype-Based Sequencing Tiling Paths, BAC Sequencing, and Gene Annotation
The physical and BES mapping data were used to order the BACs and estimate overlaps between adjacent clones. Single-nucleotide polymorphisms between overlapping BESs were used to select double-tiling paths across the region of interest representing both haplotypes present in the genomic library (i.e., ZAL2 and ZAL2m). Individual BACs were Sanger shotgun sequenced, assembled into an ordered set of contigs (Blakesley et al. 2004), and then used to generate multi-BAC assemblies representing each haplotype (GenBank accession numbers DP001173 and DP001174). Genes were annotated primarily using predicted zebra finch cDNAs or other homology search-based methods. The white-throated sparrow and zebra finch TAS2Rs were classified as intact if the open reading frame was predicted to encode seven transmembrane domains with TMHMMv2.0 (Krogh et al. 2001). TAS2Rs were classified as truncated and/or potential pseudogenes if one or more nonsense or frameshift mutations disrupted the predicted full-length open reading frame.
Amplification and Sequencing of the Inversion Break Points in Other Songbirds
PCR primers flanking the inversion break points were designed to amplify either the white-throated sparrow or zebra finch chromosomal arrangements (supplementary table 4, Supplementary Material online). The PCR amplicons were then directly sequenced and compared back with the white-throated sparrow assemblies and zebra finch genome to confirm that the orthologous locus had been amplified. The sequences from the break point amplicons have been deposited in GenBank (GenBank accession numbers GU815874–GU815877, GU815894, GU815906–GU815910, and GU815921–GU815925).
Identification of TAS2R Genes in Other Songbirds
BLAT (Kent 2002) and BlastP (Altschul et al. 1990) searches using the published chicken TAS2R proteins and four zebra finch TAS2R proteins that mapped to the cluster of interest were used to identify additional TAS2Rs in the zebra finch genome, and the sequences of those genes are included in the Supplementary Material. A series of PCR primers designed based on individual white-throated sparrow TAS2Rs as well as a pair of degenerate primers designed using HYDEN (Linhart and Shamir 2002) were used to amplify and either directly sequence or clone and sequence related TAS2Rs from other songbirds. A complete list of the primers is provided in supplementary table 5 (Supplementary Material online). The resulting sequences from each species were then clustered and classified to generate a minimal set of sequences representing each sampled gene. In particular, sequences with <2% divergence were presumed to be potential alternative alleles of the same gene and clustered together. A single sequence from each cluster was then used in the phylogenetic analyses. Note high-fidelity Taq (Platinum Taq Polymerase High Fidelity, Invitrogen) was used in the reactions to generate amplicons that were cloned before being sequenced, and the representative sequences from each cluster have been deposited in GenBank (GenBank accession numbers GU815878–GU815893, GU815895–GU815905, and GU815911–GU815920).
Phylogenetic and Sequence Evolution Analyses
A quartet-based maximum-likelihood method (10,000 puzzling steps, VT substitution model) (Schmidt et al. 2002) was used to construct a phylogeny based on amino acid sequences of the TAS2R proteins. A minimum evolution method (1,000 bootstraps, Tamura-Nei 93 substitution model with rate variation among sites) as implemented in MEGA (Kumar et al. 2004) was used to construct a phylogeny based on DNA sequences of the TAS2R genes. In the case of the protein phylogeny, the stickleback T2R1 protein, which represents a distinct clade of TAS2Rs specific to fish, was used as the outgroup (Dong et al. 2009). The naming convention for the chicken TAS2Rs follows the one used in Dong et al. (2009). Pairwise divergence (including standard error) was calculated considering synonymous sites using the method described in Yang and Nielsen (2000). The ratio of nonsynonymous (Ka) to synonymous (Ks) substitutions (ω) was estimated under various models using PAML (Yang 1997), and likelihood ratio tests (Yang 1998) were used to test for significant differences between the models (Yang et al. 2000, 2005).
DNA Samples
Genomic DNA was extracted from tissue samples provided by the Burke Museum of Natural History and Culture for the house sparrow (Passer domesticus, UWBM #79787), yellow wagtail (Motacilla flava, UWBM #78657), pine siskin (Carduelis pinus, UWBM #72966), hepatic tanager (Piranga flava, UWBM #77743), northern cardinal (Cardinalis cardinalis, UWBM #77974), Bullock’s oriole (Icterus bullockii, UWBM #55978), ovenbird (Seiurus aurocapillus, UWBM #78563), Cassin’s sparrow (Aimophila cassinii, UWBM #77699), lark bunting (Calamospiza melanocorys, UWBM #77753), white-crowned sparrow (Zonotrichia leucophrys, UWBM #73050), and Harris’s sparrow (Zonotrichia querula, UWBM #69719). The DNA from a dark-eyed junco (Junco hyemalis) was obtained from blood taken from a locally captured individual.
Results
Mapping and Sequencing of a TAS2R Gene Cluster in the White-Throated Sparrow Linked to the Break Points of an Evolutionary Chromosomal Rearrangement
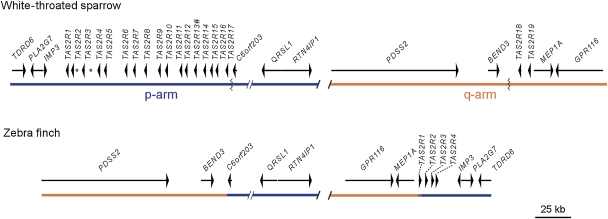
As part of an ongoing genomic effort to characterize the ZAL2/2m chromosomal polymorphism in the white-throated sparrow, cytogenetic mapping of the polymorphism identified two intervals corresponding to ∼69.8–79.6 Mb and ∼106.9–108.8 Mb on zebra finch chromosome 2 (chromosome 3 in the taeGut1 assembly, Warren et al. 2010) predicted to contain the break points of an evolutionary inversion that occurred since the MRCA of the zebra finch and white-throated sparrow 20–45 Ma (Barker et al. 2004). To further localize and ultimately sequence the inversion break points in the white-throated sparrow, we constructed a BAC-based physical map across these regions by screening a ∼25X BAC library we developed for this species (see Materials and Methods). Comparative mapping of the isolated white-throated sparrow BESs to the zebra finch genome allowed us to further refine the position of the break points (see supplementary fig. 1, Supplementary Material online), which were confirmed to map to the p- and q-arms of both the ZAL2 and ZAL2m chromosomes by fluorescence in situ hybridization (supplementary fig. 2, Supplementary Material online). BAC-based genomic sequence assemblies were then generated across the inversion break points and annotated with respect to their gene content (fig. 1).
FIG. 1.—
Comparison of the inversion break point regions and TAS2R gene clusters in the white-throated sparrow and zebra finch. The gene content of the regions flanking the inversion break points is shown for the white-throated sparrow (top) and zebra finch (bottom). The position and orientation of genes are indicated by arrows. The location of the TAS2R gene fragments identified in the white-throated sparrow, which are on the same strand as the genes in the cluster, are indicated by *. The truncated white-throated sparrow TAS2R13 is indicated marked with a #. The slashed lines indicate regions of the chromosome that are present but are not shown here. The location of one of the inversion break points in zebra finch maps between BEND3 and C6orf203, where the orange and blue lines abut (chr3:70,274,175-70,274,209 [taeGut1]). In the white-throated sparrow, the edges of that break point are indicated by jagged lines. The precise location of the other break point relative to zebra finch could only be estimated and is indicated by a short hashed line under zebra finch TAS2R1. Note that the IMP3 locus is a retrogene found only at this genomic location in birds. Also note that, at a minimum, two additional inversions encompassing one or both of the break point regions illustrated here are needed to account for the relative orientations of the white-throated sparrow and zebra finch break point intervals.
Using gene annotation and genomic sequence alignments between the white-throated sparrow and zebra finch, we were able to precisely localize the position of one of the inversion break points with respect to the zebra finch genome assembly to an ancestral CR1-J2 transposable element between BEND3 and the ortholog of the human C6orf203 (fig. 1). The exact position of the other inversion break point could not be resolved because it mapped within a cluster of TAS2Rs between MEP1A and IMP3 (fig. 1), confounding our efforts to generate a simple alignment of the orthologous zebra finch and white-throated sparrow sequence across the region. In particular, four TAS2Rs mapped to this break point region in the zebra finch, whereas in the white-throated sparrow, 16 intact, one truncated, and two 71-bp TAS2R gene fragments mapped to the q-arm break point region, and two intact TAS2Rs mapped to the p-arm break point region (fig. 1). Thus, the break points of this evolutionary chromosomal rearrangement physically colocalize with a TAS2R cluster.
Reconstructing the History of the Evolutionary Chromosomal Rearrangement
To determine in which ancestral lineage and the time frame in which the inversion between the zebra finch and white-throated sparrow occurred, we first compared the local gene order of the break point regions with the orthologous chromosomal segments in a set of other representative tetrapods for which genome assemblies are available (i.e., chicken, Anolis lizard, human, platypus, opossum, and Xenopus tropicalis). The local gene order observed in the other species was the same as in the zebra finch (data not shown), indicating the most parsimonious explanation for the structural differences between the two songbird genomes is that the inversion occurred in the white-throated sparrow lineage since the MRCA with zebra finch.
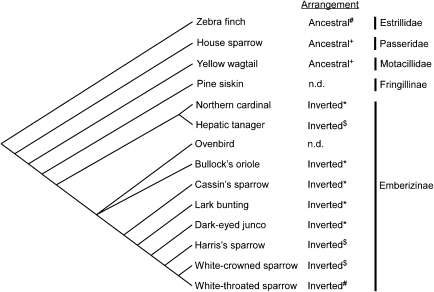
To further refine the time frame in which the inversion occurred, we developed a set of diagnostic PCR assays centered on the break points that amplified either the white-throated sparrow or zebra finch chromosomal arrangements (see Materials and Methods) and used these assays to genotype a sampling of other Emberizinae and a pair of closely related birds, the house sparrow (Passeridae), and yellow wagtail (Motacillidae) (fig. 2). Visualization of the PCR products by gel electrophoresis and direct sequencing of the amplicons indicated that the inverted arrangement found in the white-throated sparrow is present in all the successfully typed Emberizinae, whereas the house sparrow and yellow wagtail shared the ancestral arrangement with the zebra finch (fig. 2). Thus, the most parsimonious time frame for the genesis of the inversion is prior to the MRCA of the sampled Emberizinae but some time after the MRCA of the Emberizinae and Motacillidae/Passeridae.
FIG. 2.—
Phylogenetic distribution of the inversion linked to the TAS2R cluster. A cladogram representing the evolutionary relationships of a sample of related birds typed for the chromosomal arrangement based on Zink et al. (1991), Yuri and Mindell (2002), and Barker et al. (2004) is shown on the left. The evidence for the chromosomal arrangement (termed ancestral or inverted) for each species is denoted by the following symbols: # assembled genomic sequence, + PCR amplification and sequence verification corresponding to one zebra finch-specific break point, * PCR amplification and sequence verification corresponding to both white-throated sparrow arrangement-specific break points, and $ PCR amplification and sequence verification corresponding to one of the white-throated sparrow arrangement break points. In the case of the pine siskin, PCR amplification of the break points was attempted but subsequent sequence analyses indicated that we were unable to amplify the break points in that species (n.d., no data). Subfamily or family designations are shown on the far right.
Origin and Expansion of the TAS2R Gene Cluster
Duplication and losses of TAS2Rs are common among vertebrates and have resulted in differences in TAS2R repertoires between species (Go et al. 2005; Go 2006; Dong et al. 2009). As the first step toward reconstructing the differences in gene content associated with this cluster between the white-throated sparrow and zebra finch, we first sought to determine the long-term history of TAS2Rs at this position in the genome. To do so, we used existing gene annotation and homology searches of the orthologous regions in the representative sample of tetrapods to determine the phylogenetic distribution of species in which at least one TAS2R mapped to this interval. A single TAS2R was present between MEP1A and IMP3 in the chicken genome but not in the orthologous segments of the other sampled tetrapod genomes (Anolis lizard, opossum, platypus, human, and X. tropicalis), suggesting that this cluster might be specific to aves.
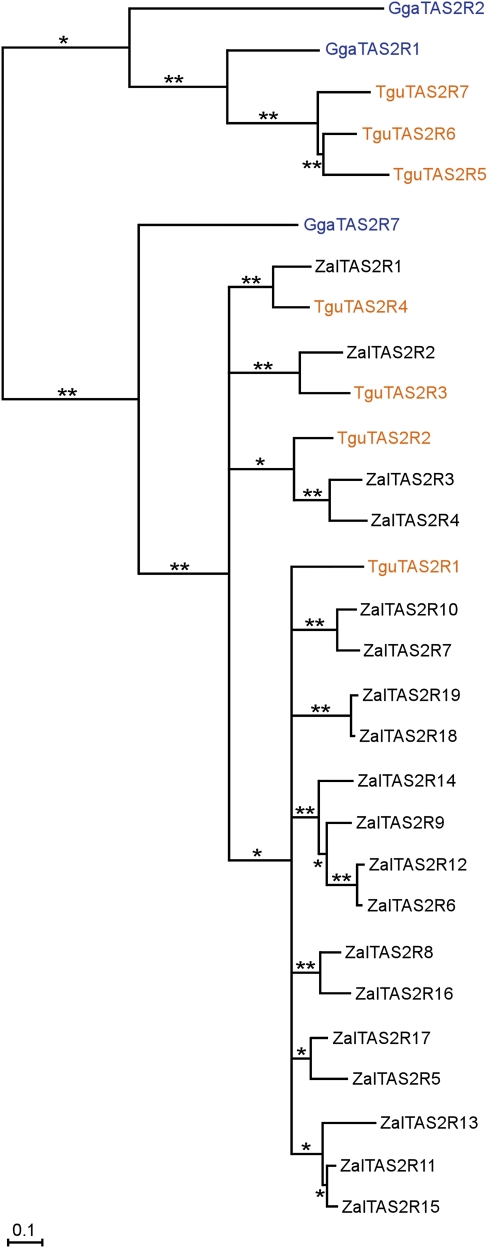
Next, to elucidate the evolutionary relationship of the TAS2Rs in this cluster to one another and to TAS2Rs found elsewhere in avian genomes, we generated a phylogeny of all intact TAS2Rs present in the chicken (n = 3) (Hillier et al. 2004) and zebra finch (n = 7, see Materials and Methods) genomes as well as the 19 intact or truncated TAS2Rs that comprise this cluster in the white-throated sparrow. The TAS2Rs broadly clustered into two main groups. The first group included all the white-throated sparrow TAS2Rs as well as the zebra finch and chicken TAS2Rs, which map to the orthologous intervals. The second group included the zebra finch and chicken TAS2Rs, which map elsewhere in the genome (fig. 3). The white-throated sparrow TAS2Rs fell into four well-supported subgroups, each represented by a single zebra finch gene and from 1 to 15 white-throated sparrow genes (fig. 2). (Also note that all four of the songbird subgroups were more closely related to one another than the single chicken TAS2R7 that maps to the orthologous interval.) Specifically, although the white-throated sparrow cluster contains single orthologs of zebra finch TAS2R3 and TAS2R4, there are two white-throated sparrow para-orthologs for zebra finch TAS2R2 and 15 para-orthologs of TAS2R1 (fig. 2). (Note that the term para-ortholog is used here to denote instances of non-1-to-1 orthology, and paralog is used to refer to the relationship between the TAS2Rs within a species.) The difference in gene number between zebra finch TAS2R2 and TAS2R1 and their white-throated sparrow para-orthologs could be due to either gene gain in the white-throated sparrow lineage, gene loss in the zebra finch lineage, or both.
FIG. 3.—
Phylogeny of the white-throated sparrow TAS2Rs. The predicted TAS2R protein sequences from the white-throated sparrow (Zal), chicken (Gga), and zebra finch (Tgu) were used to construct a phylogenetic tree. The branch lengths are scaled according to the calculated evolutionary distances (amino acid substitutions/site, VT substitution model). * and ** indicate nodes with 0.5–0.75 and >0.75 bootstrap support. Note that the fish TAS2R protein used to root the tree (see Materials and Methods) is not shown.
An Expanded Set of TAS2R Clusters Is a Common Feature of Emberizinae, Fringillidae, and Motacillidae Genomes
To better estimate when the duplications giving rise to the TAS2R gene set in the white-throated sparrow occurred, we calculated the intraspecies synonymous divergence per site (dS) within the two groups of white-throated sparrow TAS2Rs that may have expanded recently by duplication (i.e., the two para-orthologs of the zebra finch TAS2R2 and 15 para-orthologs of zebra finch TAS2R1). Compared with the interspecies dS values of the zebra finch and white-throated sparrow orthologs/para-orthologs, which ranged from 0.11 ± 0.02 to 0.19 ± 0.03, the dS values within the two groups of white-throated sparrow TAS2Rs were distributed across a range from 0.01 ± 0.01 to as high as 0.20 ± 0.03, with the divergence between the genes in different subgroups varying from 0.07 ± 0.02 to 0.20 ± 0.03 (table 1). Though these dS values may not accurately represent the true time since a duplication occurred due to factors such as gene conversion (Graur and Li 2000), because most of the dS values between white-throated sparrow paralogs exceeded the interspecies divergence between the white-throated sparrow and other birds in the genus, that is, >0.05 substitutions/site, (Zink et al. 1991; Thomas et al. 2008), these results suggested that most of the duplications were unlikely to be species specific. Thus, we predicted that an expanded TAS2R cluster similar to that seen in the white-throated sparrow would be present in other species in the Emberizinae subfamily and songbirds in other closely related families.
Table 1.
Divergence among the Recently Duplicated White-Throated Sparrow TAS2Rs
| Gene Group | dS Range Within Group | dS Range between Groups |
| TAS2R3, 4 | 0.07 ± 0.02 | NA |
| TAS2R18, 19 | 0.01 ± 0.01 | 0.12 ± 0.2 to 0.19 ± 0.03 |
| TAS2R6, 9, 12, 14 | 0.04 ± 0.01 to 0.16 ± 0.03 | 0.12 ± 0.02 to 0.20 ± 0.03 |
| TAS2R7, 10 | 0.06 ± 0.02 | 0.12 ± 0.02 to 0.19 ± 0.03 |
| TAS2R8, 16 | 0.08 ± 0.02 | 0.12 ± 0.02 to 0.20 ± 0.03 |
| TAS2R5, 17 | 0.10 ± 0.02 | 0.07 ± 0.02 to 0.19 ± 0.03 |
| TAS2R11, 15 | 0.14 ± 0.03 | 0.07 ± 0.02 to 0.17 ± 0.03 |
NOTE.—dS values were calculated for the recently duplicated and intact TAS2Rs based on pairwise alignments of ∼245 synonymous sites and grouped according the phylogeny presented in figure 3. The dS range within the group refers to minimum and maximum dS values observed within that cluster of genes and the dS range between groups refers to the minimum and maximum divergence between genes that fell into distinct clusters but were para-orthologs of zebra finch TAS2R1. NA, not applicable.
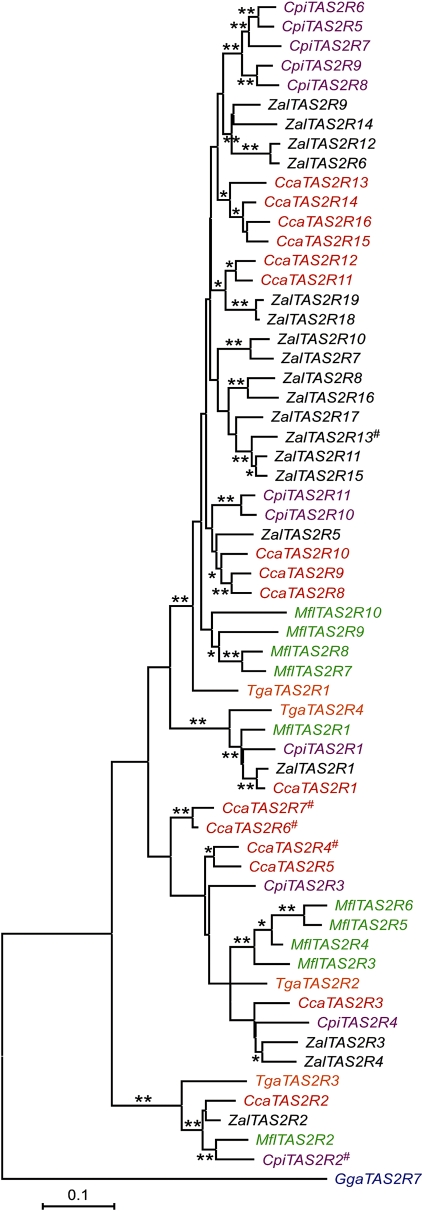
To test that hypothesis, we designed gene-specific and degenerate PCR primers based on the white-throated sparrow TAS2Rs and used them to amplify and sequence TAS2Rs in a relatively divergent Emberizinae (northern cardinal), a representative of the Fringillidae subfamily (pine siskin) and a representative from another closely related family, Motacillidae (yellow wagtail). These efforts yielded partial sequences of 13 intact and 3 truncated TAS2Rs from the northern cardinal, 10 intact and 1 truncated TAS2R from the pine siskin, and 10 intact TAS2Rs from the yellow wagtail. Due to potential PCR biases or failures related to the underlying divergence between the target species and the primers and nonexhaustive sampling of the amplicons, we do not believe this collection represents a complete set of the TAS2Rs present in this cluster in these three species. With that limitation in mind, we nonetheless generated a phylogeny that included those sequences along with all the other chicken, zebra finch, and white-throated sparrow TAS2Rs to further elucidate the timing and patterns of gene duplications and losses in this TAS2R gene cluster (fig. 4).
FIG. 4.—
Phylogeny of additional songbird TAS2Rs. The DNA sequences of the white-throated sparrow (Zal), chicken (Gga), and zebra finch (Tgu) TAS2Rs that map to this cluster, along with homologs amplified from the yellow wagtail (Mfl), pine siskin (Cpi), and cardinal (Cca), were used to construct a phylogenetic tree (minimum evolution, 1,000 bootstrap replicates). The branch lengths are scaled according to the calculated evolutionary distances (TN93). * and ** indicate nodes with 0.5–0.75 and >0.75 bootstrap support, respectively. Genes with truncating mutations are marked with a #.
As observed in the phylogeny presented in figure 3, the songbird TAS2Rs fell into four groups, each represented by one of the zebra finch TAS2R1–4 genes (fig. 4). The tree topology for the genes clustering with zebra finch TAS2R3 and TAS2R4 was either similar to, or fully recapitulated, the species phylogeny and showed no evidence of duplications or losses in any of the sampled species (fig. 4). On the other hand, the topologies associated with newly expanded sets of duplicated white-throated sparrow genes (the para-orthologs of zebra finch TAS2R1 and TAS2R2) were not a simple reflection of the species phylogeny, with the genes tending to form species-specific groups (fig. 4). In particular, with some exceptions, the TAS2Rs from the yellow wagtail, pine siskin, and northern cardinal formed species-specific monophyletic groups of 2 to 5 genes within the larger gene clusters defined by the presence of zebra finch TAS2R1 and TAS2R2 (fig. 4). Thus, although an expanded repertoire of TAS2Rs in this cluster compared with zebra finch is a common feature among songbirds closely related to the white-throated sparrow, the tendency is for the genes to form species-specific clusters within the groups defined by zebra finch TAS2R1 and TAS2R2 rather than recapitulate the species phylogeny, suggesting a complex history that likely involves common ancestral and lineage-specific gene gains and losses and/or concerted evolution.
Genetic Variation and Patterns of Evolution in the White-Throated Sparrow TAS2Rs
Genetic variation within populations in the TAS2Rs has been linked to variation in taste perception in humans and other mammals (Chandrashekar et al. 2000; Kim et al. 2003; Sandell and Breslin 2006; Wooding et al. 2006). We were therefore interested in determining whether there was any genetic evidence for potential functional variants in the TAS2Rs segregating in the white-throated sparrow population. To address that question, we compared the coding regions of the TAS2Rs between our haplotype-specific BAC-based assemblies that included the alternative TAS2R alleles present in the individual from which the genomic library was constructed (see Materials and Methods). Eleven of the genes (TAS2R1–10 and TAS2R14) encoded the identical proteins on both haplotypes, and the alleles of TAS2R11-12 and TAS2R15 differed by just one or two amino acid replacements. More significant divergence was observed between the alternative alleles of TAS2R18 and TAS2R19, which differed by 6 and 10 amino acid replacements, respectively. Finally, on one of the sequenced haplotypes, frameshift mutations that we predicted would be gene-inactivating mutations were found in TAS2R16 (c.del150_151) and TAS2R17 (c.605del). Hence, there is potential functional genetic variation, including differences in gene dosage, associated with this TAS2R cluster in the white-throated sparrow.
To compare the patterns of molecular evolution of the TAS2Rs in this cluster with those reported for the mammalian TAS2Rs, we carried out a series of analyses estimating the ratio of nonsynonymous (Ka) to synonymous (Ks) substitutions, referred to as ω, across a tree including all the intact white-throated sparrow and zebra finch TAS2Rs in this cluster (see Materials and Methods). First, we used the simplest model (M0) to estimate a ω for the entire tree and compared this with a model that allows the rate to vary on every branch (M1). The M0 model estimated ω across the entire tree to be 0.95, and this single estimate of ω was not significantly different from the model in which ω was free to vary (M0 vs. M1; 2Δl = 39.067, degrees of freedom [df] = 34, P = 0.25), suggesting that all the genes have been evolving at a relatively uniform level of functional constraint. Because the estimated ω of 0.95 was very close to the value of 1 expected for protein-coding sequence that is evolving neutrally, we tested to see whether this observed value was different from 1 by comparing the model estimating the single ω (M0) with a model in which ω was fixed at 1 (M0: ω fixed at 1). The comparison of those models indicated that the estimated ω of 0.95 for the white-throated sparrow and zebra finch genes did not differ significantly from 1 (2Δl = 0.678, df = 1, P = 0.41).
In previous studies, human and ape TAS2Rs were also found to be evolving at an overall rate near 1 (Wang et al. 2004; Fischer et al. 2005), which was attributed to a composite of heterogeneous rates of evolution across the protein and not necessarily due to uniform lack of functional constraint across the entire protein. To determine whether the zebra finch and white-throated sparrow TAS2Rs in this cluster exhibited a similar pattern of evolution, we compared a model in which a single ω was estimated for all codons (M0) to a model in which codons were partitioned into three groups, each of which is associated with a discrete estimate of ω (M3). The model in which three discrete ω values were estimated fit the data significantly better than the single-rate model (M0 vs. M3: 2Δl = 487.4, df = 4, P = 9.50 × 10−11, p0 = 0.36, p1 = 0.43, p2 = 0.20, ω0 = 0.14, ω1 = 0.98, ω2 = 3.19), and comparisons between additional models were also consistent with a subset of the codons evolving at an ω > 1 (M1a vs. M2a: 2Δl = 154.9, df = 2, P = 5.9 × 10−11 and M7 vs. M8: 2Δl = 175.1, df = 2, P = 8.9 × 10−11). In addition, as was observed previously in mammals (Shi et al. 2003; Fischer et al. 2005; Go et al. 2005), the codons falling in the high ω group were overrepresented in the extracellular domains, which are presumably involved in ligand binding and underrepresented in the intracellular portions of the protein (χ2 test, P = 9.27 × 10−5 for the subset of codons estimated to have an ω > 1 with a Bayesian posterior probability >0.95 by all three models, M3, M2a, and M8, see fig. 5). Thus, although it is not surprising that functional constraint as reflected in ω would vary across a protein, the signatures of molecular evolution observed for the songbird TAS2Rs were reminiscent of what had been seen before in humans and other mammals.
FIG. 5.—
Distribution of rapidly evolving codons in the songbird TAS2Rs. The protein domains of all the white-throated sparrow TAS2Rs were predicted using TMHMMv2.0 (Krogh et al. 2001), and the location of the domains in representative TAS2R, TAS2R1 is illustrated here (e = extracellular, m = transmembrane, and i = intracellular). Amino acids corresponding to codons associated with an ω > 1 with a Bayesian posterior probability >0.95 in all models (M3, M2a, and M8) are indicated by ^. Amino acids corresponding to codons that were excluded from the estimates of ω are marked by an x.
Discussion
TAS2R Repertoires Differ among Birds
Lineage-specific expansions and contractions of the TAS2R gene family have occurred repeatedly in tetrapods, and as a consequence, the number of TAS2R genes and pseudogenes identified in sequenced genomes varies from as few as three to as many as 66 (Dong et al. 2009). Compared with the other sampled tetrapods, the number of TAS2Rs identified in the chicken genome stood out for two reasons. First, with the exception of the platypus genome, which encodes just four intact TAS2Rs, all the other sampled tetrapod genomes encode at least 15 intact TAS2Rs. Although it has been hypothesized that the low number of TAS2Rs identified in the platypus genome is related to its minimal exposure to bitter compounds in its semiaquatic diet (Dong et al. 2009), there is no apparent dietary explanation for the low number of TAS2Rs observed in the chicken, and it was proposed that other genes in the chicken genome may have acquired the ability to function as TAS2Rs, thereby precluding the need for a TAS2R repertoire similar to other tetrapods (Hillier et al. 2004). Second, the chicken genome was the only sampled tetrapod that had no partial TAS2R sequences or pseudogenes (Dong et al. 2009). To account for both the low number of TAS2Rs and lack of pseudogenes in the chicken, a hypothesis was put forward that the limited number of chicken TAS2Rs and absence of TAS2R pseudogenes could be a consequence of the genome size reduction in the avian lineage and subsequent lack of expansion of this gene family, perhaps due to a limited potential for gene duplication in avian genomes (Go 2006).
Our results demonstrate that, although the low number of TAS2Rs in the chicken genome is remarkable, it is not an accurate reflection of all avian genomes. In particular, the results of our study on this specific TAS2R cluster in the white-throated sparrow are consistent with it having originated from an expansion from 1 to 4 genes in the Passerine lineage since the MRCA with chicken (Galliformes) ∼100 Ma (van Tuinen and Hedges 2001), followed by a subsequent expansion to 19 genes in the lineage leading to the white-throated sparrow since its MRCA with the zebra finch. Consequently, the number of intact TAS2Rs in this single gene cluster in the white-throated sparrow is within the range found in the entire genome of most other tetrapods (i.e., 15–49) (Dong et al. 2009). Moreover, we also identified a truncated TAS2R, two very short TAS2R gene fragments and null alleles for two of the TAS2Rs in the white-throated sparrow. Thus, although the avian TAS2R repertoire might have been reduced as a consequence of an ancestral genome size reduction and might have retained in that reduced state in the Galliformes lineage, it is clear that an expansion of this gene family did occur in the Passerine lineage. This inferred gene expansion and other similar examples does not support the hypothesis that avian genomes are universally limited in their duplicability (Go 2006). Although we have no direct evidence to rule out the possibility that other genes may function as bitter taste receptors in chickens or other birds, we would argue that the low number of TAS2Rs in the chicken genome is not due to a novel acquisition of bitter taste perception by other genes but is instead due to the evolutionary history of expansion and contraction in this gene family.
It should also be noted that though other specific hypotheses can explain the differences in TAS2R gene content we observed between the chicken, zebra finch, and white-throated sparrow, all of them would have to include significant gene gains, gene losses, or a combination of both, which is similar to the pattern of evolution of this gene family observed in other vertebrates (Nei et al. 2008). In particular, the interspecies differences in gene content and the presence of pseudogenes in this avian TAS2R cluster are consistent with the gene birth-and-death mode of evolution that describes the evolution of many gene families (Nei and Rooney 2005). Additionally, our partial surveys of the TAS2R content of this cluster in other species closely related to the white-throated sparrow indicated that concerted evolution mediated by gene conversion could be another potential evolutionary process common to other gene families (Nei and Rooney 2005) that has occurred in the recent past within this cluster, though additional studies will be needed to determine if this indeed true. Therefore, although our study was limited to a single gene cluster, we argue that our data support the hypothesis that, as with mammalian genomes, gene birth-and-death is a fundamental mechanism by which the functional TAS2R repertoires in birds have evolved.
Physical Linkage of the TAS2R Cluster With Inversion Break Points
Changes in gene copy number linked to chromosomal rearrangement break points have been hypothesized to be a contributing factor that leads to the selection for and eventual fixation of some new chromosomal arrangements (Larkin et al. 2009). In addition, the repair of double-strand breaks that mediate an inversion can sometimes result in the formation of duplications at inversion break points (Kehrer-Sawatzki et al. 2005; Ranz et al. 2007). Our reconstruction of the history of the inversion linked to the TAS2R gene cluster placed the origin of this chromosomal rearrangement after the MCRA of the Emberizinae and the Motacillidae/Passer but before the radiation of the Emberizinae. Because this time frame overlaps the period in which the Emberizinae began their initial migration and radiation into the New World 16–20 Ma (Barker et al. 2004), the linkage of the expanded set of TAS2Rs at the inversion break points left open the possibility of an adaptive expansion of this cluster linked directly to the emergence of the inversion at a key moment in the history of these birds. However, although some of the divergence estimates generated from comparisons of the recently duplicated TAS2Rs in the white-throated sparrow overlap the time frame in which the inversion originated, the range of divergence observed between the white-throated sparrow paralogs do not support a hypothesis in which a burst of duplications accompanied the genesis of the inversion. Furthermore, given the relative antiquity of the inversion, even with greater sampling of species to refine the time frame in which this rearrangement originated, the complicated evolutionary history of the TAS2R cluster will likely preclude an accurate reconstruction of the timing of the inversion relative to the TAS2R duplications. Nevertheless, it would still be interesting to determine the inversion status and full complement of TAS2Rs from this cluster in a greater sampling of closely related birds to assess the potential influence the inversion had on patterns of evolution in the TAS2R cluster.
Genetic Evidence for Variation in Bitter Taste Perception in the White-Throated Sparrow
Taste tests comparing bitter taste perception between humans with different TAS2R38 genotypes combined with biochemical studies have shown that one or more amino acid replacements can alter the ligand-binding sensitivity of TAS2Rs (Bufe et al. 2005). In the case of the classic taster/nontaster alleles, the three possible genotypes at this locus can lead to low (nontaster/nontaster), intermediate (taster/nontaster), and high (taster/taster) perception of certain bitter compounds (Kim et al. 2003; Bufe et al. 2005; Sandell and Breslin 2006). Although our current study did not include any direct measurements and comparisons of bitter taste perception in the white-throated sparrow nor any allele-specific ligand-binding assays, we did identify nonsynonymous variants in five of the intact white-throated sparrow TAS2Rs, suggesting that there is the potential for variation in bitter taste perception in the white-throated sparrow population. Furthermore, we also identified null alleles for two of the genes that would be obvious candidates for altering the bitter taste perception between individuals with 0, 1, or 2 functional copies of those genes.
Our study did not include an estimate of the population frequency of the nonsynonymous variants we detected in a single white-throated sparrow but there is one aspect of the allelic diversity within the TAS2Rs that is worthy of further discussion. As mentioned in the Results section, the white-throated sparrow TAS2Rs are physically located on chromosome 2, which in this species is associated with a common chromosomal polymorphism linked to variation in a spectrum of traits, including plumage, social behavior, and mate choice (Thorneycroft 1966; Tuttle 2003). Gene flow between the alternative chromosome arrangements, designated ZAL2 and ZAL2m, which differ by a minimum of two inversions, is restricted to a small segment of the chromosome near the tip of the p-arm (Thorneycroft 1975; Thomas et al. 2008). Consequently, inside the rearrangement there is a high level of divergence between the chromosome types, with the majority of differences representing fixed differences between ZAL2 and ZAL2m (Thomas et al. 2008). Based on our current cytogenetic and genetic mapping studies, we are able to place TAS2R1–17, which maps to the p-arm, outside the inverted segments and thus in the region where alleles are shared between ZAL2 and ZAL2m. In contrast, TAS2R18 and TAS2R19, which map to the q-arm, fall within the inverted segments. Thus, one or more of the nonsynonymous variants detected here between the ZAL2 and ZAL2m alleles of those two genes are likely to represent fixed differences between the chromosome arrangements and as a result could be at allele frequencies similar to the chromosomal polymorphism, which has a minor allele frequency of ∼25% (Thorneycroft 1975). Because ZAL2/ZAL2m heterozygotes and ZAL2 homozygotes are present at approximately equal frequencies in the population (Thorneycroft 1975), the nonsynonymous variants we detected between the TAS2R alleles located inside the inversion, if functional, could be a major contributor to variation in bitter taste perception in this species and might factor into the variation in male habitat occupancy that has also been linked to the chromosomal polymorphism (Knapton and Falls 1982). Future genetic, biochemical, and ecological studies will help us evaluate these hypotheses.
Implications for Bitter Taste Perception in Birds
Bitter taste perception in birds has been understudied compared with mammals, particularly humans. Indeed, bitter taste perception in birds has been described as “enigmatic” (Mason and Clark 2000) and “poorly understood” (Matson et al. 2004). Although studies of bitter taste perception in birds are limited, it is still clear that birds do in fact taste bitter compounds (Kare and Mason 1986) and can use bitter taste as a signal to avoid toxic food sources (Skelhorn and Rowe 2010). Moreover, cockatiels (Nymphicus hollandicus) are found to have a sensitivity to the bitter compound quinine that is comparable with humans and is even higher than in some mammals (Matson et al. 2004). Thus, although the tens to hundreds of taste buds birds have pale in comparison to the hundreds to thousands of taste buds found in mammals (Kare and Mason 1986), this relative deficit does not preclude birds from perceiving bitter compounds as effectively as those species with more taste buds.
It is also worth noting that the relationship between the number of functional TAS2Rs encoded in the genome and an organism’s relative sensitivity to bitter compounds as well as the number of bitter compounds an organism can detect is likely quite complex. In particular, some TAS2Rs can detect a host of seemingly structurally distinct bitter compounds, whereas the ligand binding of other TAS2Rs may be much more specific (Meyerhof et al. 2010). This observation led to the idea that expansions of a TAS2R repertoire can partition the generalist function of one or a few TAS2Rs among any number of new duplicate copies of the ancestral receptors (Meyerhof et al. 2010), thereby facilitating changes in sensitivity to specific bitter compounds that are fine-tuned to the current environment (Behrens et al. 2009; Meyerhof et al. 2010). In the context of the results of this study, this would imply that white-throated sparrows, with many more functional TAS2Rs in this particular cluster than either the chicken or zebra finch, are likely to be more finely tuned in their perception of bitter compounds that are ligands for the receptors in this cluster compared with the other two species. However, the interspecies difference in TAS2R gene content is not proof that white-throated sparrows are simply better at tasting particular bitter compounds than either chickens or zebra finch. To clarify these issues and generally enhance our understanding of bitter taste perception in birds, future biochemical studies geared toward identifying the natural ligands for the avian TAS2Rs and genetic studies correlating intra- and interspecies genetic differences in these genes with variation in bitter taste perception will be needed.
Supplementary Material
Supplementary figures S1–S2, and tables S1–S5 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
The authors thank Cheryl Strauss for technical writing comments on the manuscript, the anonymous reviewers for their comments, Greg K. Tharp for computer support, the BC Cancer Agency Genome Sciences Centre, Vancouver, Canada for generation of the BESs, and members of the NIH Sequencing Center including E. D. Green, R. Blakesley, G. Bouffard, and J. McDowell. DNA samples were provided by The Burke Museum of Natural History and Culture. This work was supported by the National Institutes of Health (NIH) (1R21MH082046 to J.K.D., J.J.L., D.L.M., C.L.M, and J.W.T.) and the NIH Intramural Sequencing Center was supported in part by the Intramural Research Program of the National Human Genome Research Institute of the NIH.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414. doi: 10.1146/annurev.nutr.26.061505.111329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci U S A. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, et al. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Behrens M, et al. The human bitter taste receptor hTAS2R50 is activated by the two natural bitter terpenoids andrographolide and amarogentin. J Agric Food Chem. 2009;57:9860–9866. doi: 10.1021/jf9014334. [DOI] [PubMed] [Google Scholar]
- Blakesley RW, et al. An intermediate grade of finished genomic sequence suitable for comparative analyses. Genome Res. 2004;14:2235–2244. doi: 10.1101/gr.2648404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, et al. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Burns KJ. Molecular systematics of tanagers (Thraupinae): evolution and biogeography of a diverse radiation of neotropical birds. Mol Phylogenet Evol. 1997;8:334–348. doi: 10.1006/mpev.1997.0430. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, et al. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger TE, et al. Solitary chemoreceptor cells in the nasal cavity serve as sentinels of respiration. Proc Natl Acad Sci U S A. 2003;100:8981–8986. doi: 10.1073/pnas.1531172100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Gilad Y, Man O, Paabo S. Evolution of bitter taste receptors in humans and apes. Mol Biol Evol. 2005;22:432–436. doi: 10.1093/molbev/msi027. [DOI] [PubMed] [Google Scholar]
- Glendinning JI. Is the bitter rejection response always adaptive? Physiol Behav. 1994;56:1217–1227. doi: 10.1016/0031-9384(94)90369-7. [DOI] [PubMed] [Google Scholar]
- Go Y. Proceedings of the SMBE Tri-National Young Investigators' Workshop 2005. Lineage-specific expansions and contractions of the bitter taste receptor gene repertoire in vertebrates. Mol Biol Evol. 2006;23:964–972. doi: 10.1093/molbev/msj106. [DOI] [PubMed] [Google Scholar]
- Go Y, Satta Y, Takenaka O, Takahata N. Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics. 2005;170:313–326. doi: 10.1534/genetics.104.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D, Li WH. Fundamentals of molecular evolution. Sunderland (MA): Sinauer Associates, Inc; 2000. [Google Scholar]
- Griffiths R, Double MC, Orr K, Dawson RJ. A DNA test to sex most birds. Mol Ecol. 1998;7:1071–1075. doi: 10.1046/j.1365-294x.1998.00389.x. [DOI] [PubMed] [Google Scholar]
- Hillier LW, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hofmann T. Identification of the key bitter compounds in our daily diet is a prerequisite for the understanding of the hTAS2R gene polymorphisms affecting food choice. Ann N Y Acad Sci. 2009;1170:116–125. doi: 10.1111/j.1749-6632.2009.03914.x. [DOI] [PubMed] [Google Scholar]
- Kare M, Mason J. The chemical senses in birds. In: Sturkie P, editor. Avian physiology. New York: Springer-Verlag; 1986. pp. 59–67. [Google Scholar]
- Kehrer-Sawatzki H, Sandig CA, Goidts V, Hameister H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet Genome Res. 2005;108:91–97. doi: 10.1159/000080806. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim UK, et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Knapton RW, Falls JB. Polymorphism in the white-throated sparrow: habitat occupancy and nest-site selection. Can J Zool. 1982;60:452–459. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Lack D. Darwin's finches. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Larkin DM, et al. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19:770–777. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann B. Receptors and transduction in taste. Nature. 2001;413:219–225. doi: 10.1038/35093032. [DOI] [PubMed] [Google Scholar]
- Linhart C, Shamir R. The degenerate primer design problem. Bioinformatics. 2002;18(Suppl 1):S172–S181. doi: 10.1093/bioinformatics/18.suppl_1.s172. [DOI] [PubMed] [Google Scholar]
- Maehashi K, et al. Bitter peptides activate hTAS2Rs, the human bitter receptors. Biochem Biophys Res Commun. 2008;65:851–855. doi: 10.1016/j.bbrc.2007.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JR, Clark L. The chemical senses in birds. In: Sturkie P, editor. Avian physiology. San Diego (CA): Academic Press; 2000. pp. 39–56. [Google Scholar]
- Matson KD, Millam JR, Klasing KC. Cockatiels (Nymphicus hollandicus) reject very low levels of plant secondary compounds. Appl Anim Behav Sci. 2004;85:141–156. [Google Scholar]
- Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2010;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–151. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoegawa K, et al. An improved approach for construction of bacterial artificial chromosome libraries. Genomics. 1998;52:1–8. doi: 10.1006/geno.1998.5423. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- Ranz JM, et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell MA, Breslin PA. Variability in a taste-receptor gene determines whether we taste toxins in food. Curr Biol. 2006;16:R792–794. doi: 10.1016/j.cub.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–814. doi: 10.1093/molbev/msg083. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Monroe BL. Distribution and taxonomy of birds of the world. New Haven (CT): Yale University Press; 1990. [Google Scholar]
- Skelhorn J, Rowe C. Birds learn to use distastefulness as a signal of toxicity. Proc Biol Sci. 2010;277:1729–1734. doi: 10.1098/rspb.2009.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, et al. Positive selection on a high-sensitivity allele of the human bitter taste receptor TAS2R16. Curr Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Sternini C, Anselmi L, Rozengurt E. Enteroendocrine cells: a site of 'taste' in gastrointestinal chemosensing. Curr Opin Endocrinol Diabetes Obes. 2008;15:73–78. doi: 10.1097/MED.0b013e3282f43a73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan RT, Morehouse CB, Thomas JW. Uprobe 2008: an online resource for universal overgo hybridization-based probe retrieval and design. Nucleic Acids Res. 2008;36:W149–W153. doi: 10.1093/nar/gkn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, et al. Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res. 2002;12:1277–1285. doi: 10.1101/gr.283202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JW, et al. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics. 2008;179:1455–1468. doi: 10.1534/genetics.108.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorneycroft HB. Chromosomal polymorphism in the white-throated Sparrow, Zonotrichia albicollis (Gmelin) Science. 1966;154:1571–1572. doi: 10.1126/science.154.3756.1571. [DOI] [PubMed] [Google Scholar]
- Thorneycroft HB. A cytogenetic study of the white-throated sparrow, Zonotrichia albicollis. Evolution. 1975;29:611–621. doi: 10.1111/j.1558-5646.1975.tb00855.x. [DOI] [PubMed] [Google Scholar]
- Tuttle EM. Alternative reproductive strategies in the white-throated sparrow: behavioral and genetic evidence. Behav Ecol. 2003;14:425–432. [Google Scholar]
- van Tuinen M, Hedges SB. Calibration of avian molecular clocks. Mol Biol Evol. 2001;18:206–213. doi: 10.1093/oxfordjournals.molbev.a003794. [DOI] [PubMed] [Google Scholar]
- Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13:2671–2678. doi: 10.1093/hmg/ddh289. [DOI] [PubMed] [Google Scholar]
- Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, et al. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding S, et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–934. doi: 10.1038/nature04655. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yuri T, Mindell DP. Molecular phylogenetic analysis of Fringillidae, “New World nine-primaried oscines” (Aves: Passeriformes) Mol Phylogenet Evol. 2002;23:229–243. doi: 10.1016/S1055-7903(02)00012-X. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- Zink RM, Dittmann DL, Rootes WL. Mitochondrial DNA variation and the phylogeny of Zonotrichia. Auk. 1991;108:578–584. [Google Scholar]