Abstract
Anthrax toxin receptor 1 (ANTXR1) / tumor endothelial marker 8 (TEM8) is one of two known proteinaceous cell surface anthrax toxin receptors. A metal ion dependent adhesion site (MIDAS) present in the integrin-like inserted (I) domain of ANTXR1 mediates the binding of the anthrax toxin subunit, protective antigen (PA). Here we provide evidence that single point mutations in the I domain can override regulation of ANTXR1 ligand-binding activity mediated by intracellular signals. A previously reported MIDAS-mutant of ANTXR1 (T118A) was found to retain normal metal ion binding and secondary structure but failed to bind PA, consistent with a locked inactive state. Conversely, mutation of a conserved I domain phenylalanine residue to a tryptophan (F205W) increased the proportion of cell-surface ANTXR1 that bound PA, consistent with a locked active state. Interestingly, the KD and total amount of PA bound by the isolated ANTXR1 I domain was not affected by the F205W mutation, indicating that ANTXR1 is preferentially found in the active state in the absence of inside-out signaling. Circular dichroism (CD) spectroscopy and 1H-15N heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) revealed that structural changes between T118A, F205W and WT I domains were minor despite a greater than 103-fold difference in their abilities to bind toxin. Regulation of toxin binding has important implications for the design of toxin inhibitors and for the targeting of ANTXR1 for anti-tumor therapies.
Keywords: Bacterial toxins; integrin; receptor regulation; receptor structure-function; anthrax toxin; anthrax toxin receptor; ANTXR; inserted domain; metal ion dependent adhesion site, MIDAS; tumor endothelial marker 8, TEM8
Bacillus anthracis produces the disease known as anthrax through the secretion of a tripartite AB type toxin. The toxin consists of a single binding moiety termed protective antigen (PA), and two catalytic subunits termed lethal factor (LF) and edema factor (EF). These constituents combine to produce lethal toxin (PA + LF) and edema toxin (PA + EF) (1, 2). Cellular intoxication begins with PA binding one of two proteinaceous cell surface receptors, termed anthrax toxin receptor 1 / tumor endothelial marker 8 (ANTXR1/TEM8) or anthrax toxin receptor 2 / capillary morphogenesis gene 2 (ANTXR2/CMG2) (3, 4). Cleavage of PA by either serum or cell–surface proteases allows oligomerization and subsequent catalytic subunit binding (5–7). The receptor–bound toxin complex is endocytosed in a receptor mediated, clathrin dependent process (8). A subsequent drop in endosomal pH yields structural rearrangement of oligomeric PA followed by pore formation and delivery of the catalytic units into the cytosol (9–12). LF, a Zn2+ dependent metalloprotease, cleaves mitogen activated protein kinase kinases (MKKs) (2, 13). EF, a Ca2+ / calmodulin dependent adenylate cyclase, yields an increase in intracellular cAMP (1, 2, 14).
The cell surface receptors responsible for PA binding, ANTXR1 and ANTXR2, are type I transmembrane proteins, with 40% overall amino acid identity, 60% identity within the extracellular I domain, and a 100% conserved metal ion dependent adhesion site (MIDAS) motif (3, 4, 15). ANTXR1 and ANTXR2 bind PA via their I domains in a MIDAS-dependent manner (3, 4), with the MIDAS-bound metal providing a direct link between the I domain and PA (15–17). The contribution of a key acidic residue from PA (D683) completes MIDAS cation coordination. Removing this charge contribution from PA (D683N) yields an abrogation or reduction in PA binding to ANTXR1 and ANTXR2, respectively (15, 17–19). Treatment with EDTA or mutation of the amino terminal MIDAS aspartate in ANTXRs abolishes cation coordination and ligand binding (15, 19, 20). These findings and subsequent co-crystal structures led us to hypothesize that PA binding to ANTXRs resembles ligand binding to α integrin I domains (16, 17, 21) (Supp. Fig. 1).
Integrin I domains exist in active or inactive states that were defined biochemically based on ability to bind ligand (21, 22). Structural analysis revealed that integrin I domains adopt two conformational states, termed open and closed, that were proposed to correspond to active and inactive states respectively (23–25). This led to a model whereby activation status is controlled through conformational switch. While inactive mutants of ANTXRs have been reported (15, 19, 26), it is unknown if the ANTXRs can adopt a closed conformation (27).
The ANTXR1 gene is expressed as three known splice variants, termed ANTXR1-sv1, -sv2, and –sv3, which encode for proteins with a long 221 amino acid cytoplasmic tail, a short 25 amino acid cytoplasmic tail, and a secreted form, respectively (4, 28). Recently, Go and colleagues demonstrated that cells expressing ANTXR1-sv2 bound four-fold more PA than cells expressing ANTXR1-sv1 when normalized for cell-surface ANTXR1 expression levels (26). The authors hypothesized that this difference in PA binding resulted from inside-out signaling in which an intracellular signal is transduced to the extracellular I domain resulting in conformational changes and alterations in ligand binding activity. Specifically, it was suggested that ANTXR1-sv1 exists as a mixture of open and closed conformations, while ANTXR1-sv2 exists primarily in the open conformation. The cytosolic tail of –sv1 directly interacts with cellular actin and a point mutation in the cytoplasmic domain (Y383C) disrupts the actin interaction and alleviates repression of PA binding (26, 29, 30). While these data are consistent with inside-out signaling, the mechanism by which alterations in the cytosolic tail lead to changes in PA binding is still unclear.
In this work we test whether mutation of conserved I domain residues can alter the cellular regulation of PA binding. We confirm that the equilibrium between active and inactive states is regulated via the long –sv1 tail, and show that control of this equilibrium may be overridden via point mutations in the I domain. In contrast to integrin I domains, the isolated extracellular ANTXR1 I domain is preferentially in an activated ligand binding state. These studies support a model whereby PA binding to ANTXR1 is regulated via inside-out signaling that is propagated through the extracellular I domain.
MATERIALS AND METHODS
Construction of ANTXR variants
QuickChange™ site directed mutagenesis was performed according to manufacturer’s protocol to introduce point mutations into ANTXR1 splice variant 1, 2 and soluble I domain (sANTXR1). The oligonucleotide 5’-CCCGTGAATGACGGCTGGCAGGCTCTGCAAGGC-3’ and its reverse complement were used to generate the F205W point mutation. The T118A mutation has been previously described (15). To generate the sANTXR1 I domain containing the T118A mutation, the I domain region from ANTXR1(T118A) was PCR amplified with primers 5’-CGGGATCCGAGGATGGGGGTCCAGCCTGCTAC-3’ and 5’-TTTTCCTTTTGCGGCCGCTTATTCAGCTGCTAGAATTTCGATGCAGGAC-3’ that introduce BamHI and NotI restriction sites and sub-cloned into the pGEX-4T-1 vector (GE Healthcare).
Reagents and Proteins
All reagents were from Sigma-Aldrich or EMD Biosciences unless otherwise noted. sANTXR1 I domain was purified as previously described (31) with the following notable exceptions: 1 L of Minimal Media (75 mM KH2PO4, 74 mM K2HPO4, 63 mM Na2HPO4, 14 mM K2SO4, 21 mM NH4Cl, 0.21 mM CaCl2-2H2O, 0.11 mM FeSO4-7H20, 0.03 mM MnCl2-4H20, 0.017 mM CoCl2-6H2O, 0.012 mM ZnSO4-7H2O, 0.009 mM CuCl2-2H2O, 0.0015 mM H3BO3, 0.001 mM (NH4)6Mo7O24-4H2O, 0.067 mM EDTA), was inoculated with a 1:167 dilution of overnight culture and grown at 37°C until OD600 reached 1.0. Culture was induced with 0.05 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology), and grown overnight at 25°C. The bacterial pellet from 5 L of culture was resuspended in 42 mL Buffer B (20 mM Tris-HCl, 5 mM EDTA, pH 7.5) and lysed via French Press. Filtered supernatant was loaded onto a Hightrap Q Fast Flow (FF) 5 mL column (GE Healthcare), the column was attached to a Bio-Rad BioLogic DuoFlow system, and protein was eluted using a 0 – 0.5 M NaCl linear gradient. Fractions containing protein of interest as determined by SDS-PAGE and Gel-Code Blue Stain Reagent (Thermo Scientific) were combined. Pooled fractions were loaded onto 5 mL Glutathione Sepharose 4 FF beads (GE Healthcare). GST-tagged protein was eluted with GST elution buffer (10 mM reduced glutathione, 50 mM Tris-HCL, pH 8.0). For removal of GST tag, 10 unit/mg thrombin was added to protein pool and cleavage reaction proceeded overnight at room temperature (RT). Cleaved protein was loaded onto Hightrap Q FF column and eluted as described above. Fractions of interest as determined by SDS-PAGE and Gel-Code Blue staining were pooled. A glutathione sepharose column was used to remove free GST and uncleaved GST tagged protein. Protein was concentrated in an Amicon Ultra-4 or 15 Centrifugal Unit with 10,000 MWCO membrane to 0.5 mL. SDS-PAGE and Gel-Code Blue staining were performed to confirm purity of protein. sANTXR1 I domain was flash frozen in liquid nitrogen and stored at −80°C.
ELISA
PA was purified as previously described (32) and was diluted in TBS (137 mM NaCl, 2.7 mM KCl, 25 mM Tris-Base, pH 7.4) to 5 µg/mL and 40 µL / well was adsorbed to a 384-well microtiter plate overnight at 4°C. As a negative control, TBS + 3 % bovine serum albumin (BSA) was utilized instead of PA. Wells were washed and blocked with TBST (TBS + 0.1% Tween 20) + 3% BSA for 1 hour at RT. GST tagged sANTXR1s were diluted in TBST + 3% BSA + cation (1 mM CaCl2, MgCl2, or MnCl2) or 10 mM EDTA and allowed to bind for 1 hour at RT. The plate was washed 3×, then 40 µL of horseradish peroxidase conjugated rabbit anti-GST (diluted 1:10,000) was added to each well and incubated 1 hour at RT. Plate was washed 3×, then 50 µL 1-Step Ultra TMB-ELISA was added to each well and incubated at RT for 20 minutes. Fifty microliters of 2 M sulfuric acid was added to each well to stop the reaction. Absorbance was measured at 450 nm.
Cell Lines / Transduction
Generation of PA receptor-deficient CHO-R1.1 cells derived from CHO-K1 cells is described elsewhere (4). ANTXR1 DNA was stably introduced into CHO-R1.1 cells with retroviral vectors as previously described (15). CHO-R1.1 cells were maintained in F12 medium with 10% fetal bovine serum (Invitrogen) and 1% penicillin-streptomycin (Sigma-Aldrich). Transduced receptor positive cells were cultured in the presence of 0.8 mg/mL G418.
Cell-Based PA Binding Assays
Because WT PA rapidly oligomerizes and forms a high avidity complex, a protease-resistant PA mutant (PASSSR) (6) was utilized in order to accurately gauge the binding interaction of monomeric PA to ANTXR1. In addition, a single lysine to cysteine mutation that does not affect PA function was introduced [PASSSR(K729C)] for site-specific labeling. PASSSR(K729C) was labeled with Alexa Fluor 647 C2 maleimide as per manufacturer’s protocol (Pierce). Subconfluent 15 cm plates of receptor negative CHO-R1.1 or ANTXR1 transduced CHO-R1.1 cells were washed with DPBS without Ca2+ or Mg2+ (Cellgro). Plates were then incubated with 3 mL of DPBS + 1 mM EDTA for 5 minutes at RT. Detached cells were then washed 3× with Media or TBS + 3% dialyzed FBS (TBS+) containing 2 mM CaCl2, MgCl2, MnCl2, or EDTA. 1 ×106 cells were incubated in the presence of media or TBS+ (with cation or EDTA) with AlexaFluor-647 labeled PASSSR(K729C). PA protein concentrations ranged from 0.2 nM to 1600 nM. Cells were incubated on ice at 4°C while rocking for 6 hours, then pelleted at 800 × g in an Eppendorf microcentrifuge for 1 min. Cells were then washed 3× in their respective solution, resuspended in 500 µL PBS + 1% formaldehyde, then analyzed by flow cytometry using a FACSCalibur (BD Biosciences). Geometric mean fluorescence values for PA binding (AlexaFluor-647) were determined for cell populations gated on FSC and SSC, or additionally on a narrow window of EGFP fluorescence (115 – 155 relative fluorescence units) using Cell Quest (BD) and FlowJo (Tree Star, Inc.) software. Binding curves were determined using GraphPad Prism 4 software (GraphPad Software, Inc.) with KD determined via non-linear regression using equation 1:
| (1) |
Circular Dichroism
The GST tag was removed from sANTXR1 proteins as described above then buffer exchanged via Zebra Spin Desalting Columns (Pierce) into CD Buffer (50 mM NaCl, 10 mM Na2HPO4-7H2O, pH 7.4). Proteins were diluted to 0.2 mg/mL and concentration verified by Bio-Rad Protein Assay Kit (Bio-Rad) and equivalent band intensity as determined by 12% SDS-PAGE and Gel-Code Blue staining (33). Two-hundred microliters of each sample were analyzed on Jasco J-715 in a 0.1 cm pathlength cuvette. Temperature was kept at a constant 25°C with a Jasco PTC-348 thermoelectrically controlled cell holder. Far-UV spectra were accumulated from 260 nm to 197 nm. CD spectra were normalized by subtraction of the background scan with buffer alone. The fraction of secondary structure was computed with the self-consistent method of Sreerama and Woody (SELCON3) (34) with 10 iterations to convergence utilizing the multiple algorithms of Hennesey & Johnson (35), Kabsch & Sander (36), and Leavitt & Greer (37) for consistency.
Inductively coupled plasma optical emission spectrometry (ICP-OES)
sANTXR1 variants were incubated with 5 mM EDTA for > 10 minutes at 25°C. EDTA was removed via Zebra Spin Desalting Columns equilibrated in PBS (137 mM NaCl, 27 mM KCl, 43 mM Na2HPO4-7H2O, 1.5 mM KH2PO4, pH 7.4). Each sample was then incubated with 1 mM MnCl2 or PBS alone for > 5 minutes to allow cation binding. All samples were then buffer exchanged again with Zebra Spin Desalting Columns into ICP grade H2O to remove free cations. Samples ranged from 0.05 – 0.2 mg total protein. An equal volume of Optima grade nitric acid (Fisher) was added to each sample and proteins were digested for 2 hours at 95°C. Samples were then diluted to a 5% HNO3 working concentration. A standard curve was generated with known concentrations of Mn2+ in 5% HNO3. Triplicate readings were taken on a TJA Radial Iris 1000 ICP-OES for each sample. The ratio of sANTXR1 to Mn2+ was then calculated utilizing sANTXR1 concentrations within range of the standard curve (0 – 4 µM). Data presented are the average of three independent experiments.
Surface Plasmon Resonance
PASSSR(K729C) was conjugated to a CM5 chip via thiol linkage as per manufacturer’s recommendation (Biacore). All setup injections were at a flow rate of 7 µL/min. In short: A 1:1 mix of N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was injected for 2 min. PDEA (80 mM 2-(2-pyridinyldithio) ethaneamine hydrochloride in 500 µL boric acid, pH 8.5) was injected for 4 min, 20 µg/mL PASSSR(K729C) in (10 mM CH3COONa-3H2O, pH 4.3) was then injected for 6 minutes, followed by L-cysteine (50 mM in 1 M NaCl, 100 mM CH3COONa-3H2O) injected for 4 minutes. Injections were repeated for flow cell 1 (FC1) without PASSSR(K729C) to blank FC1. Binding experiments were conducted at 25°C with a running buffer of HBS (10 mM HEPES, 150 mM NaCl, pH 7.4, 0.05 % P20) + 1 mM CaCl2. sANTXR1 receptor concentrations ranging from 0.05 µM – 4 µM were flowed at 30 µL/min for 100 sec, allowed to disassociate for 180 sec, and then the surface was regenerated with Regeneration Solution (0.5 M Na2CO3, pH 10.5) applied for 30 seconds with a 120 second recovery. Data were analyzed on Biacore T100 evaluation software using the steady state equilibrium affinity model derived from equation 2,
| (2) |
Req is steady state binding level, C is concentration of analyte, Rmax is analyte binding capacity at the sensor surface, KD is overall equilibrium dissociation constant, and RI is refractive index. At least 3 independent experiments were run for each sANTXR1.
Sample Preparation for 1H-15N HSQC NMR
NMR studies were performed on His6-sANTXR1 variants (residues 34 to 227 with the sequence HHHHHHLVPRGS appended to the N-terminus). Isotopically labeled proteins were produced as described above except that the minimal media utilized 15NH4Cl as the sole nitrogen source. Proteins were purified from 10 L cultures. Cell pellets were resuspended in 42 mL of Cobalt Binding Buffer (CoBB: 20 mM Na2HPO4, 0.5 M NaCl, 10 mM imidazole, pH 7.4) and lysed via French Press at ≥ 1500 PSI. The supernatant was then passed through 0.45 µm syringe filter. The filtered supernatant was then loaded onto a 1 mL HiTrap Chelating HP column equilibrated with CoCl2 (Amersham) and eluted with a linear gradient of Cobalt Elution Buffer (CoEB: CoBB + 500 mM imidazole). Samples were then separated via size exclusion chromatography (Sephacryl S-200 HR, GE Healthcare) and pure fractions, as determined via SDS-PAGE and Gel-Code Blue Stain Reagent, were pooled. Proteins were dialyzed with 12,000–14,000 MWCO Spectra/Por 2 Dialysis Membrane into NMR Buffer (50 mM NaH2PO4, 300 mM NaCl, pH 6.5) and concentrated to 500 µL via Amicon Ultra-4 or -15 Centrifugal Unit with a 10,000 MWCO membrane. Samples were then spiked with dimethyl ethyl ammonium propane sulfonate (NDSB-195) (Anatrace) to 100 mM final concentration. Final protein concentrations were 2 – 10 mg/mL. All samples contained 7% D2O.
The 1H-15N HSQC spectra, with sensitivity-enhancement, were recorded at 299 K on a CryoProbe-equipped Bruker Avance-600 MHz spectrometer. Spectra were acquired with 1024 × 128 complex data points in the 1H and 15N dimension with corresponding spectral widths of 8389 Hz and 2190 Hz, respectively. The proton carrier frequency was set to the water resonance. Data processing and analysis was performed using the programs NMRPipe and Sparky, respectively (38, 39).
RESULTS
F205W locks ANTXR1 into an active state on the cell surface
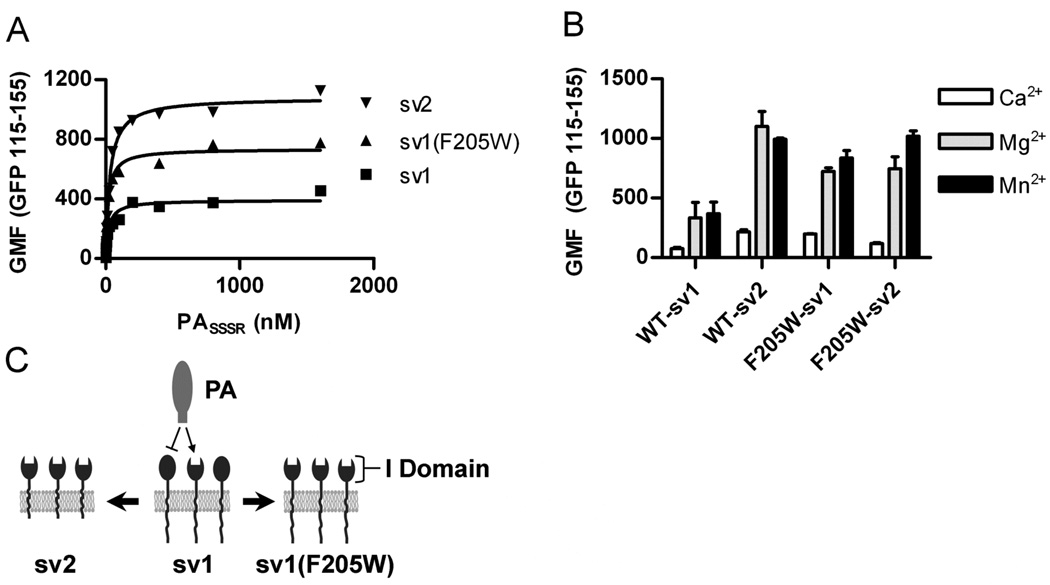
Go et al recently demonstrated that the presence of the long cytosolic tail in ANTXR1-sv1 yields a decrease in the total amount of bound PA (26). Using a flow cytometry assay in which expression of ANTXR1-EGFP fusion proteins is normalized by gating on EGFP fluorescence, we confirm Go’s results and show ANTXR1-sv2 expressing cells bind 2 – 3 fold more PA than ANTXR1-sv1 expressing cells (Fig. 1A and C). Next, a single point mutation, F205W, was introduced into ANTXR1-sv1 and -sv2. This phenylalanine is conserved across integrin I domains and ANTXR1, and substitution of a tryptophan for this conserved phenylalanine in the αM integrin overrides inside-out signaling resulting in a 2 – 3 fold increase in ligand binding (21, 25). Consistent with these findings, introduction of the F205W point mutation increased PA binding by ANTXR1–sv1 (Fig. 1). The increase in total PA binding in Ca2+ or Mn2+ reached levels comparable to WT ANTXR1–sv2 (Fig. 1B). No further increase in PA binding was observed when the F205W mutation was engineered into ANTXR1-sv2 (Fig. 1B) indicating that this mutation influences activation status of ANTXR1 rather than interacting directly with PA (Fig. 1C).
Figure 1. ANTXR1(F205W) is locked into the active conformation.
CHOR1.1 cells expressing the indicated ANTXR1-EGFP fusion proteins were incubated with AlexaFluor 647 labeled PASSSR(K729C) for 6 h on ice, and PA binding was measured by flow cytometry. ANTXR1 expression was normalized by gating on equivalent EGFP signal (115 – 155 relative fluorescence units) from each sample. (A) PA was titrated in the presence of 2 mM MgCl2 and binding measured via flow cytometry. Data shown are representative of two independent experiments with each point corresponding to the geometric mean fluorescence (GMF) of >300 individual cells. (B) The relative contribution of specific cations was determined by incubating 100 nM PA with receptor expressing cells in the presence of 2 mM CaCl2, MgCl2, or MnCl2 and analyzing as in (A). Data points represent the mean ± standard deviation (SD) for three independent experiments. (C) A model depicting the differences in ANTXR1-sv1, -sv2, or the I domain point mutant -sv1(F205W) ability to bind anthrax protective antigen (PA). Two types of receptor activation states exist, relative to PA binding ability. The membrane bound short-tailed sv2 receptors are in the active state and bind PA. The long-tailed sv1 receptors exist in an equilibrium of active and inactive receptors where active receptors are capable of binding PA (arrow) and inactive receptors do not bind PA (line-bar). The I domain point mutation F205W increases the percent of active sv1 receptor capable of binding PA and this overrides inside-out signaling through the cytoplasmic tail, which restricts binding to a proportion of the ANTXR1-sv1. Closed spheres represent inactive receptors whereas open spheres represent active receptors.
PA binding of the soluble ANTXR1 I domain is not affected by the F205W mutation
Previous studies with isolated integrin I domains demonstrate that these I domains preferentially adopt an inactive state when expressed as isolated domains (21, 24, 40). In contrast, several studies suggest that the isolated ANTXR2 I domain is fully active (16, 17, 27, 41). Therefore, to test whether the isolated ANTXR1 I domain exists in an active or inactive state we expressed WT and point mutants as soluble receptor proteins (sANTXR1). We tested F205W, which increases total PA binding (Fig. 1A and B) and the previously described I domain MIDAS variant T118A which yields a 102 – 103-fold reduction in receptor activity (15).
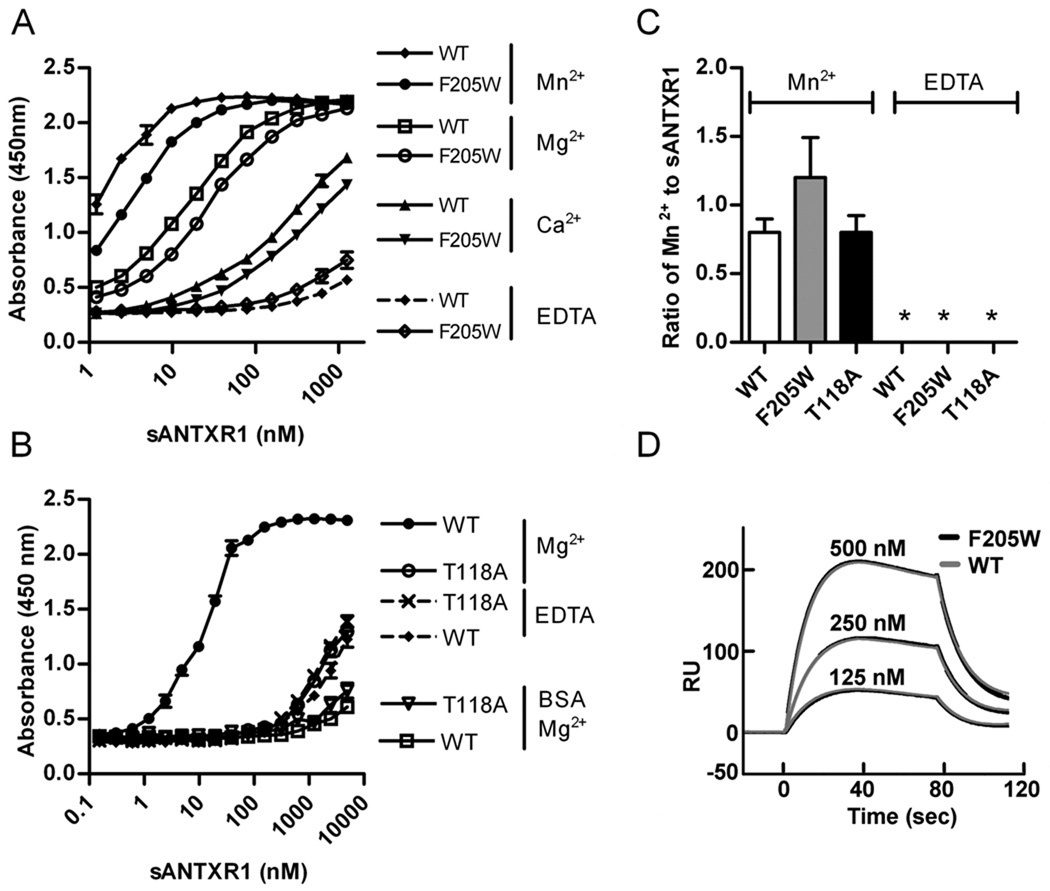
The ability of each sANTXR1 to bind PA was first assessed by ELISA. As previously reported (15), WT sANTXR1 showed dose-dependent binding to PA that was dependent on the divalent cation present, with Mn2+>Mg2+>Ca2+ (Fig. 2A). In contrast, sANTXR1(T118A) displayed a ~103-fold reduction in the ability to bind PA (Fig. 2B). Interestingly, residual PA binding by sANTXR1(T118A) appeared cation-independent, as similar binding was observed in the presence of EDTA. One possible explanation is that the T118A mutation disrupts cation coordination. However, a similar threonine to alanine mutation in the αM MIDAS was suggested to retain metal binding, though evidence for this was indirect (20, 21). To directly address this concern, we determined the metal binding capacity of WT and mutant receptors via ICP-OES. sANTXR1(WT), (F205W), and (T118A) all bound an equimolar ratio of Mn2+ indicating that the point mutations did not alter ability of receptor to bind cation (Fig. 2C) and implicating a MIDAS-dependent regulation of PA/ligand binding.
Figure 2. PA binding of the soluble ANTXR1 I domain is not affected by the F205W mutation.
(A, B) PA or BSA was adsorbed to a 384-well plate and the indicated GST-sANTXR1 proteins were titrated in the presence of 1 mM Ca2+, Mg2+, Mn2+, or EDTA. Bound GST-ANTXR1 proteins were detected using anti-GST-HRP and developed with 1-Step TMB as described in methods. Results shown are representative of at least two independent experiments performed in triplicate. Data represent mean ± the standard error (SE). (C) sANTXR1 proteins were incubated with 5 mM EDTA to remove residual cations, buffer exchanged to remove EDTA, and incubated with 1 mM MnCl2 or ddH2O at RT. Free metal was removed and proteins digested in Optima trace metal grade nitric acid. The molar ratio of bound Mn2+ per receptor molecule was determined via ICP-OES at wavelengths 257.610 nm, 259.373 nm, and 260.569 nm. An asterisk indicates a value below the detection limit. Data represent the mean ± SD for three independent experiments. (D) The F205W point mutation does not increase PA binding as measured by response units (RU) compared to WT sANTXR1 I domain. The binding curves for sANTXR1(WT) and sANTXR1(F205W) are nearly identical and overlap. PASSSR(K729C) was conjugated to a CM5 chip via thiol exchange and sANTXR1(WT) and (F205W) flowed over chip using a Biacore T-100. Sensorgrams are shown with concentrations of sANTXR1(WT) (black lines) and sANTXR1(F205W) (grey lines) indicated.
Interestingly, the ELISA binding curves were similar between sANTXR1(F205W) and WT sANTXR1 (Fig. 2A), indicating that this mutation did not affect the proportion of soluble I domain that is active. To further test PA binding ability of sANTXR, we employed surface plasmon resonance, which was previously utilized to determine the fraction of the soluble αM I domain in active versus inactive states (21, 42, 43). Consistent with ELISA results, the F205W mutation did not alter the absolute amount of equilibrium binding between sANTXR1 and PA (Fig. 2D), indicating that the proportion of sANTXR1 in the active state is similar between WT and F205W forms.
Point mutations in the soluble ANTXR1 I domain do not affect secondary structure
The increased PA binding seen with ANTXR1-sv1(F205W) and the decreased PA binding by sANTXR1(T118A) are consistent with structural changes associated with open and closed conformations reported in integrin I domains (21, 40). However, because the T118A mutation results in a large reduction in PA binding, it remained possible that this mutation affected PA binding by perturbing protein structure. Therefore, we tested whether sANTXR1(T118A) maintained proper folding and, further, whether structural differences could be detected between the active and inactive forms of sANTXR1 using CD spectroscopy.
No noticeable changes were observed in the CD spectra of sANTXR1(F205W) or sANTXR1(T118A) when compared to WT (Fig. 3). As a basis for comparison, the CD spectrum for sANTXR2, which retains 60% amino acid identity to sANTXR1, was analyzed and found to match predictions based on published X-ray crystallography structures. Interestingly, sANTXR2 exhibited a significant difference in percent of α-helix (Fig. 3; Table 1). Percent of random coil, β-sheet, and turn were similar for both sANTXR1 and -2. Thus, while small differences in secondary structure exist between sANTXR1 and -2, the differences in PA binding between WT, F205W and T118A variants of sANTXR1 are not due to misfolding or alterations in content of specific secondary structures.
Figure 3. Point mutations in the soluble ANTXR1 I domain do not affect secondary structure.
CD spectra were recorded in the far-UV range at 25°C for 0.2 mg/mL sANTXR1(WT), (F205W), and (T118A), as well as sANTXR2 (solid-black, black-dash, solid-grey, and long black dash, respectively). Analysis of the spectrum from sANTXR2 reveals different relative proportions of alpha helix compared with sANTXR1 as listed in Table 1.
Table 1.
Circular Dichroism Evaluation of the Secondary Structure of sANTXR1 and sANTXR2a
| Alpha Helix | Beta Sheet | Turn | Other | |
|---|---|---|---|---|
| sANTXR2 | 48.4 ± 3.6 | 14.7 ± 4.9 | 19.1 ± 2.7 | 19.3 ± 7.5 |
| sANTXR1(WT) | 34.5 ± 3.7 | 19.6 ± 6.3 | 21.8 ± 2.9 | 22.9 ± 9.8 |
| sANTXR1(F205W) | 34.9 ± 2.9 | 21.0 ± 7.0 | 20.8 ± 4.0 | 21.3 ± 9.6 |
| sANTXR1(T118A) | 36.9 ± 3.0 | 20.8 ± 7.2 | 20.6 ± 4.2 | 22.6 ± 10 |
The fraction of secondary structure was computed with the self-consistent method of Sreerama and Woody (SELCON3) (34) utilizing multiple algorithms.
Point mutations in the sANTXR1 I domain induce only minor shifts in tertiary structure
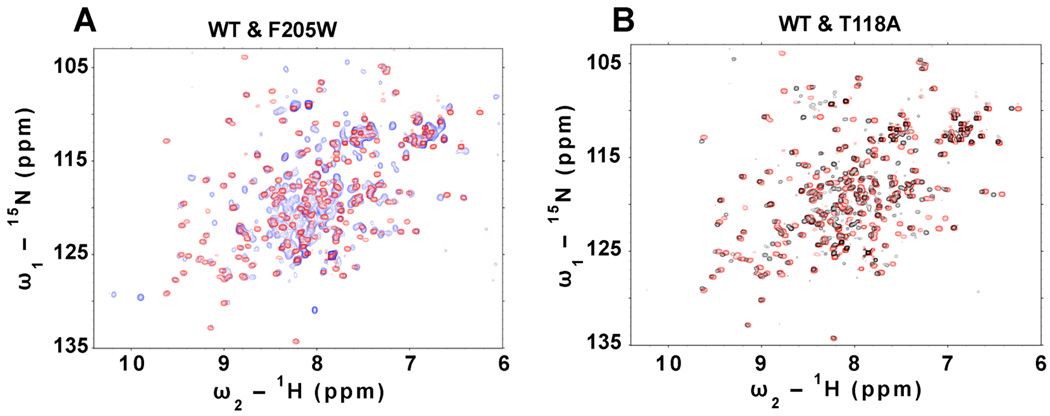
To further evaluate the effects of the F205W and T118A point mutations 1H-15N HSQC NMR spectra were recorded. Both mutant proteins are folded as judged by the linewidths and dispersion of the amide cross peaks in their NMR spectra (Fig 4). Notably, the spectrum of each mutant is generally similar to the spectrum of the wild-type protein indicating that the F205W and T118A mutations do not disrupt the global fold (Fig. 4). The F205W mutant exhibits more significant spectral changes as compared to the T118A mutant. This is presumably caused by introduction of the indole ring in this mutant which can cause significant ring current shifts. The majority of resonances in the F205W, T118A, and WT spectra are superimposable suggesting that they adopt similar conformations. Unfortunately the low solubility and yield of isotopically produced protein prohibits a more extensive analysis by NMR at this time.
Figure 4. 1H-15N HSQC NMR of sANTXR1.
1H-15N HSQC NMR indicates proper structural folding of sANTXR1 WT and mutants as judged by the linewidths and dispersion of the amide cross peaks in their NMR spectra. (A) An overlay comparison of the 1H-15N HSQC spectrum of sANTXR1(WT) and (F205W) shown in red and blue, respectively. The majority of the resonances in the spectra are superimposable, indicating similar conformations. (B) Overlay of sANTXR1(WT) and (T118A) shown in red and black, respectively. As in (A), the vast majority of resonances overlap.
ANTXR1 affinity for PA is unaffected by the F205W mutation
To address whether I domain activation states alter ANTXR1 affinity for PA, KD values were determined using the flow cytometry assay described in Fig. 1A. The KD for PA binding to cells expressing ANTXR1-sv1, in which most of the ANTXR1 is inactive, was compared to cells expressing ANTXR1–sv2, in which ~3-fold more receptor is in the active state. In addition, we measured the KD of PA binding to cells expressing ANTXR1(F205W)-sv1 and –sv2. Finally, we performed surface plasmon resonance studies to measure PA binding to WT, F205W, and T118A sANTXR1 variants. Due to slow off-rates in the presence of Mg2+, we were unable to reliably determine the affinity of the isolated I domains in the presence of cations other than Ca2+ via SPR. Therefore, in order to rule out differences in KD associated with alterations in cation usage, cell based PA binding assays were performed in TBS with 1 mM Ca2+ or Mg2+ rather than in culture media.
In both cell-based and SPR assays, affinity of ANTXR1(F205W) binding to PA was unchanged compared to WT ANTXR1 (Table 2). Consistent with the findings of Go et al (26), PA affinities for ANTXR1–sv1 and –sv2 were also similar. Binding of sANTXR1(T118A) was undetectable by SPR and therefore affinity could not be calculated (data not shown). Taken together, these data suggest that low-level PA binding to the inactive receptor does not contribute to the measured KD. Further, the increase in PA binding by ANTXR1(F205W)-sv1 is not the result of an increase in measured affinity, but may instead be due to an increase in the number of receptors in the active state.
Table 2.
SPR and Cell-based Affinity Measurementsa
| ANTXR1(WT) | KD (nM) |
|---|---|
| SPR | 1092 ± 269.0 |
| Cell Based-sv1 | 287.7 ± 28.24 |
| Cell Based-sv2 | 267.5 ± 43.09 |
| ANTXR1(F205W) | |
| SPR | 1173 ± 31.39 |
| Cell Based-sv1 | 223.5 ± 62.07 |
| Cell Based-sv2 | 400.8 ± 20.63 |
The binding affinity of sANTXR1(WT) and (F205W) to PASSSR(K729C) conjugated to a CM5 chip via thiol exchange was determined by surface plasmon resonance. Steady state equilibrium affinity model was used to calculate KD values from at least 3 independent experiments for each sANTXR. Data are the mean KD ± SE where KD is the equilibrium dissociation constant (nM). Cell-based affinity of ANTXR1-sv1 and -2 with and without F205W point mutation was determined by incubating receptor-expressing CHO cells in the presence of 1 mM CaCl2 on ice with a 0 – 1.6 µM titration of AlexaFluor 647 labeled PASSSR(K729C) for 6 hours. Geometric mean fluorescence was measured via flow cytometry and KD values determined using Prism GraphPad 4 software.
In contrast to previous reports (26, 44, 45), the KD value for the isolated WT I domain was only 3 – 5 fold higher than KD values for the full length ANTXR1-sv1 and -sv2 receptors expressed on Chinese hamster ovary cells in defined cation conditions. Therefore, removing the ANTXR1 I domain from the cell-surface receptor does not substantially alter KD with respect to PA binding.
DISCUSSION
Structural and biochemical studies show ANTXRs interact with PA in a fashion similar to known integrin I domain / ligand interactions (15–17, 19, 27). Consistent with studies on integrins (20, 21), we demonstrate that mutation of a conserved I domain phenylalanine (F205W) (Fig. 1A and B) or MIDAS threonine (T118A) (Fig. 2B) is able to override cytosolic regulation that controls the distribution of ANTXR1–sv1 between active and inactive states.
The increase in PA binding to ANTXR1(F205W)-sv1 is not the result of an increase in affinity in all receptor molecules, but rather the result of an increase in the population of activated receptor. Accordingly, engineering the F205W mutation into ANTXR1-sv2, which displays 3 – 4-fold more PA binding than ANTXR-sv1, gave no additional increase in PA binding. Conversely, the T118A mutation resulted in loss of PA binding without disrupting protein folding or metal binding. These data support the hypothesis that affinity regulation is achieved via inside-out signaling that alters the extracellular I domain activation state and that this regulation can be overcome by alterations in the I domain.
While our results are consistent with previous studies on integrins, the mechanism by which alteration of I domain residues affects PA binding are less clear. At least three models exist that may explain control of PA binding. First, as proposed for integrins, F205W and T118A may lead to stabilization of open and closed conformations respectively (19–21). Alternatively, these mutations may alter PA binding in the absence of large conformational changes. Finally, F205 and T118 may be involved in interactions with other host proteins, such as coreceptors. Each of these possibilities is discussed below.
First, I domains can adopt either an open or closed conformation, corresponding to an active or inactive state, respectively (Supp. Fig. 1A and B) (22). The aromatic side-chain of a conserved phenylalanine (F205 in ANTXR1) is buried in a MIDAS-adjacent hydrophobic pocket in the closed conformation, but is flipped out of the hydrophobic pocket and exposed to the aqueous milieu in the open conformation (21, 27, 46, 47) (Supp. Fig. 1). Li et al hypothesized that the substitution of this conserved phenylalanine by a residue with a bulky sidegroup, such as tryptophan, prevents occupancy of the hydrophobic pocket and thus “locks” the integrin I domain into an open conformation (21). The finding that ANTXR1(F205W)-sv1 displays increased PA binding is consistent with this hypothesis.
Regulation of integrin/ligand binding is additionally governed by the metal coordination status of the conserved MIDAS threonine (48). In the open conformation, the hydroxyl side chain of this threonine residue directly coordinates the MIDAS cation, though it does not directly participate in ligand binding (20, 48) (Supp. Fig. 1). In the closed conformation, the MIDAS threonine indirectly coordinates the cation. Mutation of this threonine to alanine (T118A in the ANTXRs) reduces ligand binding activity, which was hypothesized to result from preferentially stabilizing the I domain in a closed conformation (20, 21). However, the possibility of structural perturbation or loss of cation binding leading to a reduction in PA binding had not been previously addressed. We show here that the T118A mutation does not disrupt cation coordination or folding of the sANTXR1 protein. Therefore a model whereby regulation of PA binding is affected by MIDAS-associated conformational change or conformation-independent alteration of MIDAS-mediated ligand binding is the most plausible.
Lacy et al hypothesized that the ANTXR2 I domain would be unable to adopt a closed conformation based on structural characteristics of this molecule (27). The finding that the sANTXR1(T118A) and (F205W) CD and 1H-15N HSQC NMR spectra are similar to WT indicates that structural changes for this related I domain may also be limited. If true, a transition between an active and inactive ANTXR1 I domain may not require major structural rearrangements typically attributed to open and closed conformations. Further structural studies are required to address this in detail.
Finally, the differences seen in ANTXR1 activation state may be the result of receptor clustering or interaction with a coreceptor (15, 19, 26). LRP6 was reported as a coreceptor for ANTXR1 (49), though other studies question the role of this host protein in PA binding (50–52). It is possible that association of ANTXR1 with LRP6 or other unidentified coreceptor(s) could increase the affinity of the ANTXR1–PA interaction. Indeed, a coreceptor or changes in avidity resulting from receptor clustering may also explain the 3 – 5 fold increase in observed KD reported here for the full-length receptors expressed on CHO-R1.1 cells compared to sANTXR1. However, mutation of F205 results in increased PA binding, indicating that any interaction with another factor mediated by this residue would normally be inhibitory to PA binding. This conclusion is opposite of the current model of how LRP6 functions, indicating that this coreceptor is not responsible for alterations in the activation status reported here.
It was previously suggested that differences in KD values between surface-expressed (9.5 – 21 nM in complete media) and sANTXR1 (130 nM in Mg2+ buffer and 1100 nM in Ca2+ buffer), could result from misfolded protein used in the sANTXR1 studies (44), a skewing of the intoxication-based cell binding data based on overdependence on on-rates (45), or interactions with a putative coreceptor (26). Here we report that affinities vary by only 3 – 5 fold when the cation is kept constant between assay systems. Thus, a coreceptor may lead to a slight increase in affinity on the cell surface, but is not required for PA binding. Of note, our data do not address whether a coreceptor may function to influence the activation status of ANTXR1-sv1. In addition to conformational changes, activation and/or ligand binding lead to integrin clustering resulting in increased avidity (22, 53–55). It was previously shown that ANTXR1 dimerizes on the cell surface and this may be responsible increased avidity and therefore increased apparent affinity (56).
Data presented here support a model for control of ANTXR1 receptor activity via inside-out signaling which yields a MIDAS-dependent regulation of PA binding. ANTXR1 likely exists in equilibrium between active and inactive states. Control of this equilibrium is governed by the cytoplasmic domain of the receptor and may be overridden via point mutations in the I domain engineered to favor active (F205W) or inactive (T118A) states. Future attempts to define how the cell modulates ANTXR1 affinity for PA via inside-out signaling will be important for approaches to counteract anthrax toxin, as well as numerous efforts to redirect PA to target tumors.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge support of the UCLA DOE biochemistry instrumentation facility, UCLA Jonsson Comprehensive Cancer Center (JCCC), the UCLA AIDS Institute and the flow cytometry, virology, mucosal immunology cores, which are supported by the NIH awards CA-16042 and AI-28697 (UCLA-CFAR grant), the UCLA AIDS Institute, and the David Geffen School of Medicine at UCLA.
ABBREVIATIONS
- NMR
nuclear magnetic resonance
- HSQC
heteronuclear single quantum coherence
- CD
circular dichroism
- ANTXR
anthrax toxin receptor
- sANTXR
soluble anthrax toxin receptor I domain
- ANTXR1-sv1 and -sv2
anthrax toxin receptor 1 splice variant 1 and 2
- PA
protective antigen
- EF
edema factor
- LF
lethal factor
- TEM8
tumor endothelial marker 8
- CMG2
capillary morphogenesis gene 2
- MIDAS
metal ion dependent adhesion site
- SEM
standard error of the mean
- SD
standard deviation
Footnotes
This work was funded by National Institute of Health (NIH) awards AI057870 to K.A.B., AI52217 to R.T.C., and F31GM075564 to V.A.V.
SUPPORTING INFORMATION
Supporting Information Available. Supplementary Figure 1 compares I domains of αM integrin and ANTXR2. Conserved residues discussed in this manuscript are identified. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Young JA, Collier RJ. Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu Rev Biochem. 2007;76:243–265. doi: 10.1146/annurev.biochem.75.103004.142728. [DOI] [PubMed] [Google Scholar]
- 2.Banks DJ, Ward SC, Bradley KA. New insights into the functions of anthrax toxin. Expert Rev Mol Med. 2006;8:1–18. doi: 10.1017/S1462399406010714. [DOI] [PubMed] [Google Scholar]
- 3.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 5.Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 6.Klimpel KR, Molloy SS, Thomas G, Leppla SH. Anthrax toxin protective antigen is activated by a cell surface protease with the sequence specificity and catalytic properties of furin. Proc Natl Acad Sci U S A. 1992;89:10277–10281. doi: 10.1073/pnas.89.21.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman DL, Zeng W, Rivera J, Nakouzzi A, Casadevall A. Human serum contains a protease that protects against cytotoxic activity of Bacillus anthracis lethal toxin in vitro. Clin Vaccine Immunol. 2008;15:970–973. doi: 10.1128/CVI.00064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abrami L, Liu S, Cosson P, Leppla SH, van der Goot FG. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J Cell Biol. 2003;160:321–328. doi: 10.1083/jcb.200211018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krantz BA, Melnyk RA, Zhang S, Juris SJ, Lacy DB, Wu Z, Finkelstein A, Collier RJ. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrami L, Leppla SH, van der Goot FG. Receptor palmitoylation and ubiquitination regulate anthrax toxin endocytosis. J Cell Biol. 2006;172:309–320. doi: 10.1083/jcb.200507067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller CJ, Elliott JL, Collier RJ. Anthrax protective antigen: prepore-to-pore conversion. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 12.Rainey GJ, Wigelsworth DJ, Ryan PL, Scobie HM, Collier RJ, Young JA. Receptor-specific requirements for anthrax toxin delivery into cells. Proc Natl Acad Sci U S A. 2005;102:13278–13283. doi: 10.1073/pnas.0505865102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klimpel KR, Arora N, Leppla SH. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 14.Leppla SH. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci U S A. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradley KA, Mogridge J, Jonah G, Rainey A, Batty S, Young JA. Binding of anthrax toxin to its receptor is similar to alpha integrin-ligand interactions. J Biol Chem. 2003;278:49342–49347. doi: 10.1074/jbc.M307900200. [DOI] [PubMed] [Google Scholar]
- 16.Lacy DB, Wigelsworth DJ, Melnyk RA, Harrison SC, Collier RJ. Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc Natl Acad Sci U S A. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santelli E, Bankston LA, Leppla SH, Liddington RC. Crystal structure of a complex between anthrax toxin and its host cell receptor. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 18.Shimaoka M, Xiao T, Liu JH, Yang Y, Dong Y, Jun CD, McCormack A, Zhang R, Joachimiak A, Takagi J, Wang JH, Springer TA. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scobie HM, Young JA. Divalent metal ion coordination by residue T118 of anthrax toxin receptor 2 is not essential for protective antigen binding. PLoS ONE. 2006;1:e99. doi: 10.1371/journal.pone.0000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamata T, Wright R, Takada Y. Critical threonine and aspartic acid residues within the I domains of beta 2 integrins for interactions with intercellular adhesion molecule 1 (ICAM-1) and C3bi. J Biol Chem. 1995;270:12531–12535. doi: 10.1074/jbc.270.21.12531. [DOI] [PubMed] [Google Scholar]
- 21.Li R, Rieu P, Griffith DL, Scott D, Arnaout MA. Two functional states of the CD11b A-domain: correlations with key features of two Mn2+-complexed crystal structures. J Cell Biol. 1998;143:1523–1534. doi: 10.1083/jcb.143.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo BH, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimaoka M, Lu C, Palframan RT, von Andrian UH, McCormack A, Takagi J, Springer TA. Reversibly locking a protein fold in an active conformation with a disulfide bond: integrin alphaL I domains with high affinity and antagonist activity in vivo. Proc Natl Acad Sci U S A. 2001;98:6009–6014. doi: 10.1073/pnas.101130498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimaoka M, Lu C, Salas A, Xiao T, Takagi J, Springer TA. Stabilizing the integrin alpha M inserted domain in alternative conformations with a range of engineered disulfide bonds. Proc Natl Acad Sci U S A. 2002;99:16737–16741. doi: 10.1073/pnas.252633099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimaoka M, Shifman JM, Jing H, Takagi J, Mayo SL, Springer TA. Computational design of an integrin I domain stabilized in the open high affinity conformation. Nat Struct Biol. 2000;7:674–678. doi: 10.1038/77978. [DOI] [PubMed] [Google Scholar]
- 26.Go MY, Chow EM, Mogridge J. The cytoplasmic domain of anthrax toxin receptor 1 affects binding of the protective antigen. Infect Immun. 2009;77:52–59. doi: 10.1128/IAI.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lacy DB, Wigelsworth DJ, Scobie HM, Young JA, Collier RJ. Crystal structure of the von Willebrand factor A domain of human capillary morphogenesis protein 2: an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2004;101:6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Leppla SH. Cell surface tumor endothelium marker 8 cytoplasmic tail-independent anthrax toxin binding, proteolytic processing, oligomer formation, and internalization. J Biol Chem. 2003;278:5227–5234. doi: 10.1074/jbc.M210321200. [DOI] [PubMed] [Google Scholar]
- 29.Garlick KM, Mogridge J. Direct interaction between anthrax toxin receptor 1 and the actin cytoskeleton. Biochemistry. 2009;48:10577–10581. doi: 10.1021/bi9015296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner E, Kowalczyk AP, Faundez V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem. 2006;281:23227–23236. doi: 10.1074/jbc.M603676200. [DOI] [PubMed] [Google Scholar]
- 31.Ding Z, Bradley KA, Amin Arnaout M, Xiong JP. Expression and purification of functional human anthrax toxin receptor (ATR/TEM8) binding domain from Escherichia coli. Protein Expr Purif. 2006;49:121–128. doi: 10.1016/j.pep.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Maldonado-Arocho FJ, Bradley KA. Anthrax edema toxin induces maturation of dendritic cells and enhances chemotaxis towards macrophage inflammatory protein 3beta. Infect Immun. 2009;77:2036–2042. doi: 10.1128/IAI.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruger NJ. The Bradford method for protein quantitation. Methods Mol Biol. 1994;32:9–15. doi: 10.1385/0-89603-268-X:9. [DOI] [PubMed] [Google Scholar]
- 34.Sreerama N, Woody RW. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 35.Hennessey JP, Jr, Johnson WC., Jr Information content in the circular dichroism of proteins. Biochemistry. 1981;20:1085–1094. doi: 10.1021/bi00508a007. [DOI] [PubMed] [Google Scholar]
- 36.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 37.Levitt M, Greer J. Automatic identification of secondary structure in globular proteins. J Mol Biol. 1977;114:181–239. doi: 10.1016/0022-2836(77)90207-8. [DOI] [PubMed] [Google Scholar]
- 38.Goddard TD, D.G K. Sparky NMR Analysis Software. San Francisco: University of California; 2001. [Google Scholar]
- 39.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 40.Lu C, Shimaoka M, Ferzly M, Oxvig C, Takagi J, Springer TA. An isolated, surface-expressed I domain of the integrin alphaLbeta2 is sufficient for strong adhesive function when locked in the open conformation with a disulfide bond. Proc Natl Acad Sci U S A. 2001;98:2387–2392. doi: 10.1073/pnas.041606398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wigelsworth DJ, Krantz BA, Christensen KA, Lacy DB, Juris SJ, Collier RJ. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J Biol Chem. 2004;279:23349–23356. doi: 10.1074/jbc.M401292200. [DOI] [PubMed] [Google Scholar]
- 42.McCleverty CJ, Liddington RC. Engineered allosteric mutants of the integrin alphaMbeta2 I domain: structural and functional studies. Biochem J. 2003;372:121–127. doi: 10.1042/BJ20021273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong JP, Li R, Essafi M, Stehle T, Arnaout MA. An isoleucine-based allosteric switch controls affinity and shape shifting in integrin CD11b A-domain. J Biol Chem. 2000;275:38762–38767. doi: 10.1074/jbc.C000563200. [DOI] [PubMed] [Google Scholar]
- 44.Liu S, Leung HJ, Leppla SH. Characterization of the interaction between anthrax toxin and its cellular receptors. Cell Microbiol. 2007;9:977–987. doi: 10.1111/j.1462-5822.2006.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scobie HM, Marlett JM, Rainey GJ, Lacy DB, Collier RJ, Young JA. Anthrax toxin receptor 2 determinants that dictate the pH threshold of toxin pore formation. PLoS ONE. 2007;2:e329. doi: 10.1371/journal.pone.0000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JO, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 47.Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 48.Leitinger B, Hogg N. From crystal clear ligand binding to designer I domains. Nat Struct Biol. 2000;7:614–616. doi: 10.1038/77895. [DOI] [PubMed] [Google Scholar]
- 49.Wei W, Lu Q, Chaudry GJ, Leppla SH, Cohen SN. The LDL receptor-related protein LRP6 mediates internalization and lethality of anthrax toxin. Cell. 2006;124:1141–1154. doi: 10.1016/j.cell.2005.12.045. [DOI] [PubMed] [Google Scholar]
- 50.Young JJ, Bromberg-White JL, Zylstra C, Church JT, Boguslawski E, Resau JH, Williams BO, Duesbery NS. LRP5 and LRP6 are not required for protective antigen-mediated internalization or lethality of anthrax lethal toxin. PLoS Pathog. 2007;3:e27. doi: 10.1371/journal.ppat.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryan PL, Young JA. Evidence against a human cell-specific role for LRP6 in anthrax toxin entry. PLoS ONE. 2008;3:e1817. doi: 10.1371/journal.pone.0001817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abrami L, Kunz B, Deuquet J, Bafico A, Davidson G, van der Goot FG. Functional interactions between anthrax toxin receptors and the WNT signalling protein LRP6. Cell Microbiol. 2008;10:2509–2519. doi: 10.1111/j.1462-5822.2008.01226.x. [DOI] [PubMed] [Google Scholar]
- 53.Bazzoni G, Hemler ME. Are changes in integrin affinity and conformation overemphasized? Trends Biochem Sci. 1998;23:30–34. doi: 10.1016/s0968-0004(97)01141-9. [DOI] [PubMed] [Google Scholar]
- 54.Carman CV, Springer TA. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr Opin Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Dustin ML, Bivona TG, Philips MR. Membranes as messengers in T cell adhesion signaling. Nat Immunol. 2004;5:363–372. doi: 10.1038/ni1057. [DOI] [PubMed] [Google Scholar]
- 56.Go MY, Kim S, Partridge AW, Melnyk RA, Rath A, Deber CM, Mogridge J. Self-association of the transmembrane domain of an anthrax toxin receptor. J Mol Biol. 2006;360:145–156. doi: 10.1016/j.jmb.2006.04.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.