Abstract
V3 loop is a major neutralizing determinant of the HIV-1 gp120. Using 3D structures of cholera toxin B subunit (CTB), complete V3 in the gp120 context and V3 bound to a monoclonal antibody (mAb) we designed two V3-scaffold immunogen constructs (V3-CTB). The full-length V3-CTB presenting the complete V3 in a structural context mimicking gp120, was recognized by the large majority of our panel of 24 mAbs. The short V3-CTB presenting a V3 fragment in the conformation observed in the complex with the 447-52D Fab, exhibited high affinity binding to this mAb. The immunogens were evaluated in rabbits using DNA-prime/protein-boost protocol. Boosting with the full-length V3-CTB induced high anti-V3 titers in sera that potently neutralize multiple HIV virus strains. The short V3-CTB was ineffective. The results suggest that very narrow antigenic profile of an immunogen is associated with poor Ab response. An immunogen with broader antigenic activity elicits robust Ab response.
Keywords: Immunogen design, HIV-1, gp120, v3 loop, cholera toxin B subunit, neutralizing antibody, 447-52D, HIV vaccine
Introduction
Development and use of an HIV-1 (HIV) vaccine is arguably the most effective and efficient means to stop the spread of the AIDS pandemic. However, numerous attempts to elicit protective immunity to HIV have met with limited or no success (Hanke et al. 2007, Francis et al. 2003, Rerks-Ngarm et al. 2009). Elicitation of broadly reactive, cross-clade neutralizing antibodies (Abs) with vaccine constructs has been particularly difficult despite the fact that it has long been recognized that the envelope glycoprotein (Env) is the target of anti-HIV neutralizing Abs (Zolla-Pazner 2005, Lasky et al. 1986, Matthews et al. 1986, Scheid et al. 2009, Corti et al. 2010).
Env spikes on the surface of the virion are trimers of gp120/gp41 complexes, with three non-covalently assembled gp41 subunits(Chan et al. 1997) anchored in the virus lipid membrane and three gp120 subunits assembled around the gp41 core. The exact arrangement of these subunits in the spike remains to be resolved(Liu et al. 2008). Functionally, gp41 drives the membrane fusion process, while gp120 mediates interactions with cellular receptors. The majority of Abs generated against gp120 are non-neutralizing, either because their binding does not prevent virus/cellular receptor interactions and subsequent fusion or because their epitopes are inaccessible on the trimeric spike structure (Parren et al. 1997, Parren, Burton, and Sattentau 1997, Kwong et al. 2002). Therefore, focusing the immune response on the regions of gp120 that are known to bind neutralizing Abs may improve the efficacy of prophylactic vaccines. An additional hurdle to vaccine development is the diversity of the virus and the required induction of Abs that recognize a broad spectrum of viruses in the many subgroups (clades) that make up the HIV group M family that is causing the pandemic (Barouch 2008, McBurney and Ross 2008).
The V3 region of gp120, while variable in sequence, possesses conserved structural and immunologic features that induce neutralizing Abs (Gorny et al. 1993, Gorny et al. 2002, Stanfield et al. 2006, Bell et al. 2008, Wu et al. 2006). Numerous human anti-V3 mAbs have been produced and characterized, and while many of these mAbs are narrow in their focus, several have broad cross-clade neutralizing activity (Gorny et al. 2004, Gorny et al. 2006, Gorny et al. 2002, Binley et al. 2004, Bell et al. 2008, Pantophlet et al. 2008, Zolla-Pazner et al. 2008, Zolla-Pazner et al. 2004). Approximately 30% of viruses can be neutralized by anti-V3 Abs (Binley et al. 2004, Gorny et al. 2006, Pantophlet et al. 2007, Corti et al. 2010, Hioe et al. 2010). This may be because the exposure of the V3 in the native gp120 trimer is limited due to masking by other parts of the protein, such as V2 (Krachmarov et al. 2006, Krachmarov et al. 2005, Barnett et al. 2001, Walker et al. 2009, Honnen et al. 2007, Nyambi et al. 2008, Wei et al. 2003), but V3 may also be “unmasked” by binding of certain ligands or antibodies (Mbah et al. 2001, Wu et al. 2008, Hioe et al. 2009). A specially designed V3-based immunogen that could induce high titers of Abs with binding modes and epitope specificities similar to those of known broadly neutralizing anti-V3 mAbs may, therefore be valuable as a component of a vaccine against HIV infection.
Cholera Toxin subunit B (CTB) and a family of closely related bacterial proteins such as E. coli enterotoxin are homo-pentamers composed of relatively small subunits (~100 amino acids). The protein is highly immunogenic and has been used in fusion constructs to enhance immunogenicity of target proteins (Martin et al. 2001, Gonzalez et al. 1993, Yamamoto et al. 1997, Eriksson et al. 2003, Matoba et al. 2006). The crystallographic structure of CTB has been solved in the free state as well as in complex with oligosaccharides (Merritt et al. 1997, Zhang et al. 1995). CTB is capable of inducing mucosal immunity(McKenzie and Halsey 1984, Czerkinsky et al. 1989, Lipscombe et al. 1991), which is an uncommon and highly desirable feature for an HIV vaccine because infection typically occurs via mucosal route.
GM1 ganglioside binding is believed to be a major factor in the immunogenic properties of CTB and related toxins. X-ray structures of CTB reveal the oligosaccharide binding sites. The availability of this structural information allows protein design without disruption of the GM1 binding site, therefore preserving immunogenicity. Moreover, CTB is non-toxic without the A-subunit present in the native toxin, and thus CTB is used as a component of an anti-cholera vaccine approved for use in humans.
The pentameric structure of CTB results in the presentation of five copies of each epitope on its surface. It has been suggested that repetitive presentation of an epitope on an immunogen can induce a stronger immune response, probably because such immunogens can trigger oligomerization of B-cell receptors recognizing the epitope (Cruz et al. 2004, Cruz et al. 2004). Thus, a combination of the attractive immunogenic features of CTB and the gp120 V3 loop could result in an immunogen that would induce a strong, broadly neutralizing immune response against the HIV. Indeed, V3-CTB immunogen constructs have been described previously, but they induced only a very modest anti-V3 immune response (Backstrom et al. 1994, Bckstrom et al. 1995). However, this previously reported design did not benefit from the wealth of structural and modeling data on gp120, the V3 loop, and V3 in complex with mAbs that have become available more recently(Stanfield et al. 2004, Stanfield et al. 2006, Dhillon et al. 2008, Kwong et al. 2000, Jiang et al. 2010). Furthermore, our preliminary analysis suggested that the short V3 fragments used for these previously reported constructs may have left out important antigenic determinants, and that the insertion position on the CTB scaffold may not have allowed the V3 fragment to form the hairpin conformations recognized by several anti-V3 mAbs.
In the present study we designed, expressed, and characterized two V3-scaffold immunogen constructs based on CTB and the V3 loop (V3-CTB) in which the V3 loop was integrated into the CTB scaffold sequence. We investigated two strategies for constructing these V3-CTB scaffold immunogens. In the first approach, a full-length 35 amino-acid V3 loop was engrafted into CTB and used as an immunogen to focus the immune response on V3. In the second approach, we attempted to narrow further the focus of the immune response on the epitope of anti-V3 mAb 447-52D. mAb 447-52D, like essentially all other human anti-V3 mAbs, recognizes a region in the crown of the V3 loop composed of ~14 amino-acids(Stanfield et al. 2004, Stanfield et al. 2006, Bell et al. 2008, Dhillon et al. 2008, Sharon et al. 2003, Burke et al. 2009, Jiang et al. 2010). In addition to the relatively straightforward restriction of the polypeptide graft to the V3 fragment that is known to interact with this mAb, the “short V3-CTB” construct was designed to induce preferentially the conformation of the V3 fragment that is compatible with its binding to this mAb, i.e. the conformation observed in the V3/mAb 447-52D complex (Sharon et al. 2003, Stanfield et al. 2004, Dhillon et al. 2008, Burke et al. 2009). The design of these two immunogens allowed the direct comparison of the effects of focusing the response on the entire V3 immunogenic region versus focusing on the much narrower epitope recognized by a particular mAb.
Results
Direct insertion, as opposed to the commonly used strategy of fusion via a linker, allows an immunogen scaffold to impose restraints on the termini of a loop. Appropriate choice of the insert length and position may be used to preferentially induce the conformations of a loop that expose conserved epitopes. We reasoned that exposure of the conformationally correct conserved epitopes of V3 should induce a broadly neutralizing anti-V3 Ab response.
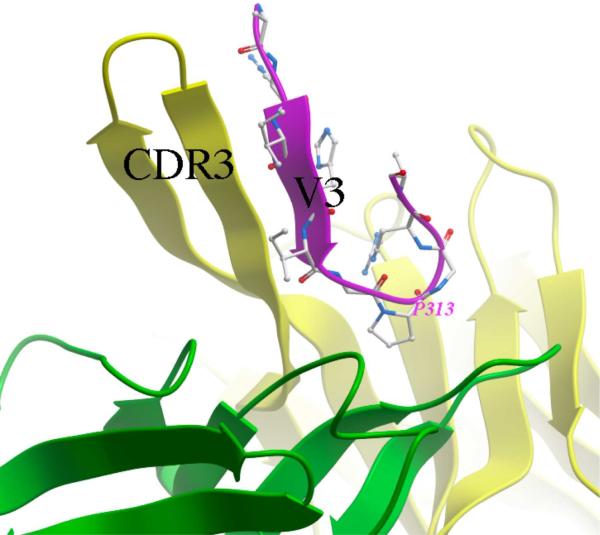
Available structural data on V3 complexes with neutralizing mAbs suggest two different binding modes as well as the epitopes associated with broad neutralization: 1) The complex of a V3 peptide with mAb447-52D (PDB accession code 1Q1J)(Stanfield et al. 2004) indicates that the epitope consists primarily of the backbone atoms of the crown of the V3 loop, which forms a beta-sheet structure together with two strands of the elongated hypervariable CDR3 loop of the mAb heavy chain (Figure 1). 2) The complex of V3 with another anti-V3 mAb, 2219 (PDB accession code 2B1H)(Stanfield et al. 2006), shows that this mAb contacts mostly side-chain atoms of V3, but the amino acids involved are highly conserved in the V3 sequence, including two positively charged side-chains and four side-chains that form a hydrophobic cluster. In the present work we investigated presentation of the entire V3 or focusing the antigen on the epitope of the first type.
Figure 1.
The V3 peptide fragment (shown in magenta ribbon representation and sticks) bound to the broadly neutralizing mAb 447-52D (light and heavy chains are shown as green and yellow ribbons, respectively). Formation of a three-strand beta-sheet composed of two strands of the heavy chain CDR3 hairpin and one V3 strand, as well as tight binding of the conserved residue P313 in the GPGR motif at the tip of the loop can be observed. All molecular graphics images were prepared in Molsoft ICM-Pro.
Design of a full-length V3 insert in the CTB scaffold
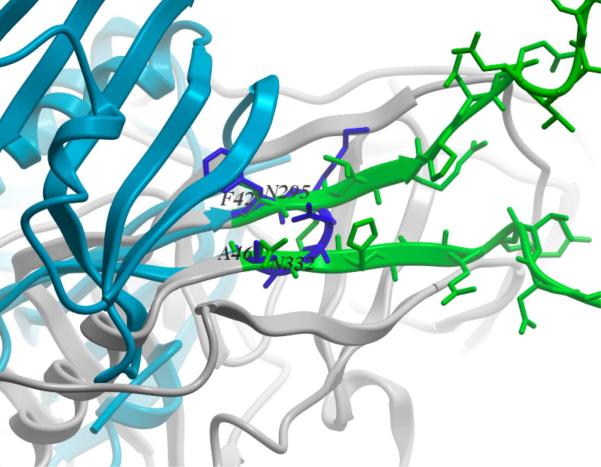
We scanned the polypeptide backbone in the X-ray structure of the CTB scaffold (PDB accession code 3CTB)(Merritt et al. 1998) for residue positions that would allow low RMSD superposition and clash-free match with the termini of the V3 loop as observed in the gp120 X-ray structure (PDB accession code 2B4C)(Huang et al. 2005). The best clash-free match had an RMSD of 0.37Å for the C, Cα, Cβ and N atoms of V3 residues N295 and N332, and the corresponding CTB residues, F42 and A46 (Figure 2). Using the CTB and gp120 structures superimposed on these residue pairs, a chimeric structure was created that included the complete V3 loop with V3 terminal cysteines C296 and C331 (Figure 3A) fused at the match points to the CTB, excluding the CTB turn residues K43-G45. Constrained minimization of the chimera model resulted in a structure that was essentially strain-free and differed by only 0.19Å backbone RMSD from the templates. Finally, a complete pentamer was reconstructed by superimposing five copies of the chimera model onto the different monomers in the X-ray structure of pentameric CTB. The final model was inspected to ensure that the grafts were not interfering with pentamer assembly, that the V3 loops projected into different segments of space around the scaffold, limiting the likelihood of undesirable V3-V3 interactions, and that the V3 inserts did not interfere with the GM1 binding site.
Figure 2.
Match of the CTB scaffold (blue) and V3 loop base (green) in the gp120 structure (grey). Close correspondence of the backbone traces of the two structures at the F42CTB/N295gp120 − A46CTB/N332gp120 junction can be observed.
Figure 3.
A) Full-length V3-CTB amino-acid sequence (V3 loop insert is in bold); B) Short V3-CTB where only the V3 crown (shown in bold) is inserted. Additional mutations are introduced to enhance conformational stability of the insert (mutated positions are underlined).
Design of a V3 crown insert in a conformation preferentially bound by mAb 447-52D
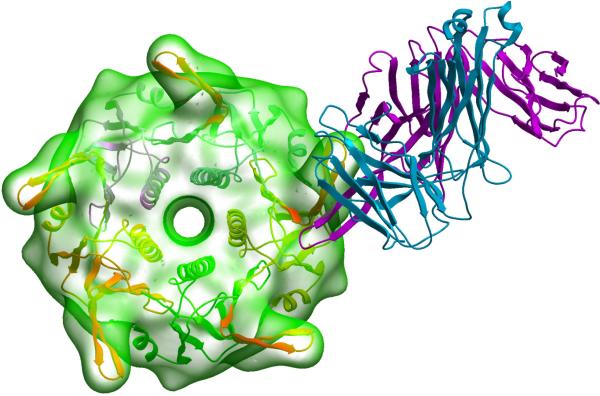
We first performed conformational searches of the loops comprising, from N- to C-terminus: (1) a single flanking phenylalanine on the N-terminus corresponding to the CTB scaffold's F42; (2) the V3 loop fragment K305-A316 observed to bind to the Fab fragment of 447-52D in the X-ray structure; (3) varying number of additional V3 residues, and (4) CTB scaffold's G45A46 on the C-terminus. When two additional V3 residues were included, a low-energy conformation similar to the experimental mAb-bound V3 peptide structure was found. This conformation and loop sequence (Figure 3B) was used to construct an initial model of the designed V3 crown-CTB protein (hereafter referred to as “short V3-CTB”) and to recreate in silico its putative complex with Fab 447-52D. Analysis of this initial complex model suggested several needed point mutations in the construct: CTB's K23 and E79 side chains were clashing with the hypervariable loop of the mAb and were changed to smaller serine residues; F317 of the V3 insert was replaced with a glutamate to reduce the exposed lipophilic surface and to stabilize the loop via salt bridging with K305 on the adjacent V3 strand; finally, CTB's A46 was mutated to a methionine in an attempt to fill in a hydrophobic pocket that formed at the base of the loop insert. After constrained minimization, a final, refined model of the short V3-CTB construct/Fab complex was obtained (Figure 4). The model's V3 residues R306-R315 that participate in Fab interaction could be superimposed onto the X-ray structure of the peptide/Fab complex with a backbone RMSD of 0.42Å (0.87Å for all heavy atoms). The pentameric complex model was also constructed to ensure that mAb interaction with each of the five V3 grafts can occur simultaneously and independently.
Figure 4.
Model of the short V3-CTB construct (ribbon and transparent surface) in complex with the Fab fragment of mAb 447-52D (magenta and blue ribbons for heavy and light chains, respectively).
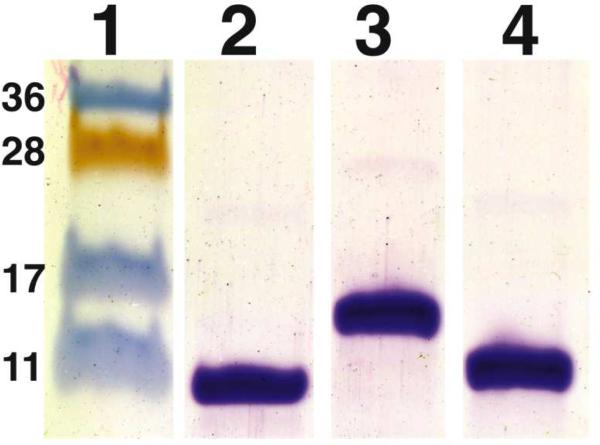
The use of a small oligomeric scaffold and direct insertion of the V3 fragment into the scaffold's tertiary structure resulted in constructs that had an exceptionally high fraction of their surface presenting relevant epitopes: the V3 surface constituted 51% and 26% of the total solvent accessible surface of the full-length V3-CTB and of the short V3-CTB immunogens, respectively. We postulated that this high proportion of V3 epitopes on the immunogen surface would result in a highly focused Ab response. The two immunogens were expressed in E. coli and purified (Figure 5).
Figure 5.
Analysis of purity of recombinant CTB immunogens. Purified recombinant immunogens were analyzed by SDS/PAGE and stained with Coomassie. Lane 1: molecular weight markers with corresponding molecular weights (kDa); Lane 2: wide type CTB. Lane 3: full-length V3-CTB. Lane 4: short V3-CTB.
Reactivity of the two V3-CTB constructs with a panel of 24 anti-V3 mAbs
The results from ELISA binding assays in which anti-V3 mAbs were titrated from 0.01 to 10 μg/ml and tested for reactivity with full-length, short and wild type V3-CTB are presented in Table I. For the full-length V3 construct, very high (50% binding at ≤ 0.01μg/ml) to high (50% binding at ≤0.1μg/ml) affinity for 9 of 10 clade B-derived mAbs and 7 of 14 non-clade B-derived mAbs was observed. Lack of reactivity or weak binding by the remaining 7 mAbs could be attributed to their specificity for the GPGQ motif that is frequently present in non-clade B V3, whereas the V3-CTB constructs used here carried the GPGR V3 motif characteristic of clade B. These results suggested that the full-length V3-CTB construct bears the relevant epitope(s) recognized by the majority of anti-V3 mAbs and might be capable of eliciting a broad range of anti-V3 Abs.
Table I.
ELISA titrations of mAb binding to two V3-CTB immunogen constructs and the wild type CTB control. mAbs listed in red were derived from non-clade B-infected subjects; those listed in blue were derived from clade-B infected individuals. Each was tested at concentrations of 10 to 0.01 μg/ml on plates coated with 1 μg/ml of the designated form of CTB. Optical densities from ELISA plates are shown and color-coded to designate strong (red) to no (white) binding. An irrelevant anti-parvovirus mAb (1418) does not bind to any of these constructs (data not shown).
| Full-length V3-CTB | Short V3-CTB | CTB WT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mAb | 10.0 | 1.0 | 0.1 | 0.01 | 10.0 | 1.0 | 0.1 | 0.01 | 10.0 | 1.0 | 0.1 | 0.01 |
| 2182 | 2.8 | 2.8 | 2.6 | 2.0 | 2.7 | 2.8 | 2.7 | 1.5 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2557 | 2.6 | 2.7 | 2.4 | 1.7 | 2.6 | 2.2 | 1.3 | 0.4 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2558 | 2.8 | 2.7 | 2.1 | 1.3 | 2.6 | 1.5 | 0.7 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2601 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3019 | 2.7 | 2.6 | 1.8 | 1.0 | 0.4 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3074 | 2.6 | 2.1 | 1.4 | 0.8 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3694 | 2.7 | 2.2 | 1.3 | 0.7 | 0.8 | 0.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3697 | 2.7 | 2.5 | 1.8 | 1.2 | 0.5 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3791 | 0.1 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3792 | 2.3 | 1.3 | 0.4 | 0.2 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3869 | 2.4 | 2.0 | 1.1 | 0.6 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3881 | 0.4 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3904 | 0.1 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 3906 | 0.4 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.2 |
| 257 | 2.8 | 2.7 | 2.3 | 0.9 | 1.9 | 0.8 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| 391–95 | 2.6 | 2.6 | 2.3 | 1.2 | 2.6 | 2.5 | 2.6 | 1.5 | 0.1 | 0.1 | 0.1 | 0.1 |
| 419 | 2.6 | 2.0 | 0.9 | 0.2 | 0.4 | 0.2 | 0.1 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| 447 | 2.7 | 2.7 | 2.4 | 1.2 | 2.7 | 2.7 | 2.4 | 0.8 | 0.1 | 0.1 | 0.1 | 0.1 |
| 694/98 | 2.6 | 2.4 | 1.7 | 0.6 | 2.7 | 2.7 | 2.2 | 0.7 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2456 | 2.5 | 2.2 | 1.4 | 0.5 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 1006-15D | 2.6 | 2.4 | 2.3 | 1.4 | 1.7 | 0.7 | 0.3 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2191 | 2.6 | 2.6 | 2.1 | 1.0 | 0.5 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2219 | 2.7 | 2.7 | 2.2 | 1.2 | 1.6 | 0.5 | 0.3 | 0.2 | 0.1 | 0.1 | 0.1 | 0.1 |
| 2442 | 2.7 | 2.5 | 2.0 | 0.9 | 2.7 | 2.8 | 2.5 | 0.8 | 0.1 | 0.1 | 0.1 | 0.1 |
In contrast, the structurally focused short V3-CTB construct exhibited a much more narrow binding profile. Importantly, the affinity of mAb 447-52D for the short and full-length V3-CTB constructs were similar, which validated the design of the former which had been guided by the X-ray structure of the complex of Fab 447-52D with the V3 peptide fragment. A few other mAbs, mostly clade B-derived (391–95, 694/98, 2442), also retained virtually the same affinities as those that they had for the full-length V3-CTB construct, but the large majority of the mAbs in the panel showed little or no reactivity with the short V3-CTB construct. While X-ray structures are not available for the other mAbs that exhibit high affinity to the short V3-CTB construct, we can hypothesize that they utilize a binding mode similar to that of mAb 447-52D. Thus, the full-length and short constructs represent successful rational designs, correctly presenting the complete V3 loop and the epitope recognized by mAb 447-52D, respectively.
Immunogenicity of the V3-CTB constructs
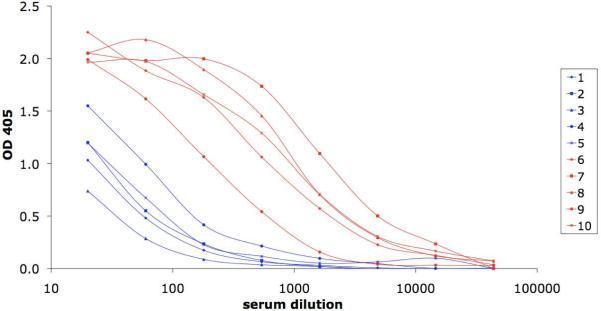
The V3-CTB constructs were evaluated for immunogenicity using an immunization protocol described previously (Zolla-Pazner et al. 2008, Zolla-Pazner et al. 2009) in which rabbits were primed three times with clade B gp120 DNA and boosted twice with either the full-length or the short V3-CTB construct. Sera from immunized rabbits were tested for V3 binding by ELISA using a V3 fusion protein consisting of the clade B V3 consensus sequence linked to the Fc fragment of rabbit IgG (Davis et al. 2009). Sera were titrated at dilutions ranging from 1:20 to 1:50,000. Sigmoidal curves were generated using sera from animals boosted with the full-length V3-CTB construct, and a geometric mean titer for 50% binding (GMT50) of 1:535 was achieved by the sera from the five animals in this group (Figure 6). By contrast, a plateau of binding was not achieved with the sera from any of the animals boosted with short V3-CTB, even when tested at 1:20, and thus GMT50 could not be calculated. The curves, shown in Figure 6 suggest that the GMT50 for the animals boosted with short V3-CTB would be approximately two orders of magnitude less than that for the animals boosted with full-length V3-CTB.
Figure 6.
ELISA titration curves of immune sera vs. V3 fused to the Fc fragment of rabbit IgG. Immune sera were obtained two weeks after the second protein boost from rabbits designated as #6–10, boosted with the full length V3-CTB (red), or from rabbits #1–5, boosted with the short V3-CTB (blue).
Virus neutralization
In agreement with V3 binding data from ELISA experiments, sera from animals immunized with the full-length V3-CTB construct displayed stronger neutralizing Ab responces than animals immunized with short V3-CTB. In Table II, data are shown for the 50% neutralizing titers (NT50) of the SF162 pseudovirus (psV) and chimeric psVs carrying the consensus V3 sequences of clades B, F, A/E, A1, AG, H or C in the HIV-1SF162 envelope backbone. The data, generated using the CD4+CCR5+CXCR4+U87 cell line as target cells, demonstrate that the Ab response when full-length V3-CTB was used as a boost induced Abs that could recognize and neutralize the psVs bearing the V3 loops from all of these different clades. In contrast, the sera from rabbits boosted with the short V3-CTB construct only neutralized the psV carrying the homologous clade B V3 loop, and at titers approximately two orders or magnitude less than those in the sera from the full-length V3-CTB-boosted animals.
Table II.
Reciprocal 50% neutralization titres (NT50) of sera from rabbits boosted with the short V3-CTB construct (rabbits 1–5) or the full-length V3-CTB construct (rabbits 6–10). Pseudoviruses tested include the wild type SF162 and chimeric pseudoviruses carrying V3 sequences corresponding to the consensus sequences of clades B, F, A1, AE, AG, H and C.
| DNA Prime | Protein Boost | Rabbit # | SF162wt | B | F | A/E | A1 | AG | H | C |
|---|---|---|---|---|---|---|---|---|---|---|
| JR-FL gp120 | Short V3-CTB | 1 | <10 | 316 | <10 | <10 | <10 | <10 | <10 | <10 |
| 2 | <10 | 332 | <10 | <10 | 21 | <10 | <10 | <10 | ||
| 3 | <10 | 410 | <10 | <10 | <10 | <10 | <10 | <10 | ||
| 4 | <10 | 1,638 | <10 | 16 | 31 | <10 | <10 | <10 | ||
| 5 | <10 | 512 | <10 | <10 | <10 | <10 | <10 | <10 | ||
|
| ||||||||||
| JRFL gp120 | Full-length V3-CTB | 6 | <10 | 24,970 | 34 | 56 | 32 | <10 | <10 | <10 |
| 7 | 516 | 64,250 | 820 | 159 | 620 | 1,022 | 17 | 209 | ||
| 8 | 495 | 37,987 | 613 | 648 | 296 | 190 | 24 | 67 | ||
| 9 | 222 | 8,521 | 124 | 33 | 130 | 53 | 10 | 22 | ||
| 10 | 510 | 50,912 | 1,022 | 559 | 686 | 317 | 111 | 104 | ||
Sera from the animals boosted with full-length V3-CTB were also tested for their ability to neutralize psVs from the standard multi-clade Tier 1 panel (Li et al. 2005) using the CD4+CCR+TZM.bl cell line as target cells. A pool of pre-bleed sera from these animals showed no neutralizing activity (50% neutralizing titer <1:20) against the four psVs tested. In contrast, sera derived from each animal two weeks after the second boost with full-length V3-CTB neutralized three of the four psVs tested (MW965.26, Bx08.16, and BaL.26), and two of five sera displayed neutralizing titers against the fourth psV (SS11961.1) (Table III). Fifty percent neutralizng titers ranged from 1:22 to 1:4,437. The immune sera were also tested for their ability to neutralize a panel of primary isolates from clades B, A, and AG (Table IV). Sera from rabbits immunized with the short V3-CTB construct had low neutralization titers and failed to neutralize most viruses tested. For example, the only virus that was convincingly neutralized was BZ167, where 4 of 5 sera displayed a GMT50 of 1:18. In contrast, 5 of 5 rabbits boosted with full-length V3-CTB neutralized BZ167 with a GMT50 value 1:113. Neutralizing titers in sera of animals boosted with the full-length V3-CTB were detected against five additional viruses in this panel. None of the sera in either groups neutralized any clade C primary isolates (data not shown). Thus, the full-length V3-CTB induced a stronger and broader neutralizing Ab response and induced cross-clade neutralizing Abs.
Table III.
Reciprocal 50% neutralization titers (NT50) of sera from rabbits boosted with the full-length V3-CTB construct. Pseudoviruses tested include those from the standard multi-clade Tier 1 panel as well as a negative control psV carrying the envelope of murine leukemia virus (MuLV).
| Clade C | Clade B | Clade B | Clade B | ||
|---|---|---|---|---|---|
| Rabbit # | MW965.26 | Bx08.16 | BaL.26 | SS11961.1 | MuLV |
| 6 | 28 | 105 | 125 | 22 | <20 |
| 7 | 4,437 | 129 | 171 | 25 | <20 |
| 8 | 451 | 58 | 63 | <20 | <20 |
| 9 | 159 | 62 | 59 | <20 | <20 |
| 10 | 1,206 | 103 | 148 | <20 | <20 |
Table IV.
Primary isolates neutralization. The serum from each animal was tested against each of the seven primary isolates listed. The 50% neutralizing titer for each serum/virus combination is shown where 50% neutralization was achieved at a dilution of 1:10 or higher; all empty cells in the table denote little or no neutralization at a serum dilution of 1:10 . The yellow, green and blue boxes denote group GMT50 values of 1:10–50,1:51–100, and >1:100, respectively.
| DNA Prime | Protein Boost | Clade B | Clade A | Clade AG | |||||
|---|---|---|---|---|---|---|---|---|---|
| Rabbit # | BX08 | BZ167 | CA5 | VI313 | CA1 | DJ263 | NYU6525 | ||
| gp120JR-FL | Short V3-CTB | 1 | 11 | ||||||
| 2 | 12 | ||||||||
| 3 | 23 | 10 | |||||||
| 4 | 34 | ||||||||
| 5 | |||||||||
|
| |||||||||
| gp120JR-FL | Full-length V3-CTB | 6 | 58 | 114 | 28 | ||||
| 7 | 54 | 602 | 49 | 10 | 438 | ||||
| 8 | 49 | 493 | 12 | 20 | 30 | ||||
| 9 | 53 | 26 | 14 | 54 | 22 | ||||
| 10 | 21 | 21 | 111 | ||||||
T-helper epitopes in the two immunogens
In order to inform future immunogen design efforts, we investigated further the potential determinants of immunogenicity for the two constructs. We applied epitope prediction algorithms available from the Immune Epitope Database and Analysis Resource (Peters et al. 2005). MHC-II binding predictions were performed on both immunogen constructs against multiple MHC alleles. Consensus percentile ranks of three different prediction methods were utilized to score putative epitopes (Bui et al. 2005, Nielsen, Lundegaard, and Lund 2007, Sturniolo et al. 1999). Hits with low consensus percentile ranks that correspond to the likelihood of T cell recognition are listed in Table V. An epitope inherited from the CTB scaffold was found in both immunogens. A known T-cell epitope from the HIV V3 loop (Bergmann, Stohlmann, and McMillan 1993) was detected in the full-length V3-CTB construct. This epitope was missing in the short V3-CTB construct, however the short V3-CTB construct contained two additional predicted T-cell epitopes: one emerged at the scaffold-graft junction and the other was introduced serendipitously with the point mutations intended to stabilize the construct and improve Ab binding. In summary, evaluation of the existence of predicted T-cell epitopes suggests that both constructs contain multiple T-cell epitopes and comparable levels of T-help can be expected. Because evaluation of T cell activity in rabbits is problematic, T cell immunogenicity was not evaluated with cells from the immunized rabbits.
Table V.
Result of T-cell epitiope predictions for the sequences of two immunogenic constructs. Epitopes with lowest Consensus Percentile Ranks (CPR) are shown.
| Construct | Sequence fragment | Location | MHC Allele | C.P.R. |
|---|---|---|---|---|
| Full-length | YTTGEIIGDIRQAHC | V3 | DRB1-0301 | 0.9% |
| Full-length and short | EKLCVWNNKTPRAIA | Scaffold | DRB1-1302 | 0.9% |
| Full-length and short | Same as above | Scaffold | DRB1-0802 | 1.1% |
| Short | TQIHTLNNSITSYTE | Mutated Scaffold | DRB1-1302 | 0.1% |
| Short | REMAIITFKRIHIGP | Junction | DRB1-1501 | 0.29% |
Immunogenicity of the scaffold
Given that multiple factors, such as protein stability and binding to cellular receptors may have affected the overall immunogenicity of the constructs, we compared the anti-CTB scaffold Ab levels in the sera of the two sets of immunized rabbits. ELISA data from sera binding to CTB indicated that very similar levels of Abs against CTB were elicited in the two groups: reciprocal GMT50 titers were 1.9 ×104 and 1.8 × 104 for the short and full-length V3-CTB immunized groups, respectively. Therefore, it appears that the two constructs have comparable overall immunogenicity.
Discussion
Extensive immunologic and viral studies have previously shown that many anti-V3 mAbs display cross-reactivity between V3 peptides and gp120 proteins from diverse viruses of the different clades of HIV-1 (Gorny et al. 1993, Gorny et al. 2002, Binley et al. 2004, Pantophlet et al. 2008). These studies demonstrated that, although by definition V3 is highly variable in its sequence, this region of gp120 contains immunologically conserved elements. Immunologic data are further supported by findings that, despite its sequence variability, V3 must retain certain conserved structures in order to interact with the chemokine receptors on the surface of target cells (Shioda, Levy, and Cheng-Mayer 1992, Trkola et al. 1996, Hill et al. 1997, Labrosse et al. 2001, Cardozo et al. 2007). Moreover, the conformational conservation of V3 is confirmed by crystallographic (Stanfield et al. 2006, Stanfield et al. 2004, Bell et al. 2008, Dhillon et al. 2008, Burke et al. 2009, Jiang et al. 2010) and NMR(Sharon et al. 2003) studies. This extensive literature was the basis of our initial use of the V3 region of gp120 as an epitope for inducing Abs with broad immunologic and antiviral activity.
In the present work, recombinant chimeric V3-CTB immunogens were successfully designed using structural data, molecular modeling, and protein engineering. The short V3-CTB form bound to mAb 447-52D, whose epitope it was intended to mimic optimally. The full-length V3-CTB bound to most anti-V3 mAbs, demonstrating the success of the design in presenting the V3 epitopes as exposed, correctly folded, Ab-accessible conformations. Use of these new immunogens to boost the immune response of rabbits showed that the full-length V3-CTB construct was able to induce V3-binding Abs and Abs that display cross-clade neutralizing activity against psVs and primary isolates. The full-length V3-CTB immunogen induced a much stronger and broader Ab response than did the short V3-CTB.
The two V3-scaffold constructs that we have designed and tested were based on the V3 loop found in clade B viruses. These were used because much of the immunologic and structural data was based on studies of the clade B V3 loop. However, the clade B V3 loop is relatively unusual among the HIV-1 group M virus clades because it contains a GPGR rather than a GPGQ motif at its center (Leitner T 2005). Use of the clade B V3 as the epitope in both the short and full-length V3-CTB constructs studied here resulted in immunogens that were relatively limited in their ability to induce Abs that neutralize psVs and viruses carrying V3 loops that contain the GPGQ motif. Thus, for example, the neutralizing titers of the sera from animals boosted with full-length V3B-CTB, against the chimeric psV carrying the homologous V3B were 2–3 orders of magnitude higher than those against chimeric psVs carrying heterologous V3 loops bearing the GPGQ motif (Table II). Our previously published work (Zolla-Pazner et al. 2009) suggests that follow-up studies with full-length V3-CTB immunogens where V3B is replaced by V3 sequences containing the GPGQ motif or other rationally designed V3 loops will give rise to more broadly reactive and perhaps higher titers of anti-V3 Abs.
As noted above, the use of CTB as a scaffold for the HIV-1 V3 epitope was based on scans of the Protein Data Base for molecules with surface-exposed beta-turns that could be structurally matched to the beta-hairpin structure at the base or in the crown of the V3 loop. Among the proteins found to have a suitable beta-turn, we further looked for proteins that would form high-order oligomers, could be easily expressed in bacteria, and for which some immunogenicity data was available. Wild type CTB emerged as the preferred scaffold because it has been used extensively as a component of vaccine in humans to protect against cholera (marketed as Dukoral (Lopez-Gigosos et al. 2007)), and has been tested as a scaffold for other immunogens (Backstrom et al. 1994, Matoba et al. 2006).
We have previously shown that using an immunization regimen in which animals are primed with gp120 DNA and boosted with a scaffold immunogen carrying only the V3 epitope of gp120 is able to focus the immune response on this neutralizing epitope and induce cross-clade neutralizing Abs (Zolla-Pazner et al. 2008, Zolla-Pazner et al. 2009). In the previous studies, the V3-scaffold immunogen consisted of various V3 loops fused to the C-terminus of a truncated form of the murine leukemia virus gp70 (Kayman et al. 1994). This construct carried one copy of V3 per molecule of gp70. In contrast, the V3-CTB immunogens designed and tested in this study carries five copies of V3 per pentamer of CTB. Although direct comparisons cannot be made because immunizations were not conducted in parallel using exactly the same immunization regimen, it would appear that full-length V3-CTB is a better immunogen than full length V3-gp70, giving Ab responses of greater breadth and potency. The most direct comparison can be made by analyzing the responses of rabbits #6–10, immunized in this study with clade B gp120 DNA and boosted with the full-length V3B-CTB (B/V3B-CTB) to those immunized in an earlier study with the gp120 DNA from a clade A strain carrying a V3 loop with the GPGR motif (Ar) and boosted with V3B-gp70 (Ar/V3B-gp70)(Zolla-Pazner et al. 2008). The GMT50 for neutralization of V3 chimeric psVs averaged 10-fold higher in the rabbits receiving the B/V3B-CTB vs. the Ar/V3B-gp70 regimen. This may be due to the difference in the V3 valency of the immunogens (one for V3-gp70 vs. five for V3-CTB), and/or to differences in modes of antigen presentation and induction of B cell maturation due to the differential binding of these immunogens to cell receptors: CTB binds to ganglioside GM1, mammalian cell wall glycosphingolipid widely distributed in all tissues, whereas gp70 binds to mouse cationic amino acid transporter (mCAT-1) (Albritton et al. 1993). Another difference between the gp70 and CTB carriers is that the gp70-V3 proteins contained N-linked glycans at the base of the V3 loop and at the internal glycosylation site at position 6 of the V3 loop. The proximity of this position to residues known to be included in V3 epitopes might affect immunogenicity.
The inability of the short V3-CTB construct to elicit significant Ab titers underscores the challenge of designing effective recombinant immunogens that direct the immune response towards a highly restricted singular three-dimensional epitope. The poor performance of the short V3-CTB construct may be explained by several possible causes: 1) the rabbit Ab repertoire may not contain genes that are appropriate to develop 447-52D-like Abs, or 2) flexibility of an epitope loop may be required for immunogenicity. Further investigation would be needed to establish the minimal essential epitope(s) within the complete V3 that are sufficient for a robust immune response, although it is clear from previous studies that the length of V3 is not the only critical variable that contributes to the induction of neutralizing Abs (Yang et al. 2004). The limited efficacy of previously described V3-CTB constructs and other HIV epitope-scaffold immunogens for eliciting neutralizing Abs (Bckstrom et al. 1995, Backstrom et al. 1994, Law et al. 2007, Muster et al. 1994, Eckhart et al. 1996) also highlights the challenge of constructing effective recombinant immunogens that focus the immune response on neutralizing epitopes.
In contrast, the immunogenicity data obtained after boosting with the full-length V3-CTB underscores the potential that optimally designed immunogens can have in focusing the immune response on a neutralizing epitope. The full-length V3-CTB induced cross-clade neutralizing Abs in rabbits. Because the immunogen was rationally designed, this result serves as an important initial point for immunogen optimization for achieving the desired breadth and potency for a protective Ab response. Importantly, a variety of V3 loop sequences and structures can be placed on this scaffold to optimize the breadth and potency of the Ab response. Moreover, this approach may serve as a platform for designing other epitope-scaffold immunogens that will induce Abs specific for additional HIV-1 neutralizing epitopes and/or for protective epitopes against other pathogens.
Materials and Methods
Design of the antigen constructs: Full-length V3 graft
Conformation of the complete V3 loop in the gp120 context as revealed by the X-ray structure (PDB code 2B4C (Huang et al. 2005)) was used as a prototype to identify grafting site(s) on the scaffold. Coordinates of a pair of gp120 residues, N295 (immediately preceding the cysteine bridge C296–C331) and residue N332 (immediately following the bridge) were used to scan the CTB structure (PDB code 3CHB (Merritt et al. 1998)) for a pair of residues closely matching the query in 3D configuration. Low RMSD matches were next subjected to a scaffold clash test: gp120 N295 and N332 were superimposed onto identified residue pairs in CTB, and the resulting position of the V3 loop was checked for any clashes with the rest of the CTB scaffold. When low RMSD and clash-free superposition was achieved, the complete model of the chimeric protein was constructed in ICM using a regularization procedure which threads an idealized polypeptide chain through the template structures (Abagyan, Totrov, and Kuznetsov 1994).
Design of the antigen constructs: Short V3 crown graft
The fragment of V3 loop observed in the mAb 447-52D/peptide complex structure (PDB) consists of a GPGR beta-turn and a beta-strand segment that is N-terminal to the turn. Although the C-terminal segment of the V3 crown is disordered in this X-ray structure, experimental evidence from other structures (Sharon et al. 2003) suggests that it has a strong propensity to form another strand, thus completing a beta-hairpin. We reconstructed a low-energy hairpin structure by BPMC global energy minimization in ICM(Abagyan and Totrov 1994). Initially unstructured N- and C-terminal polypeptide segments of varying length were added to the residues observed in the X-ray structure of the bound peptide (PDB code 1Q1J) and subjected to Monte-Carlo sampling. Residues corresponding to I307-R316 observed in the X-ray structure and engaged in mAb interaction were kept rigid. Terminal residues of the loop were harmonically constrained to the positions of F42 and A46 in CTB scaffold.
Preparation of the recombinant V3-CTB constructs
Wild type (WT) CTB (as control), the full-length V3-CTB and the short V3-CTB genes were chemically synthesized and cloned into pSUMO plasmids. Amino-terminally SUMO-tagged forms of CTB and the two immunogen constructs were produced by induction of T7 RNA polymerase in E. coli strain BL21(DE3) containing pSUMO-CTB, pSUMO-full-length V3-CTB, or pSUMO-short V3-CTB, respectively. After adding IPTG to a log-phase culture grown in Luria-Bertani (LB) medium, the cells were pelleted, resuspended, and lysed with a French press. SUMO-tagged CTB and the two immunogen constructs were purified from each of the resulting cultures by affinity chromatography on a Ni-nitrilotriacetate column. SUMO tags were cleaved from the fusion proteins by a SUMO protease and removed by a Ni-NTA column. Proteins were collected in the flow-through and dialyzed overnight. The purified proteins were stored at −80°C.
Evaluation of binding of immunogen constructs to anti-V3 mAbs
Binding of the immunogen constructs to various anti-V3 mAbs was evaluated in an ELISA assay as previously described (Gorny et al. 1997). Briefly, immunogen constructs or wild type CTB were coated onto plastic plates at 1.0 ug/ml and incubated overnight at 4°C. The next day the plates were washed three times with wash buffer (1× PBS with 0.05% Tween-20, pH 7.4) before incubation for 1.5 h at 37°C with human mAbs at concentrations between 0.01 and 10.0 ug/ml. After washing, the plates were incubated with alkaline phosphatase-conjugated goat anti-human IgG (Fc-specific) for another 1.5 h at 37°C. Plates were washed again, and the substrate, p-nitrophenyl phosphate in 10% diethanolamine, was added for 30 min. Plates were read at 410 nm. Negative controls consisted of immunogen-coated wells reacted with an irrelevant human mAb (anti-parvovirus).
Determination of anti-V3 rabbit serum ELISA titers
Titration of rabbit sera against a fusion protein consisting of V3 from clade B isolate JR-CSF fused to the N-termini of rabbit Fc fragments (V3-rFc), was performed as previously described (Davis et al. 2009, Zolla-Pazner et al. 2009). Briefly, V3-rFc was coated onto plastic plates at 2.0 ug/ml and incubated for 2 h. followed by blocking with 100 ul/well of 2.5% dry milk in phosphate-buffered saline for 1 h. Serial two-fold dilutions of rabbit sera were prepared in 2.5% dry milk/PBS and, after 2 h incubation at 37° C, bound serum Abs were detected with goat-anti-rabbit Fab-specific, alkaline phosphatase-conjugated secondary Abs (Zymed). Finally, the substrate, p-nitrophenyl phosphate (Sigma) in 10% diethanolamine, pH = 9.8, was added, and the plates were read at 405 nm.
Rabbit immunization
Immunizations were performed using a prime-boost protocol previously described (Zolla-Pazner et al. 2008, Wang et al. 2006). Briefly, female New Zealand White (NZW) rabbits 6–8 weeks old (with a body weight of ~2 kg) were purchased from Millbrook Farm (Amherst, MA) and housed in the animal facility managed by the Department of Animal Medicine at the University of Massachusetts Medical School in accordance with an IACUC-approved protocol. Groups of rabbits first received three DNA immunizations at weeks 0, 2, and 4 using a Bio-Rad Helios gene gun (Bio-Rad Laboratories, Hercules, CA). The gp120 DNA vaccine plasmid or the negative control pJW4303 vector plasmid was coated onto 1.0-μm gold beads at a ratio of 2 μg of DNA per mg of gold. Each gene gun shot delivered 1 μg of DNA to a total of 36 non-overlapping sites on the shaved abdominal skin of each rabbit at each of the three priming immunizations. The animals then received two boosts with individual V3-CTB proteins at weeks 10 and 14. A total of 100 μg per injection of the V3-CTB immogen was administered intramuscularly with IFA. Blood was collected prior to immunization and 2 weeks after each immunization.
Generation and neutralization of pseudoviruses
The expression vectors for chimeric forms of SF162 env with various consensus V3 sequences were generated by introducing the modifications sequentially by QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA), as described (Krachmarov et al. 2006). The infectious pseudotyped viruses were generated by co-transfection of 293 cells with an env expression vector and with the complementing vector pNL4-3.Luc.R-E-(NIH AIDS RRRP, catalog # 3418, donated by Dr. Nathaniel Landau). Transfections were performed in tissue culture dishes using TransIT-LT1 Reagent (Mirus Bio Corporation, Madison, WI) according to the manufacturer's protocol.
Neutralizing activity was determined as previously described (Krachmarov et al. 2001) in a single-cycle infectivity assay using virions pseudotyped with the molecularly cloned HIV env of interest. In brief, psVs were incubated with 2-fold serial dilutions of heat-inactivated sera, starting at a dilution of 1:10, from immunized rabbits for 1.5 h at 37 °C, and then added to 10,000–12,000 U87-T4-CCR5 target cells/well in 96-well plates in the presence of polybrene (10 μg/ml). After 24 h, cells were re-fed with RPMI medium containing 10% FBS and 10 μg/ml polybrene, followed by an additional 24-48 h incubation. Luciferase activity was determined 48–72 h post-infection with a microtiter plate luminometer (HARTA, Inc., Gaithersburg, MD) using assay reagents from Promega, Inc. (Madison, WI). Geometric mean titers for 50% neutralization (GMT50) were determined by interpolation from neutralization curves and are averages of at least two independent assays.
A second psV neutralization assay was performed as previously described (Li et al. 2005, Seaman et al. 2007). Similar to the U87 assay described above, 2-fold serial dilutions of heat-inactivated sera were prepared starting at a dilution of 1:20. The serum/psV mixtures were then incubated with the TZM.bl target cells and luciferase activity measured 48 hr later. A pool of pre-bleed sera were tested as neative controls, and all sera were also tested against the negative control psV carrying the envelope of murine leukemia virus (MuLV).
Neutralization of HIV primary isolates
Neutralization of primary isolates grown in human PBMCs was measured as the reduction in luc reporter gene expression after a single round of virus infection using TZM-bl cells as previously described (Zolla-Pazner et al. 2008)(Li et al., 2005). Briefly, 200 TCID50 of virus were incubated with various dilutions of test serum samples for 1 h at 37 °C in a total volume of 150 μl of growth medium in 96-well flat-bottom culture plates. Freshly trypsinized cells (1 × 104 cells) were added to each well and maintained in culture medium containing 1 μM indinavir sulfate and, also containing DEAE-dextran (25 μg/ml) when needed for efficient viral growth. Controls contained cells only, and cells plus virus. After a 48 h incubation, 50 μl of Bright Glo reagent (Promega, Madison, WI) was added, and after a 2 min incubation, well contents were transfer to 96-well black solid plates and luminescence measured. The percent neutralization was calculated relative to the effect of pre-immune serum from the same rabbit at the same dilution. The 50% neutralizing titers (NT50) were determined using the method of Least Squares.
Acknowledgements
This work was made possible with generous support from Collaboration for AIDS Vaccine Development (CAVD) program of the Bill and Melinda Gates Foundation, NIH grants AI 36085 and AI 27742, and research funds from the Department of Veterans Affairs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Immune epitope Database and Analysis Resource. In.
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abagyan R, Totrov M, Kuznetsov D. ICM-A new method for protein modeling and design: Applications to. J Comp Chem. 1994;15:488–506. [Google Scholar]
- Albritton LM, Kim JW, Tseng L, Cunningham JM. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom M, Lebens M, Schodel F, Holmgren J. Insertion of a HIV-1-neutralizing epitope in a surface-exposed internal region of the cholera toxin B-subunit. Gene. 1994;149:211–217. doi: 10.1016/0378-1119(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Barnett SW, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, Wang S, Mboudjeka I, Leung L, Lian Y, Fong A, Buckner C, Ly A, Hilt S, Ulmer J, Wild CT, Mascola JR, Stamatatos L. The ability of an oligomeric human immunodeficiency virus type 1 (HIV-1) envelope antigen to elicit neutralizing antibodies against primary HIV-1 isolates is improved following partial deletion of the second hypervariable region. J Virol. 2001;75:5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH. Challenges in the development of an HIV-1 vaccine. Nature. 2008;455:613–619. doi: 10.1038/nature07352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bckstrom M, Holmgren J, Schodel F, Lebens M. Characterization of an internal permissive site in the cholera toxin B-subunit and insertion of epitopes from human immunodeficiency virus-1, hepatitis B virus and enterotoxigenic Escherichia coli. Gene. 1995;165:163–171. doi: 10.1016/0378-1119(95)00444-b. [DOI] [PubMed] [Google Scholar]
- Bell CH, Pantophlet R, Schiefner A, Cavacini LA, Stanfield RL, Burton DR, Wilson IA. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J Mol Biol. 2008;375:969–978. doi: 10.1016/j.jmb.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Stohlmann SA, McMillan M. An endogenously synthesized decamer peptide efficiently primes cytotoxic T cells specific for the HIV-1 envelope glycoprotein. Eur J Immunol. 1993;23:2777–2781. doi: 10.1002/eji.1830231109. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothe BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304–314. doi: 10.1007/s00251-005-0798-y. [DOI] [PubMed] [Google Scholar]
- Burke V, Williams C, Sukumaran M, Kim SS, Li H, Wang XH, Gorny MK, Zolla-Pazner S, Kong XP. Structural basis of the cross-reactivity of genetically related human anti-HIV-1 mAbs: implications for design of V3-based immunogens. Structure. 2009;17:1538–1546. doi: 10.1016/j.str.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, Zolla-Pazner S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS research and human retroviruses. 2007;23:415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Corti D, Langedijk JP, Hinz A, Seaman MS, Vanzetta F, Fernandez-Rodriguez BM, Silacci C, Pinna D, Jarrossay D, Balla-Jhagjhoorsingh S, Willems B, Zekveld MJ, Dreja H, O'Sullivan E, Pade C, Orkin C, Jeffs SA, Montefiori DC, Davis D, Weissenhorn W, McKnight A, Heeney JL, Sallusto F, Sattentau QJ, Weiss RA, Lanzavecchia A. Analysis of memory B cell responses and isolation of novel monoclonal antibodies with neutralizing breadth from HIV-1-infected individuals. PLoS ONE. 2010;5:e8805. doi: 10.1371/journal.pone.0008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz LJ, Iglesias E, Aguilar JC, Cabrales A, Reyes O, Andreu D. Different immune response of mice immunized with conjugates containing multiple copies of either consensus or mixotope versions of the V3 loop peptide from human immunodeficiency virus type 1. Bioconjugate chemistry. 2004;15:1110–1117. doi: 10.1021/bc049944u. [DOI] [PubMed] [Google Scholar]
- Cruz LJ, Iglesias E, Aguilar JC, Gonzalez LJ, Reyes O, Albericio F, Andreu D. A comparative study of different presentation strategies for an HIV peptide immunogen. Bioconjugate chemistry. 2004;15:112–120. doi: 10.1021/bc034119j. [DOI] [PubMed] [Google Scholar]
- Czerkinsky C, Russell MW, Lycke N, Lindblad M, Holmgren J. Oral administration of a streptococcal antigen coupled to cholera toxin B subunit evokes strong antibody responses in salivary glands and extramucosal tissues. Infect Immun. 1989;57:1072–1077. doi: 10.1128/iai.57.4.1072-1077.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Bibollet-Ruche F, Li H, Decker JM, Kutsch O, Morris L, Salomon A, Pinter A, Hoxie JA, Hahn BH, Kwong PD, Shaw GM. Human immunodeficiency virus type 2 (HIV-2)/HIV-1 envelope chimeras detect high titers of broadly reactive HIV-1 V3-specific antibodies in human plasma. J Virol. 2009;83:1240–1259. doi: 10.1128/JVI.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon AK, Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structure determination of an anti-HIV-1 Fab 447-52D-peptide complex from an epitaxially twinned data set. Acta Crystallogr D Biol Crystallogr. 2008;D64:792–802. doi: 10.1107/S0907444908013978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhart L, Raffelsberger W, Ferko B, Klima A, Purtscher M, Katinger H, Ruker F. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J Gen Virol. 1996;77(Pt 9):2001–2008. doi: 10.1099/0022-1317-77-9-2001. [DOI] [PubMed] [Google Scholar]
- Eriksson K, Fredriksson M, Nordstrom I, Holmgren J. Cholera toxin and its B subunit promote dendritic cell vaccination with different influences on Th1 and Th2 development. Infect Immun. 2003;71:1740–1747. doi: 10.1128/IAI.71.4.1740-1747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DP, Heyward WL, Popovic V, Orozco-Cronin P, Orelind K, Gee C, Hirsch A, Ippolito T, Luck A, Longhi M, Gulati V, Winslow N, Gurwith M, Sinangil F, Berman PW. Candidate HIV/AIDS vaccines: lessons learned from the World's first phase III efficacy trials. AIDS. 2003;17:147–156. doi: 10.1097/01.aids.0000050786.28043.62. [DOI] [PubMed] [Google Scholar]
- Gonzalez RA, Sanchez J, Holmgren J, Lopez S, Arias CF. Immunological characterization of a rotavirus-neutralizing epitope fused to the cholera toxin B subunit. Gene. 1993;133:227–232. doi: 10.1016/0378-1119(93)90643-h. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The v3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol. 2004;78:2394–2404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Hioe C, Israel ZR, Michael NL, Conley AJ, Williams C, Kessler JA, 2nd, Chigurupati P, Burda S, Zolla-Pazner S. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J Immunol. 1997;159:5114–5122. [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Cohen S, Polonis VR, Honnen WJ, Kayman SC, Krachmarov C, Pinter A, Zolla-Pazner S. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76:9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang XH, Burda S, Kimura T, Konings FA, Nadas A, Anyangwe CA, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J Virol. 2006;80:6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Karwowska S, Buchbinder A, Zolla-Pazner S. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol. 1993;150:635–643. [PubMed] [Google Scholar]
- Hanke T, Goonetilleke N, McMichael AJ, Dorrell L. Clinical experience with plasmid DNA- and modified vaccinia virus Ankara-vectored human immunodeficiency virus type 1 clade A vaccine focusing on T-cell induction. J Gen Virol. 2007;88:1–12. doi: 10.1099/vir.0.82493-0. [DOI] [PubMed] [Google Scholar]
- Hill CM, Deng H, Unutmaz D, Kewalramani VN, Bastiani L, Gorny MK, Zolla-Pazner S, Littman DR. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Visciano ML, Kumar R, Liu J, Mack EA, Simon RE, Levy DN, Tuen M. The use of immune complex vaccines to enhance antibody responses against neutralizing epitopes on HIV-1 envelope gp120. Vaccine. 2009;28:352–360. doi: 10.1016/j.vaccine.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hioe CE, Wrin T, Seaman MS, Yu X, Wood B, Self S, Williams C, Gorny MK, Zolla-Pazner S. Anti-V3 monoclonal antibodies display broad neutralizing activities against multiple HIV-1 subtypes. PLoS ONE. 2010;5:e10254. doi: 10.1371/journal.pone.0010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnen WJ, Krachmarov C, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J Virol. 2007;81:1424–1432. doi: 10.1128/JVI.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, Kong X-P. Conserved Structural Elements in the V3 Crown of HIV-1 gp120. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1861. [DOI] [PubMed] [Google Scholar]
- Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J Virol. 1994;68:400–410. doi: 10.1128/jvi.68.1.400-410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov C, Pinter A, Honnen WJ, Gorny MK, Nyambi PN, Zolla-Pazner S, Kayman SC. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J Virol. 2005;79:780–790. doi: 10.1128/JVI.79.2.780-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Honnen WJ, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80:7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krachmarov CP, Kayman SC, Honnen WJ, Trochev O, Pinter A. V3-specific polyclonal antibodies affinity purified from sera of infected humans effectively neutralize primary isolates of human immunodeficiency virus type 1. AIDS research and human retroviruses. 2001;17:1737–1748. doi: 10.1089/08892220152741432. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- Labrosse B, Treboute C, Brelot A, Alizon M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J Virol. 2001;75:5457–5464. doi: 10.1128/JVI.75.12.5457-5464.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science. 1986;233:209–212. doi: 10.1126/science.3014647. [DOI] [PubMed] [Google Scholar]
- Law M, Cardoso RM, Wilson IA, Burton DR. Antigenic and immunogenic study of membrane-proximal external region-grafted gp120 antigens by a DNA prime-protein boost immunization strategy. J Virol. 2007;81:4272–4285. doi: 10.1128/JVI.02536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner T FB, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B, editors. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; NM, LA-UR 06-0680: 2005. HIV Sequence Compendium. [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe M, Charles IG, Roberts M, Dougan G, Tite J, Fairweather NF. Intranasal immunization using the B subunit of the Escherichia coli heat-labile toxin fused to an epitope of the Bordetella pertussis P.69 antigen. Mol Microbiol. 1991;5:1385–1392. doi: 10.1111/j.1365-2958.1991.tb00785.x. [DOI] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gigosos R, Garcia-Fortea P, Reina-Dona E, Plaza-Martin E. Effectiveness in prevention of travellers' diarrhoea by an oral cholera vaccine WC/rBS. Travel Med Infect Dis. 2007;5:380–384. doi: 10.1016/j.tmaid.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Martin M, Hajishengallis G, Metzger DJ, Michalek SM, Connell TD, Russell MW. Recombinant antigen-enterotoxin A2/B chimeric mucosal immunogens differentially enhance antibody responses and B7-dependent costimulation of CD4(+) T cells. Infect Immun. 2001;69:252–261. doi: 10.1128/IAI.69.1.252-261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba N, Geyer BC, Kilbourne J, Alfsen A, Bomsel M, Mor TS. Humoral immune responses by prime-boost heterologous route immunizations with CTB-MPR(649–684), a mucosal subunit HIV/AIDS vaccine candidate. Vaccine. 2006;24:5047–5055. doi: 10.1016/j.vaccine.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Matthews TJ, Langlois AJ, Robey WG, Chang NT, Gallo RC, Fischinger PJ, Bolognesi DP. Restricted neutralization of divergent human T-lymphotropic virus type III isolates by antibodies to the major envelope glycoprotein. Proc Natl Acad Sci USA. 1986;83:9709–9713. doi: 10.1073/pnas.83.24.9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbah HA, Burda S, Gorny MK, Williams C, Revesz K, Zolla-Pazner S, Nyambi PN. Effect of soluble CD4 on exposure of epitopes on primary, intact, native human immunodeficiency virus type 1 virions of different genetic clades. J Virol. 2001;75:7785–7788. doi: 10.1128/JVI.75.16.7785-7788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney SP, Ross TM. Viral sequence diversity: challenges for AIDS vaccine designs. Expert Rev Vaccines. 2008;7:1405–1417. doi: 10.1586/14760584.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie SJ, Halsey JF. Cholera toxin B subunit as a carrier protein to stimulate a mucosal immune response. J Immunol. 1984;133:1818–1824. [PubMed] [Google Scholar]
- Merritt EA, Kuhn P, Sarfaty S, Erbe JL, Holmes RK, Hol WG. The 1.25 A resolution refinement of the cholera toxin B-pentamer: evidence of peptide backbone strain at the receptor-binding site. J Mol Biol. 1998;282:1043–1059. doi: 10.1006/jmbi.1998.2076. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Sarfaty S, Jobling MG, Chang T, Holmes RK, Hirst TR, Hol WG. Structural studies of receptor binding by cholera toxin mutants. Protein Sci. 1997;6:1516–1528. doi: 10.1002/pro.5560060716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Guinea R, Trkola A, Purtscher M, Klima A, Steindl F, Palese P, Katinger H. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J Virol. 1994;68:4031–4034. doi: 10.1128/jvi.68.6.4031-4034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyambi P, Burda S, Urbanski M, Heyndrickx L, Janssens W, Vanham G, Nadas A. Neutralization patterns and evolution of sequential HIV type 1 envelope sequences in HIV type 1 subtype B-infected drug-naive individuals. AIDS research and human retroviruses. 2008;24:1507–1519. doi: 10.1089/aid.2008.0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Aguilar-Sino RO, Wrin T, Cavacini LA, Burton DR. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology. 2007;364:441–453. doi: 10.1016/j.virol.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Wrin T, Cavacini LA, Robinson JE, Burton DR. Neutralizing activity of antibodies to the V3 loop region of HIV-1 gp120 relative to their epitope fine specificity. Virology. 2008;381:251–260. doi: 10.1016/j.virol.2008.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren PW, Burton DR, Sattentau QJ. HIV-1 antibody--debris or virion? Nat Med. 1997;3:366–367. doi: 10.1038/nm0497-366d. [DOI] [PubMed] [Google Scholar]
- Parren PW, Gauduin MC, Koup RA, Poignard P, Fisicaro P, Burton DR, Sattentau QJ. Relevance of the antibody response against human immunodeficiency virus type 1 envelope to vaccine design. Immunol Lett. 1997;57:105–112. doi: 10.1016/s0165-2478(97)00043-6. [DOI] [PubMed] [Google Scholar]
- Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger SP, Stewart S, Surko P, Way S, Wilson S, Sette A. The design and implementation of the immune epitope database and analysis resource. Immunogenetics. 2005;57:326–336. doi: 10.1007/s00251-005-0803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Seaman MS, Leblanc DF, Grandpre LE, Bartman MT, Montefiori DC, Letvin NL, Mascola JR. Standardized assessment of NAb responses elicited in rhesus monkeys immunized with single- or multi-clade HIV-1 envelope immunogens. Virology. 2007;367:175–186. doi: 10.1016/j.virol.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon M, Kessler N, Levy R, Zolla-Pazner S, Gorlach M, Anglister J. Alternative conformations of HIV-1 V3 loops mimic beta hairpins in chemokines, suggesting a mechanism for coreceptor selectivity. Structure. 2003;11:225–236. doi: 10.1016/s0969-2126(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Shioda T, Levy JA, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Williams C, Zolla-Pazner S, Wilson IA. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12:193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Stanfield RL, Gorny MK, Zolla-Pazner S, Wilson IA. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80:6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nature biotechnology. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Miiro G, Serwanga J, Pozniak A, McPhee D, Manigart O, Mwananyanda L, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker LG, Pitisuttithum P, Paris R, Allen S, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. Broad and Potent Neutralizing Antibodies from an African Donor Reveal a New HIV-1 Vaccine Target. Science. 2009 doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Pal R, Mascola JR, Chou TH, Mboudjeka I, Shen S, Liu Q, Whitney S, Keen T, Nair BC, Kalyanaraman VS, Markham P, Lu S. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350:34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wu L, Yang ZY, Xu L, Welcher B, Winfrey S, Shao Y, Mascola JR, Nabel GJ. Cross-clade recognition and neutralization by the V3 region from clade C human immunodeficiency virus-1 envelope. Vaccine. 2006;24:4995–5002. doi: 10.1016/j.vaccine.2006.03.083. [DOI] [PubMed] [Google Scholar]
- Wu X, Sambor A, Nason MC, Yang ZY, Wu L, Zolla-Pazner S, Nabel GJ, Mascola JR. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology. 2008;380:285–295. doi: 10.1016/j.virol.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Kiyono H, Yamamoto M, Imaoka K, Fujihashi K, Van Ginkel FW, Noda M, Takeda Y, McGhee JR. A nontoxic mutant of cholera toxin elicits Th2-type responses for enhanced mucosal immunity. Proc Natl Acad Sci U S A. 1997;94:5267–5272. doi: 10.1073/pnas.94.10.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZY, Chakrabarti BK, Xu L, Welcher B, Kong WP, Leung K, Panet A, Mascola JR, Nabel GJ. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J Virol. 2004;78:4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RG, Scott DL, Westbrook ML, Nance S, Spangler BD, Shipley GG, Westbrook EM. The three-dimensional crystal structure of cholera toxin. J Mol Biol. 1995;251:563–573. doi: 10.1006/jmbi.1995.0456. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S. Improving on nature: focusing the immune response on the V3 loop. Human antibodies. 2005;14:69–72. [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen S, Pinter A, Krachmarov C, Wrin T, Wang S, Lu S. Cross-clade neutralizing antibodies against HIV-1 induced in rabbits by focusing the immune response on a neutralizing epitope. Virology. 2009;392:82–93. doi: 10.1016/j.virol.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cohen SS, Krachmarov C, Wang S, Pinter A, Lu S. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology. 2008;372:233–246. doi: 10.1016/j.virol.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, Gorny MK. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS research and human retroviruses. 2004;20:1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]