Abstract
Many fundamental questions about sleep remain unanswered. The presence of sleep across phyla suggests that it must serve a basic cellular and/or molecular function. Microarray studies, performed in several model systems, have identified classes of genes that are sleep-state regulated. This has led to the following concepts: first, a function of sleep is to maintain synaptic homeostasis; second, sleep is a stage of macromolecule biosynthesis; third, extending wakefulness leads to downregulation of several important metabolic pathways; and, fourth, extending wakefulness leads to endoplasmic reticulum stress. In human studies, microarrays are being applied to the identification of biomarkers for sleepiness and for the common debilitating condition of obstructive sleep apnea.
High-throughput approaches to study gene expression
The search for genes involved in regulation by, and of, sleep and wakefulness and the quest to understand the functions of sleep at the molecular level began a decade before the advent of microarrays. Several candidate genes were studied to elucidate their roles in sleep and wakefulness (for a review, see [1]). Subtractive hybridization carried out on brain mRNA from sleep-deprived rats, performed in the early 1990s, was the first ‘high-throughput’ attempt to identify such genes [2]. Later, the application of differential display [3] broadened the candidate gene and subtractive hybridization approaches to screen for genes whose functions were poorly characterized or previously unknown.
As a result of sequencing efforts, there has been an exponential growth in the amount of information available about the DNA sequence of the human genome (e.g., see [4]) as well as the genomes of various model organisms (e.g., see [5,6]). Consequently, thousands of genes have been discovered, including many novel genes for which sequences were unavailable previously. The role of many of these genes is unknown. This facilitated the advent of microarray approaches.
A microarray contains tens of thousands of genes and is a tool for simultaneous measurement of their expression. Significant advances in gene annotations, progress in development and standardization of microarray platforms, as well as the expansion of data analysis tools, including new and powerful statistical approaches to assess expression data (reviewed in [7,8]), make a microarray approach the ideal way to initially determine the changes in transcription of the genome in response to, or as a consequence of, sleep and/or wake state. However, microarrays describe what might happen rather than what does happen in a cell or tissue because changes in a given mRNA might not translate into changes in the relevant protein. Microarrays are a tool of discovery and, as such, results are hypothesis-generating. Therefore, hypotheses arising from microarray studies need to be further assessed. In particular, it is important to distinguish whether changes in gene expression are being ‘driven by sleep’ versus ‘driving sleep or wakefulness’. Novel pathways are, however, being identified and, if confirmed, will provide new targets for therapeutic intervention to address problems with sleep.
Microarrays in sleep research
Microarrays offer a new window on the difference between the sleeping and awake brain and hold the promise of facilitating answers to some of the major questions in sleep biology (Box 1; see also [9–11]).
Box 1. Major questions in sleep biology.
What are the functions of sleep?
Which genes regulate sleep and wakefulness?
What molecular changes set the duration of wakefulness that can be sustained without impairment?
What are the molecular consequences of sleep deprivation?
What is the basis of individual differences in response to sleep deprivation and other sleep characteristics?
Are there biomarkers for sleep drive and for specific sleep disorders?
These questions are relevant to the pervasive problem of sleep deprivation in industrialized societies and to the high prevalence of several sleep disorders [12]. In this review, we describe what we have learned from microarray experiments designed to address these questions and discuss future opportunities for the application of these and related approaches.
Microarray studies addressing sleep function have focused on the brain in various species [13–19], although a recent study examined other organs [18]. Assessment of the transcriptome in model organisms has demonstrated changes in the level of transcripts of many genes between sleep and wakefulness; as many as 10% of genes in mouse brain change their expression between these two behavioral states [17]. Microarray studies performed to date have used different platforms and different study designs (Figure 1). Some studies have evaluated genes changing expression between sleep and wakefulness using fold changes (e.g., see [13]), whereas others have used false-discovery rate strategies (e.g. see [17]). Despite these differences, there is a similarity in the results with respect to pathways in the brain that are affected by sleep and wakefulness (Table 1). There remains debate as to the number of genes that are expressed differentially between sleep and wakefulness (e.g., see [13,14,17,19]). This probably reflects different study designs and, in particular, the sample sizes (power) used in different studies [13,14,17,19].
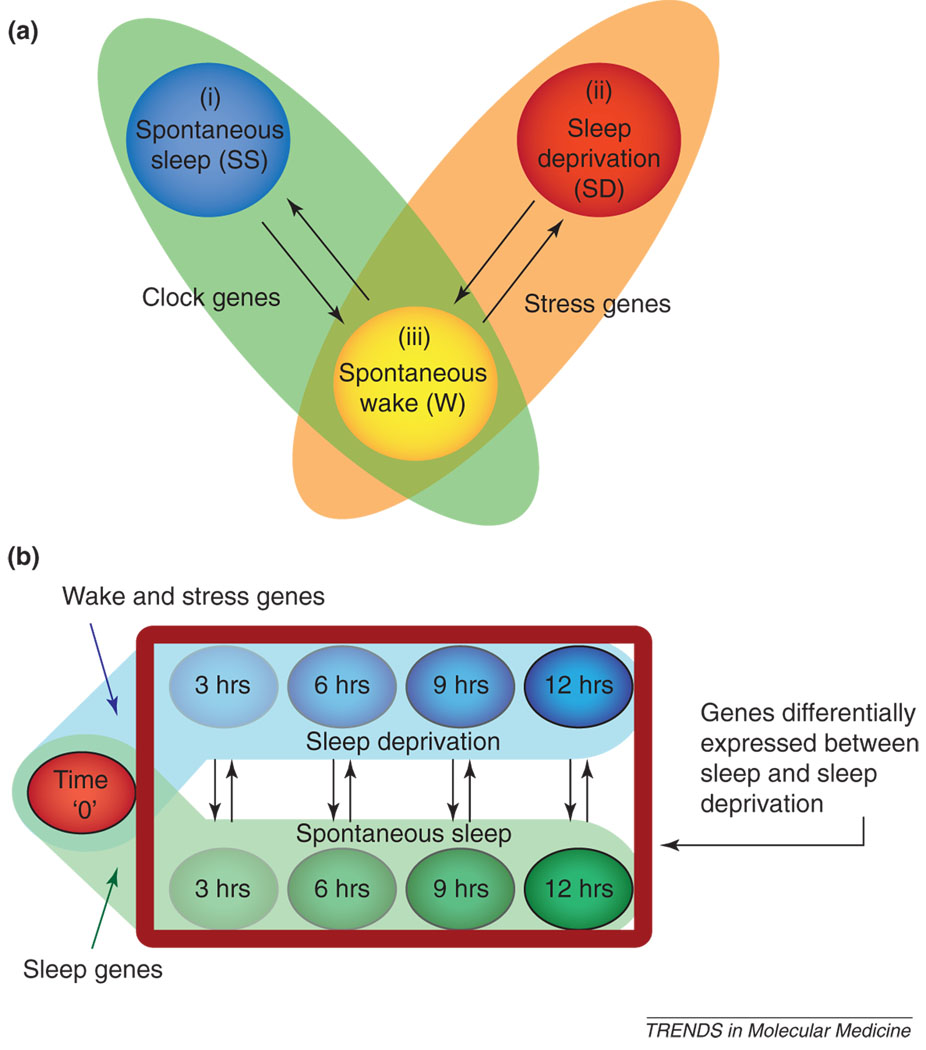
Figure 1.
Examples of experimental strategies in microarray research to elucidate the identity of genes whose expression exhibits differences between bouts of sleep and wakefulness. These complementary strategies address the confounding effects of sleep deprivation-induced systemic stress and the circadian clock on gene expression. (a) This approach [13] used three groups of rats: (i) a group of spontaneously sleeping (SS) animals who were required to sleep before sacrifice in the lights-on condition at least 75% of the previous 8 h; (ii) a group of sleep-deprived (SD) animals kept awake continuously by gentle handling and sacrificed at the same diurnal time as the SS group; and (iii) a group of animals that had a period of spontaneous wakefulness for 70% of the previous 8 h during the lights-out condition (W). Genes whose expression is higher during sleep, yet similar (lower) during sleep deprivation and the spontaneously awake condition (SS > SD = W) are called ‘sleep genes’, whereas genes conforming to the condition SS < SD = W are called ‘wake genes’. Although this design controls for stress and circadian confounders, it does not assess the direction of change in gene expression changes (i.e. up- or downregulation) or identify dynamic changes in transcriptional response. (b) This longitudinal study design [17] compares gene expression in animals allowed to sleep (green circles) before sacrifice with animals that are sleep deprived (blue circles) for different durations but sacrificed at the same time of day to control for circadian influences. An additional group of animals at ‘time zero’ (red circle) enables estimation of the direction of change for genes found to be expressed differentially between sleep and sleep deprivation. In this case, the experimental design controls for circadian influences in the estimation of temporal changes but does not take into account the effects of stress on gene expression in the group of animals subjected to sleep deprivation. Temporal changes in expression in the sleep groups are, however, not affected by stress because these animals are undisturbed. In both (a) and (b), black arrows indicate key comparisons; the strategy described in (a) relies on the fold-change, whereas the strategy in (b) relies on false-discovery rates as ‘filtering’ tools to identify differentially expressed genes. The false-discovery rate is a statistical procedure to control the number of mistakes made when performing multiple hypothesis tests, such as those performed during a search for differentially expressed genes in microarray experiments [102].
Table 1.
Microarray approaches to study gene expression during sleep, wakefulness or sleep deprivation and the key functional categories of genes expressed differentially among behavioral statesa
| Animal model | Refsb | Number of genesc |
Concepts that emerged from the studyd | ||||
|---|---|---|---|---|---|---|---|
| Response to cellular stress |
Synaptic plasticity |
Energy metabolism |
Lipid metabolism or membrane trafficking |
Downregulation of metabolism |
|||
| Rat | [13] | 651 | ✓e | ✓ | ✓ | ✓ | N/Af |
| Fruit fly | [14] | 133 | ✓ | ✓ | - | ✓ | N/Af |
| Fruit fly | [19] | 252 | - | - | - | ✓ | ✓ |
| Mouse | [17] | 3988 | ✓ | ✓ | ✓ | ✓ | ✓ |
| Mouse | [18] | 188g | ✓ | ✓ | - | ✓ | - |
| White-crowned sparrow |
[16] | ~19h | ✓ | - | ✓ | - | N/Af |
Note that the criteria for what comprise a significant change in the gene expression are not similar among studies listed in Table 1. Although two major approaches to the experimental design and data analyses are discussed briefly here, the aim of this review was not to evaluate the intricacies of experimental designs or statistical approaches to data analysis; such evaluation is beyond the scope of this review. Unavailability of the ‘raw’ microarray data for the majority of studies listed above precludes data re-analysis using a standardized approach. Thus, this review relies on the conclusions drawn by authors in each microarray paper.
Studies listed in this table are those that used the array platforms containing a complete set of probes – that is, the platforms that enabled the full assessment of transcriptional changes. Transcriptomics approaches that use subtractive hybridization [2], differential display [98,99] or ‘focused’ array platforms [49,99,100] have also been applied in sleep research. However, owing to their more limited scope, these studies are not discussed in detail here. Experimental designs and statistical approaches to data analyses vary considerably among the studies shown, although typically the duration of sleep deprivation was less than 12 h. The study with the largest sample size found the largest number of differentially expressed genes [17]. Changes in gene expression after a long duration of sleep deprivation [101] and generation of lesions of the wake-active neurons of the locus coeruleus have also been performed [15]. Microarray experiments in the rat [13] used the cerebral cortex and cerebellum and, in mice, the cerebral cortex and hypothalamus [17] or the whole brain [18] was studied. The fruit-fly experiments were performed on either whole heads [14] or isolated brains [19]; studies in the White-crowned sparrow utilized the telencephalon [16].
Number of genes identified as changing expression (either down- or upregulated) between sleep and wakefulness or sleep deprivation.
Concepts that emerged from the study are based on the functional categories of genes expressed differentially between sleep and wakefulness or the temporal pattern of gene expression.
Checkmarks indicate that among up- or downregulated genes there is a prevailing functional category of genes (such as ‘response to cellular stress’) present in the study.
The study design did not enable identification of the direction of change and hence downregulation of genes.
The number of significant genes and the functional categories differs with the genotype of the mouse studied, for example, the BDNF gene showed significant increase transcript level after sleep deprivation in the AKR/J and DBA/2J mice but not in C57BL/6J mice [18]. Since the synaptic-plasticity-related gene Homer1a increases uniformly across different strain of mice, the synaptic plasticity was checked (✓) accordingly.
The number is an estimate based on authors’ statement that 0.11% of genes change expression as a result of sleep restriction.
Model systems for the study of sleep
In recent years, an important advance in sleep research has been the identification of a sleep state in non-mammalian model systems: namely, the fruit fly (Drosophila melanogaster) [20–22], the zebrafish (Danio rerio) [23–25] and, most recently, the nematode (Caenorhabditis elegans) [26] (see also [27–29]). Crucial to the identification of sleep in these model systems has been the use of behavioral rather than electrophysiological criteria to define the sleep state. The main behavioral criteria, in addition to quiescence (lack of movement), are the following: first, elevated arousal threshold (i.e. when asleep, it takes a larger stimulus to make the animal move or a longer time to respond to a fixed stimulus); second, homeostasis (i.e. following sleep deprivation the animal returns to sleep faster, has increased durations of sleep bouts, often considered a measure of sleep depth, and can sleep at inappropriate circadian times); and, third, the timing of sleep propensity correlates with the molecular clock or rhythmic expression of clock genes. Not only are the behavioral aspects of sleep conserved among the models but there is conservation of both neurotransmitter systems and the components of the molecular signaling pathways that regulate sleep [30]. For example, signaling mechanisms involving cyclic AMP, which promotes wakefulness [31,32], and epidermal growth factor, which promotes sleep [33,34], have the same role in different model systems (for review, see [30]).
Model systems provide a great opportunity for investigation by microarrays. Microarrays have been applied to the study of sleep in fruit flies [14,19], rats [13,15] and mice [17,18]. In addition, microarray studies have been conducted recently in the white-crowned sparrow (Zonotrichia leukophrys) [16], albeit with a limited sample size. This sparrow, like many long-distance avian migrants, has the ability to go without sleep during its migration. An area ripe for future investigation is microarray studies of changes in gene expression in the nematode or zebrafish. Microarray studies that have been conducted in flies, rodents and birds show that similar molecular pathways are altered by sleep, wakefulness and sleep deprivation (Table 1).
What microarrays are teaching us about the molecular functions of sleep
Analyses of the functional categories of genes changing expression between sleep and wakefulness, as revealed by microarray studies, have led to new hypotheses about the functions of sleep and are now the focus of hypothesis-driven research. Two major theories have emerged that are not mutually exclusive: first, that sleep and wakefulness regulate synaptic strength, with up-scaling during wakefulness and down-scaling during sleep [35,36]; and, second, that sleep is a stage of macromolecule biosynthesis [17]. In the following sections, we explore each of these theories.
Sleep and wakefulness and synaptic scaling
Microarray studies conducted in rats [13] identified numerous genes involved in the acquisition and potentiation of synaptic plasticity (a term relating to the ability of synapses to change in strength); these genes expressed increased transcript levels during wakefulness. Conversely, the levels of expression of genes involved in synaptic consolidation or depression increase during sleep [13]. These observations have been incorporated within a hypothesis of sleep function referred to as the ‘synaptic homeostasis theory’ of sleep–wake control [35] (for a review of this theory, see [36]). It is postulated that wakefulness is accompanied by synaptic potentiation of cortical networks through brain-derived neurotrophic factor (BDNF)-dependent and other signaling mechanisms. Such synaptic potentiation is achieved through activities and learning during wakefulness (commonly termed as experience) and occurs in neuronal circuits that are activated by the relevant experience. Indeed, stimulation of whiskers in the rat modifies the local electroencephalogram (EEG) pattern (the EEG is a measure of electrical activity produced by the brain) [37,38]. This observation supports the notion that there is a potentiation of synaptic strength in the barrel cortex that receives and processes tactile information derived from the contralateral face of the animal. It is postulated that an increase in overall synaptic potentiation during wakefulness will require more resources (energy, space) to maintain this level of potentiation. It has been further suggested that synaptic potentiation is linked causally to the intensity of EEG slow-wave activity during subsequent sleep and that, during slow-wave sleep, there is synaptic down-scaling. Such down-scaling of synaptic strength has a beneficial effect on neuronal function by returning the system to an overall pre-wakefulness ‘balance’, although this leaves traces of experiences that occurred during wakefulness and increases synapse signal-to-noise ratios.
Sleep and wakefulness and macromolecule biosynthesis
Microarray studies have also identified other classes of genes whose patterns of expression change during sleep [17]. In mouse cerebral cortex and, to a lesser extent, hypothalamus there is upregulation during sleep of genes encoding proteins in various, predominantly intermediary, biosynthetic pathways for heme, protein and lipid [17]. A significant number of genes encoding the structural constituents of the ribosomes, translation-regulation activity and formation of transfer RNA (tRNA; a small RNA that transfers a specific amino acid to a growing polypeptide during protein synthesis) and ribosome biogenesis are also upregulated during sleep. Thus, synthesis of proteins and other macromolecules seems likely to occur during sleep, more so than during wakefulness.
Genes whose expression increases progressively during sleep include genes encoding multiple enzymes of the cholesterol-synthesis pathway, proteins involved in cholesterol uptake and transport and relevant transcription factors and chaperones responsible for transcriptional regulation of cholesterol-related genes [17]. Thus, membrane cholesterol is expected to increase during sleep. Cholesterol has an important role in membrane stability and is the key structural component of membrane microdomains called lipid rafts [39]. The transcript levels of several genes encoding lipid-raft-resident proteins, such as flotillin, also increase during sleep [17]. Rafts bring together neurotransmitter receptors with other signaling molecules and, by so doing, alter the strength of signaling [39]. The reassembling of lipid rafts during sleep would alter signaling of several neurotransmitters [40–43]. This might be in preparation for subsequent wakefulness, when there is enhanced release of various neurotransmitters on arousal from sleep. Reassembly of lipid rafts during sleep would enable maximal signaling on awakening. It is unclear how an increase in cholesterol synthesis during sleep would impact on synaptic down-scaling during sleep that is proposed in the synaptic homeostasis theory of sleep–wake control [35].
In addition, during sleep there is an upregulation of genes encoding proteins involved in the functioning of vesicle pools, as well as in the functioning of all antioxidant enzymes and components of intracellular transport. These observations suggest that sleep is a stage of rebuilding and repair in preparation for subsequent wakefulness (Figure 2).
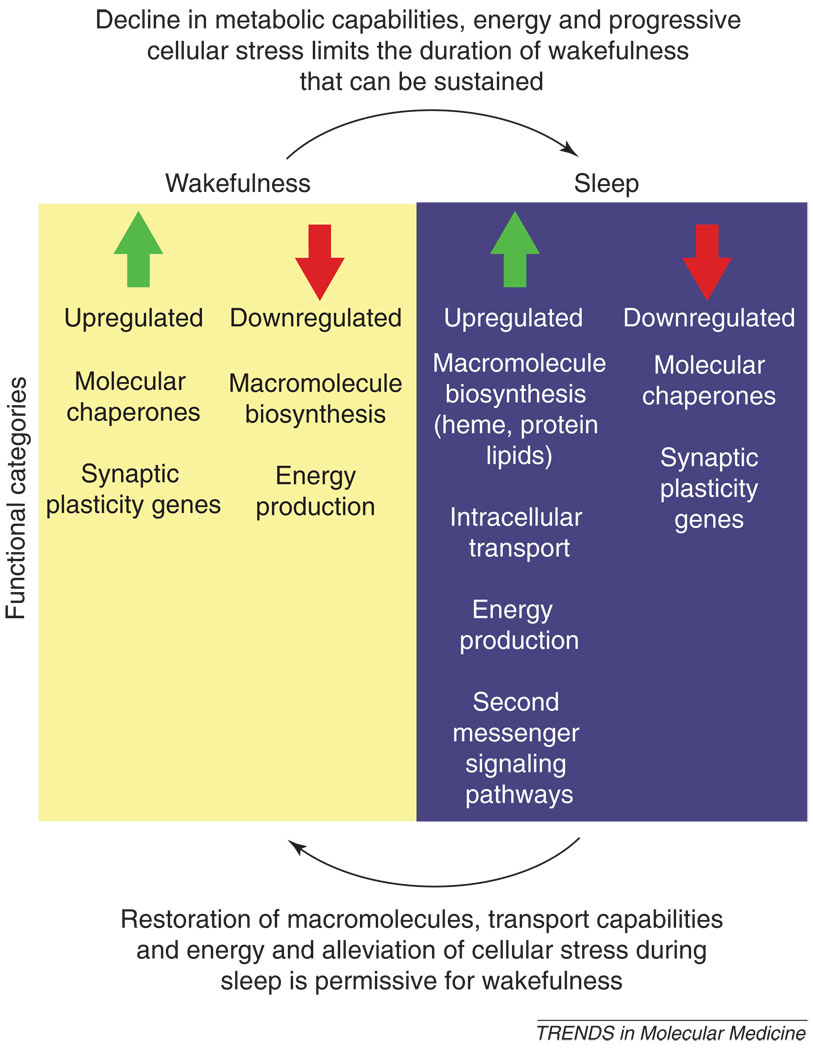
Figure 2.
During wakefulness, upregulation of genes involved in synaptic plasticity occurs and, with extended wakefulness, upregulation of molecular chaperones follows. If wakefulness is extended, a major mechanism promoting sleep might be the progressive decline in processes that promote wakefulness, thereby enabling sleep to occur. During sleep, there is a downregulation of genes involved in synaptic plasticity and those encoding molecular chaperones. Macromolecular biosynthesis during sleep is evidenced by the elevated expression of genes involved in the synthesis of lipids, protein, heme and other molecules. There is also increased expression of genes involved in intracellular transport, second-messenger signaling pathways and some genes involved in energy regulation.
What microarrays are teaching us about mechanisms limiting the duration of wakefulness
In humans, wakefulness can be sustained for 16 h without impairment of performance [44]. In mice, the duration of the longest episodes of sustained wakefulness is much shorter, being in the order of 3 h [45,46]. Accordingly, there must be a cost to extending wakefulness and there is likely to be molecular mechanisms that limit the duration of wakefulness. If so, extending wakefulness beyond these limits, as in acute sleep deprivation, should be deleterious. Microarray studies have revealed two key concepts: first that extending wakefulness leads to endoplasmic reticulum (ER) stress; and, second, that multiple genes in key cellular pathways are downregulated with prolonged wakefulness.
Extended wakefulness leads to cellular stress
Studies of gene expression have revealed that expression of the HSPA5 (heat shock 70 kDA protein 5) gene, which encodes the molecular chaperone commonly known as BiP, increases with extended wakefulness in different species and different brain regions [13,16,17,47] (see also [48,49]). These findings suggest that, with extended wakefulness, there is stress in the ER. In rodents, other genes encoding heat-shock proteins and molecular chaperones are also upregulated during sleep deprivation [17,49]. Increased expression of the Hspa5 gene occurs as part of the signaling pathway called the unfolded-protein response (UPR). This is an adaptive response that enables cells to survive stress in the ER, resulting from perturbations in calcium homeostasis, redox status, elevated secretory protein synthesis, misfolded proteins, glucose deprivation or altered glycosylation [50]. The UPR helps to restore normal ER function by reducing protein translation and by upregulating the expression of chaperones to increase the ER capacity for folding or to promote degradation of misfolded proteins (for reviews, see [51,52]). All components that are activated as part of the UPR are elevated in mouse cerebral cortex after 6 h of sleep deprivation [48]. In this study, sleep deprivation was performed at the beginning of the lights-on period, thus prolonging wakefulness that normally occurs in the lights-off period. A recent microarray study indicates that expression of the Hspa5 gene increases with sleep deprivation not only in brain but also in liver [18]. Thus, ER stress might be a general response to sleep deprivation that is not specific to brain. Hence, the prevailing ethos that sleep is ‘of the brain, by the brain and for the brain’ [53] is challenged by recent microarray data [18]. There is, however, limited information currently on how sleep and prolonged wakefulness impact on gene expression in peripheral tissues – an area ripe for future study.
Downregulation of genes for multiple cellular processes as wakefulness is prolonged
In both fruit-fly brains and mouse cerebral cortex and hypothalamus, the largest class of genes that are expressed differentially between sleep-deprived animals and sleeping controls are those that show decline in expression with prolonged wakefulness [17,19]. As the duration of wakefulness progresses, there is a reduction in the expression of genes involved in multiple physiological processes; reduction in these processes might act together to limit wakefulness. In the brain of the fruit fly, multiple genes involved in different steps in the protein-production pathway are downregulated with extended wakefulness [19]. In mouse cerebral cortex and hypothalamus, there is a reduction during extended wakefulness in the expression of genes encoding proteins involved in the main pathways of carbohydrate, energy, tricarboxylic acid (TCA) anabolism and various metabolic pathways, such as lipid, aldehyde and amine synthesis [17]. Thus, a mechanism promoting sleep might be the decline in processes that help sustain wakefulness, thereby enabling sleep to occur (Figure 2).
Genetic perturbations can test the functional roles of genes identified using microarrays
One disadvantage of microarray strategies is that one does not know whether the genes identified as changing expression with behavioral states are doing so as a consequence of the state (they could be part of the function of sleep) or whether they could be involved in regulating the sleep state. These possibilities are not mutually exclusive.
This disadvantage has led researchers to use the forward-genetic strategy of screening mutants for an altered sleep phenotype. If a mutant animal with an altered phenotype is identified, then it can be assumed that the gene that is mutated is involved in regulating the process. This strategy, applied in D. melanogaster [54–56], has identified two genes involved in regulating sleep – Sh, encoding the shaker K+ channel [54] and the gene encoding extracellular GPI-anchored protein termed Sleepless [55] – and, when applied in C. elegans, has identified a cyclic-GMP-dependent protein kinase [26,57].
Using currently available resources, however, particularly in the fruit fly, an investigator can determine quickly whether the altered expression of genes identified in microarray studies will affect sleep. In the fruit fly, there are lines available with transposable elements that disrupt gene function by insertion into identified genes [58–60]. Also available is a library of RNA interference (RNAi) lines that target 88% of protein-coding genes [61]. These transposon-insertion lines and the RNAi lines can lead to reduced gene function. The power of available D. melanogaster genetic tools is that spatial and temporal control of gene expression are also possible using two- and three-component transgenic systems (for a review, see [62]). Gene expression can be altered in both the positive and negative direction and can disregulate the gene of interest identified in microarray studies. It is, therefore, feasible to test whether a gene indentified in a microarray experiment also regulates sleep.
This technique was used in the study of the molecular chaperone gene Hspa5 in the fruit fly. The expression of Hspa5 increases with sleep deprivation in fruit flies [47], mice [17], rats [13] and birds [16]. Although numerous studies indicate that extended wakefulness leads to increased expression of the molecular chaperone Hspa5, it has been shown recently through genetic manipulation of Drosophila that alteration of Hspa5 levels does affect the amount of recovery sleep following sleep deprivation. There is no alteration in the amount of baseline sleep or wakefulness. Increased expression of Hspa5 increases the amount of sleep recovery following sleep deprivation [63]. By contrast, increased expression of a dominant-negative form of Hspa5 decreases the amount of sleep recovery after sleep deprivation [63]. These observations are compatible with two possible explanations. Hspa5 itself could be a sleep-promoting molecule with higher levels of Hspa5 leading to more recovery sleep. Alternatively, and more likely, is that higher levels of Hspa5 delay activation of the UPR, as revealed by in vitro studies [64,65]. Hence, there is a need for more recovery sleep, the final mechanism of defense. Thus, processes controlling the baseline amounts of sleep versus wake and those controlling recovery sleep following sleep deprivation can be separated at the molecular level. The Hspa5 protein is involved in regulating recovery sleep. The role of Hspa5 protein, which was identified by microarrays as important to sleep–wake function, was not understood fully until gene expression was altered experimentally.
Microarrays and quantitative trait loci and the genetic basis of inter-individual differences
Aspects of human sleep, such as timing [66] and sleep duration [67], are heritable. Similarly, many sleep-related traits are heritable in mice [45,46,68] (for review, see [69,70]). Using mouse recombinant-inbred strains and a quantitative trait locus (QTL) approach, a region on chromosome 13 involved in the response to sleep deprivation was identified [45]. QTL data were further characterized by considering genes in the region that were expressed differentially during sleep and wakefulness in microarray studies [17,18]. By combining haplotype analysis with data from gene-expression profiling by microarrays, hundreds of genes located within the QTL interval were narrowed down to a few genes [71]. However, only one of these genes, as determined by expression profiling [i.e. Homer1a (a splice variant of the Homer1 gene)], has a differential increase in expression with sleep deprivation among the recombinant inbred strains used for QTL mapping [18,71]. Moreover, there is a polymorphism in the regulatory region of the Homer1 gene that probably impacts on the transcript level observed in microarray profiling [71]. The promoter region of the Homer1 gene contains multiple CRE sites that will bind the cyclic-AMP response element-binding protein (CREB) [72], and the polymorphism present in this promoter might impact CREB binding. Mice with low levels of CREB owing to a deletion of the α and δ isoforms of CREB have reduced wakefulness during the early part of their night-time active period [31]. Whether this reduction in wakefulness is mediated, at least in part, by reduced increases in Homer1a with wakefulness in CREB hypomorph mice is currently unknown.
Thus, microarray studies support the hypothesis that a variation in Homer1a expression explains differences in response to sleep deprivation [18,71].
Microarrays and molecular signatures of sleep deprivation and sleep disorders
Microarrays are also powerful tools to identify the molecular signatures of diseases (for reviews, see [73,74]). However, they are only just beginning to be applied to the study of sleep deprivation and sleep disorders [75–78].
In D. melanogaster, a microarray study identified that the expression of the gene encoding amylase increases with prolonged wakefulness [75]. In humans, amylase increases in saliva with sleep deprivation [75]. Thus, assessment of amylase provides a potential biomarker of sleep loss. The importance of evaluating whether there are biomarkers for sleep homeostasis (sleepiness) has been emphasized previously [79–81]. Such biomarkers will provide assessments of sleepiness that could be used in various settings and provide a means for distinguishing individuals who are particularly sensitive to the effects of sleep deprivation.
Molecular signatures might also be developed for sleep disorders, in particular, the common condition known as obstructive sleep apnea (for a review, see [82]). In this condition, breathing stops repetitively during sleep, resulting in repeated interruption of sleep and in cyclical repeated hypoxic episodes [83]. These hypoxic episodes result in free-radical production and activation of the transcription factor NFκB and inflammatory pathways (for review, see [83]). Microarrays have shown that, in patients with obstructive sleep apnea, there are changes in transcript level in several genes involved in modulation of reactive-oxygen species (ROS), including heme oxygenase, superoxide dismutase and catalase [77]. These changes in obstructive sleep apnea patients are suggestive of the activation of mechanisms to modulate, and adapt to, increased ROS developing in response to the frequent episodes of intermittent hypoxia [77]. Thus, temporal changes in molecular-pathway components across the sleep period might provide a signature of the presence of this and perhaps other sleep disorders [84].
Concluding remarks
Here, we have argued that microarrays are a discovery strategy that has had a major impact in generating hypotheses about fundamental questions in sleep biology and is beginning to identify biomarkers for both the effects of sleep deprivation and for specific sleep disorders. Recent data suggest that extending wakefulness leads to ER stress and several different strategies, including the use of specific pharmacological agents that modify the process [85], support this concept. Whether sleep deprivation leads to ER stress in the brain in humans is unknown currently, although it seems likely that this will be the case because this has been demonstrated in all species studied to date. Given that ER stress induced by sleep deprivation is also found in the liver, it is conceivable that ER stress might be demonstrated in peripheral leukocytes in humans – an area for further study.
The concept of modulation of synaptic plasticity with sleep or wake state, arising from microarray studies, is also supported by investigation of phosphorylation of the glutamate receptors calcium/calmodulin-dependent protein kinase II (CaMKII) and glycogen synthase kinase 3β (GSK3β) [86]. However, other approaches, at least in developing animals [87], suggest that strengthening of synaptic connections occurs during sleep, not wakefulness, which is compatible with the improvements in performance of specific tasks that occur in humans from before to after sleep (for review, see [88]). Thus, further study of this concept is required because the situation might be more complex. It is conceivable that different neuronal groups respond differently with respect to sleep or wake changes in synaptic plasticity.
The concept that sleep is a stage of macromolecular synthesis is relatively recent, although compatible with earlier data that protein synthesis occurs during sleep [89,90]. Further studies to validate this are required. In particular, it will be important to determine the molecular basis for the switch from energy resources being used during wakefulness to support neuronal firing to those being used during sleep for synthesis of key molecules.
Although much has been accomplished, much more remains to be done (Box 2). Microarray studies of changes in gene expression during the cycle of sleep and wakefulness in the nematode and zebrafish should further clarify what functions of sleep are conserved across phylogeny. Sleep and wakefulness, unlike the circadian clock, which can function as a cell-autonomous molecular negative-feedback loop, are controlled by interacting neuronal circuits (for review, see [53,91–93]). Thus, understanding sleep and wakefulness at the level of transcription requires the study of changes in gene expression within identified populations of neuronal cells. Identification and isolation of particular neuronal cell populations would then be the basis of a microarray-based examination of gene expression in model organisms. This is possible with laser microdissection, high-throughput in situ hybridization or the use of flow cytometry for cell separation (for a review of these methods, see [94]). The first attempt to assess changes in the transcript level in a specific neuronal population was made by Maret et al., who used microarrays to study RNA isolated from neurons expressing Homer1a gene [18]. In this experiment, a poly-A binding protein (PAPB) was expressed in transgenic mice under the control of the promoter of the Homer1 gene, followed by the purification of the PAPB and the bound mRNA. This study identified several transcripts with changing expression in Homer1a-expressing neurons after sleep loss, including, as predicted, the transcript of the Homer1a gene [18].
Box 2. Future applications of microarrays in sleep research.
To identify conserved gene expression changes in sleep–wake periods in additional model organisms to identify key conserved sleep functions.
To determine changes in gene expression during sleep–wake periods and on sleep deprivation in identified neuronal populations.
To determine changes in gene expression during sleep–wake and sleep deprivation in peripheral organs; separating effects of sleep–wake and sleep deprivation from clock influences.
To develop molecular signatures for diagnostic applications and for identifying individuals particularly at risk of adverse outcomes stemming from sleep deprivation and sleep disorders.
Recent advances in sequencing technologies, such as short-read high-throughput sequencing (RNA-Seq), make it clear that current methods, including microarrays, are not identifying all of the transcriptional landscape of mammalian cells [95]. RNA-Seq detects as many as 25% more gene transcripts than microarrays. Thus, high-throughput sequencing of the transcriptome during sleep and wakefulness will provide additional information concerning changes in gene expression between these behavioral states.
Although the control of sleep and wakefulness originates in the brain and many functions of sleep are centered on the brain, lack of sleep affects peripheral organs, as seen by the effects of sleep deprivation on metabolism [96] and cardiac health [97]. Therefore, microarray studies also need to extend to peripheral organs so that the effect of sleep and wakefulness, as well as sleep deprivation on gene expression in heart, lung and kidney, among others, can be assessed.
In further investigations in different species, different neuronal groups and different organs, study designs need to be used to separate the effects of sleep or wake from circadian clock influences, as has been done in recent studies [18].
One of the goals of clinical research is to identify the earliest possible stage of progressive diseases, so that treatment can be initiated. Thus, although the searches for predictive biomarkers of sleep disorders and the deleterious effects of sleep deprivation have only just begun, microarray-based strategies are already beginning to identify pathways to provide both diagnostic information about the presence of a specific sleep disorder and prognostic information.
Acknowledgements
We are grateful to Daniel Barrett and Jennifer Montoya for their help in preparation of this manuscript. The original research was supported by NIH grants HL60287, AG17628 and HL66611, and by the NIH/NHGRI Ruth L. Kirchstein Postdoctoral Fellowship HG003968 to K.R.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mackiewicz M, Pack AI. Functional genomics of sleep. Respir. Physiol. Neurobiol. 2003;135:207–220. doi: 10.1016/s1569-9048(03)00045-4. [DOI] [PubMed] [Google Scholar]
- 2.Rhyner TA, et al. Molecular cloning of forebrain mRNAs which are modulated by sleep deprivation. Eur. J. Neurosci. 1990;2:1063–1073. doi: 10.1111/j.1460-9568.1990.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 3.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 5.Gupta BP, Sternberg PW. The draft genome sequence of the nematode Caenorhabditis briggsae, a companion to C. elegans. Genome Biol. 2003;4:238. doi: 10.1186/gb-2003-4-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 7.Quackenbush J. Computational analysis of microarray data. Nat. Rev. Genet. 2001;2:418–427. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 8.Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet. Res. 2001;77:123–128. doi: 10.1017/s0016672301005055. [DOI] [PubMed] [Google Scholar]
- 9.Mignot E. Why we sleep: the temporal organization of recovery. PLoS Biol. 2008;6:e106. doi: 10.1371/journal.pbio.0060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirelli C, Bushey D. Sleep and wakefulness in Drosophila melanogaster. Ann. N. Y. Acad. Sci. 2008;1129:323–329. doi: 10.1196/annals.1417.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirelli C, Tononi G. Is sleep essential? PLoS Biol. 2008;6:e216. doi: 10.1371/journal.pbio.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colten HR, Altevogt BM. Sleep Disorders and Sleep Deprivation: an Unmet Public Health Problem. The National Academies Press; 2006. [PubMed] [Google Scholar]
- 13.Cirelli C, et al. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 14.Cirelli C, et al. Sleep and wakefulness modulate gene expression in Drosophila. J. Neurochem. 2005;94:1411–1419. doi: 10.1111/j.1471-4159.2005.03291.x. [DOI] [PubMed] [Google Scholar]
- 15.Cirelli C, Tononi G. Locus ceruleus control of state-dependent gene expression. J. Neurosci. 2004;24:5410–5419. doi: 10.1523/JNEUROSCI.0949-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones S, et al. Molecular correlates of sleep and wakefulness in the brain of the white-crowned sparrow. J. Neurochem. 2008;105:46–62. doi: 10.1111/j.1471-4159.2007.05089.x. [DOI] [PubMed] [Google Scholar]
- 17.Mackiewicz M, et al. Macromolecule biosynthesis: a key function of sleep. Physiol. Genomics. 2007;31:441–457. doi: 10.1152/physiolgenomics.00275.2006. [DOI] [PubMed] [Google Scholar]
- 18.Maret S, et al. Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20090–20095. doi: 10.1073/pnas.0710131104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerman JE, et al. Multiple mechanisms limit the duration of wakefulness in Drosophila brain. Physiol. Genomics. 2006;27:337–350. doi: 10.1152/physiolgenomics.00030.2006. [DOI] [PubMed] [Google Scholar]
- 20.Hendricks JC, et al. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 21.Shaw PJ, et al. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman JE, et al. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 2008;31:371– 376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokogawa T, et al. Characterization of sleep in zebrafish and insomnia in hypocretin receptor mutants. PLoS Biol. 2007;5:e277. doi: 10.1371/journal.pbio.0050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prober DA, et al. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J. Neurosci. 2006;26:13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhdanova IV, et al. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001;903:263–268. doi: 10.1016/s0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- 26.Raizen DM, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–572. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 27.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr. Biol. 2008;18:R670–R679. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Youngsteadt E. Genetics. Simple sleepers. Science. 2008;321:334–337. doi: 10.1126/science.321.5887.334. [DOI] [PubMed] [Google Scholar]
- 29.Olofsson B, de Bono M. Sleep: dozy worms and sleepy flies. Curr. Biol. 2008;18:R204–R206. doi: 10.1016/j.cub.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Zimmerman JE, et al. Conservation of sleep: insights from non-mammalian model systems. Trends Neurosci. 2008;31:371–376. doi: 10.1016/j.tins.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Graves LA, et al. Genetic evidence for a role of CREB in sustained cortical arousal. J. Neurophysiol. 2003;90:1152–1159. doi: 10.1152/jn.00882.2002. [DOI] [PubMed] [Google Scholar]
- 32.Hendricks JC, et al. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat. Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 33.Foltenyi K, et al. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- 34.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat. Neurosci. 2007;10:1300–1307. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 35.Tononi G, Cirelli C. Sleep and synaptic homeostasis: a hypothesis. Brain Res. Bull. 2003;62:143–150. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 36.Tononi G, Cirelli C. Sleep function and synaptic homeostasis. Sleep Med. Rev. 2006;10:49–62. doi: 10.1016/j.smrv.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Vyazovskiy V, et al. Unilateral vibrissae stimulation during waking induces interhemispheric EEG asymmetry during subsequent sleep in the rat. J. Sleep Res. 2000;9:367–371. doi: 10.1046/j.1365-2869.2000.00230.x. [DOI] [PubMed] [Google Scholar]
- 38.Vyazovskiy VV, et al. Regional pattern of metabolic activation is reflected in the sleep EEG after sleep deprivation combined with unilateral whisker stimulation in mice. Eur. J. Neurosci. 2004;20:1363–1370. doi: 10.1111/j.1460-9568.2004.03583.x. [DOI] [PubMed] [Google Scholar]
- 39.Simons K, Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 40.Butchbach ME, et al. Association of excitatory amino acid transporters, especially EAAT2, with cholesterol-rich lipid raft microdomains: importance for excitatory amino acid transporter localization and function. J. Biol. Chem. 2004;279:34388–34396. doi: 10.1074/jbc.M403938200. [DOI] [PubMed] [Google Scholar]
- 41.Eroglu C, et al. Glutamate-binding affinity of Drosophila metabotropic glutamate receptor is modulated by association with lipid rafts. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10219–10224. doi: 10.1073/pnas.1737042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hering H, et al. Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J. Neurosci. 2003;23:3262–3271. doi: 10.1523/JNEUROSCI.23-08-03262.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schrattenholz A, Soskic V. NMDA receptors are not alone: dynamic regulation of NMDA receptor structure and function by neuregulins and transient cholesterol-rich membrane domains leads to disease-specific nuances of glutamate-signalling. Curr. Top. Med. Chem. 2006;6:663–686. doi: 10.2174/156802606776894519. [DOI] [PubMed] [Google Scholar]
- 44.Van Dongen HP, et al. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–126. doi: 10.1093/sleep/26.2.117. [DOI] [PubMed] [Google Scholar]
- 45.Franken P, et al. The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 2001;21:2610–2621. doi: 10.1523/JNEUROSCI.21-08-02610.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franken P, et al. Genetic determinants of sleep regulation in inbred mice. Sleep. 1999;22:155–169. [PubMed] [Google Scholar]
- 47.Shaw PJ, et al. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 48.Naidoo N, et al. Sleep deprivation induces the unfolded protein response in mouse cerebral cortex. J. Neurochem. 2005;92:1150–1157. doi: 10.1111/j.1471-4159.2004.02952.x. [DOI] [PubMed] [Google Scholar]
- 49.Terao A, et al. Differential increase in the expression of heat shock protein family members during sleep deprivation and during sleep. Neuroscience. 2003;116:187–200. doi: 10.1016/s0306-4522(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 50.Kaufman RJ. Orchestrating the unfolded protein response in health and disease. J. Clin. Invest. 2002;110:1389–1398. doi: 10.1172/JCI16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Y, Hendershot LM. The mammalian endoplasmic reticulum as a sensor for cellular stress. Cell Stress Chaperones. 2002;7:222–229. doi: 10.1379/1466-1268(2002)007<0222:tmeraa>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 53.Hobson JA. Sleep is of the brain, by the brain and for the brain. Nature. 2005;437:1254–1256. doi: 10.1038/nature04283. [DOI] [PubMed] [Google Scholar]
- 54.Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 55.Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu MN, et al. A genetic screen for sleep and circadian mutants reveals mechanisms underlying regulation of sleep in Drosophila. Sleep. 2008;31:465–472. doi: 10.1093/sleep/31.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raizen DM, et al. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics. 2006;173:177–187. doi: 10.1534/genetics.106.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Metaxakis A, et al. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics. 2005;171:571–581. doi: 10.1534/genetics.105.041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 60.Spradling AC, et al. The Berkeley Drosophila Genome Project gene disruption project: single P element insertions mutating 25% of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 62.McGuire SE, et al. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- 63.Naidoo N, et al. A role for the molecular chaperone protein BiP/GRP78 in Drosophila sleep homeostasis. Sleep. 2007;30:557–565. doi: 10.1093/sleep/30.5.557. [DOI] [PubMed] [Google Scholar]
- 64.Dorner AJ, et al. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992;11:1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertolotti A, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 66.Kerkhof GA, Van Dongen HP. Morning-type and evening-type individuals differ in the phase position of their endogenous circadian oscillator. Neurosci. Lett. 1996;218:153–156. doi: 10.1016/s0304-3940(96)13140-2. [DOI] [PubMed] [Google Scholar]
- 67.Gottlieb DJ, et al. Genome-wide association of sleep and circadian phenotypes. BMC Med. Genet. 2007;8((Suppl 1)):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Franken P, et al. Genetic variation in EEG activity during sleep in inbred mice. Am. J. Physiol. 1998;275:R1127–R1137. doi: 10.1152/ajpregu.1998.275.4.R1127. [DOI] [PubMed] [Google Scholar]
- 69.Franken P, Tafti M. Genetics of sleep and sleep disorders. Front. Biosci. 2003;8:e381–e397. doi: 10.2741/1084. [DOI] [PubMed] [Google Scholar]
- 70.Tafti M, Franken P. Invited review: genetic dissection of sleep. J. Appl. Physiol. 2002;92:1339–1347. doi: 10.1152/japplphysiol.00834.2001. [DOI] [PubMed] [Google Scholar]
- 71.Mackiewicz M, et al. Analysis of the QTL for sleep homeostasis in mice: Homer1a is a likely candidate. Physiol. Genomics. 2008;33:91–99. doi: 10.1152/physiolgenomics.00189.2007. [DOI] [PubMed] [Google Scholar]
- 72.Bottai D, et al. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J. Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kittleson MM, Hare JM. Molecular signature analysis: using the myocardial transcriptome as a biomarker in cardiovascular disease. Trends Cardiovasc. Med. 2005;15:130–138. doi: 10.1016/j.tcm.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 74.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol. Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 75.Seugnet L, et al. Identification of a biomarker for sleep drive in flies and humans. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19913–19918. doi: 10.1073/pnas.0609463104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Irwin MR, et al. Sleep deprivation and activation of morning levels of cellular and genomic markers of inflammation. Arch. Intern. Med. 2006;166:1756–1762. doi: 10.1001/archinte.166.16.1756. [DOI] [PubMed] [Google Scholar]
- 77.Hoffmann MS, et al. Microarray studies of genomic oxidative stress and cell cycle responses in obstructive sleep apnea. Antioxid. Redox Signal. 2007;9:661–669. doi: 10.1089/ars.2007.1589. [DOI] [PubMed] [Google Scholar]
- 78.Khalyfa A, et al. Genome-wide gene expression profiling in children with non-obese obstructive sleep apnea. Sleep Med. 2008 doi: 10.1016/j.sleep.2007.11.006. ; Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 79.Dawson D, McCulloch K. Managing fatigue: it’s about sleep. Sleep Med. Rev. 2005;9:365–380. doi: 10.1016/j.smrv.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 80.Jones CB, et al. Fatigue and the criminal law. Ind. Health. 2005;43:63–70. doi: 10.2486/indhealth.43.63. [DOI] [PubMed] [Google Scholar]
- 81.Van Dongen HP, et al. Individual differences in adult human sleep and wakefulness: Leitmotif for a research agenda. Sleep. 2005;28:479–496. doi: 10.1093/sleep/28.4.479. [DOI] [PubMed] [Google Scholar]
- 82.Pack AI. Advances in sleep-disordered breathing. Am. J. Respir. Crit. Care Med. 2006;173:7–15. doi: 10.1164/rccm.200509-1478OE. [DOI] [PubMed] [Google Scholar]
- 83.McNicholas WT, Ryan S. Obstructive sleep apnoea syndrome: translating science to clinical practice. Respirology. 2006;11:136–144. doi: 10.1111/j.1440-1843.2006.00824.x. [DOI] [PubMed] [Google Scholar]
- 84.Arnardottir E, et al. Molecular signatures of obstructive sleep apnea in adults: a review and perspective. Sleep. doi: 10.1093/sleep/32.4.447. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Methippara MM, et al. Salubrinal, an inhibitor of protein synthesis, promotes deep slow wave sleep. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008 doi: 10.1152/ajpregu.90765.2008. DOI:10.1152/ajpregu.90765.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vyazovskiy VV, et al. Molecular and electrophysiological evidence for net synaptic potentiation in wake and depression in sleep. Nat. Neurosci. 2008;11:200–208. doi: 10.1038/nn2035. [DOI] [PubMed] [Google Scholar]
- 87.Frank MG, et al. Sleep enhances plasticity in the developing visual cortex. Neuron. 2001;30:275–287. doi: 10.1016/s0896-6273(01)00279-3. [DOI] [PubMed] [Google Scholar]
- 88.Stickgold R. Sleep-dependent memory consolidation. Nature. 2005;437:1272–1278. doi: 10.1038/nature04286. [DOI] [PubMed] [Google Scholar]
- 89.Nakanishi H, et al. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur. J. Neurosci. 1997;9:271–279. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 90.Ramm P, Smith CT. Rates of cerebral protein synthesis are linked to slow wave sleep in the rat. Physiol. Behav. 1990;48:749–753. doi: 10.1016/0031-9384(90)90220-x. [DOI] [PubMed] [Google Scholar]
- 91.Pace-Schott EF, Hobson JA. The neurobiology of sleep: genetics, cellular physiology and subcortical networks. Nat. Rev. Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 92.Saper CB, et al. Homeostatic, circadian, and emotional regulation of sleep. J. Comp. Neurol. 2005;493:92–98. doi: 10.1002/cne.20770. [DOI] [PubMed] [Google Scholar]
- 93.Saper CB, et al. The sleep switch: hypothalamic control of sleep and wakefulness. Trends Neurosci. 2001;24:726–731. doi: 10.1016/s0166-2236(00)02002-6. [DOI] [PubMed] [Google Scholar]
- 94.Nelson SB, et al. The problem of neuronal cell types: a physiological genomics approach. Trends Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 95.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 96.Knutson KL, et al. The metabolic consequences of sleep deprivation. Sleep Med. Rev. 2007;11:163–178. doi: 10.1016/j.smrv.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ayas NT, et al. A prospective study of sleep duration and coronary heart disease in women. Arch. Intern. Med. 2003;163:205–209. doi: 10.1001/archinte.163.2.205. [DOI] [PubMed] [Google Scholar]
- 98.Cirelli C, Tononi G. Differences in gene expression between sleep and waking as revealed by mRNA differential display. Brain Res. Mol. Brain Res. 1998;56:293–305. doi: 10.1016/s0169-328x(98)00057-6. [DOI] [PubMed] [Google Scholar]
- 99.Cirelli C, Tononi G. Gene expression in the brain across the sleep-waking cycle. Brain Res. 2000;885:303–321. doi: 10.1016/s0006-8993(00)03008-0. [DOI] [PubMed] [Google Scholar]
- 100.Terao A, et al. Gene expression in the rat brain during sleep deprivation and recovery sleep: an Affymetrix GeneChip study. Neuroscience. 2006;137:593–605. doi: 10.1016/j.neuroscience.2005.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cirelli C, et al. Changes in brain gene expression after long-term sleep deprivation. J. Neurochem. 2006;98:1632–1645. doi: 10.1111/j.1471-4159.2006.04058.x. [DOI] [PubMed] [Google Scholar]
- 102.Storey J. A direct approach to false discovery rates. J. Roy. Stat. Soc. B. 2002;64:479–498. [Google Scholar]