Abstract
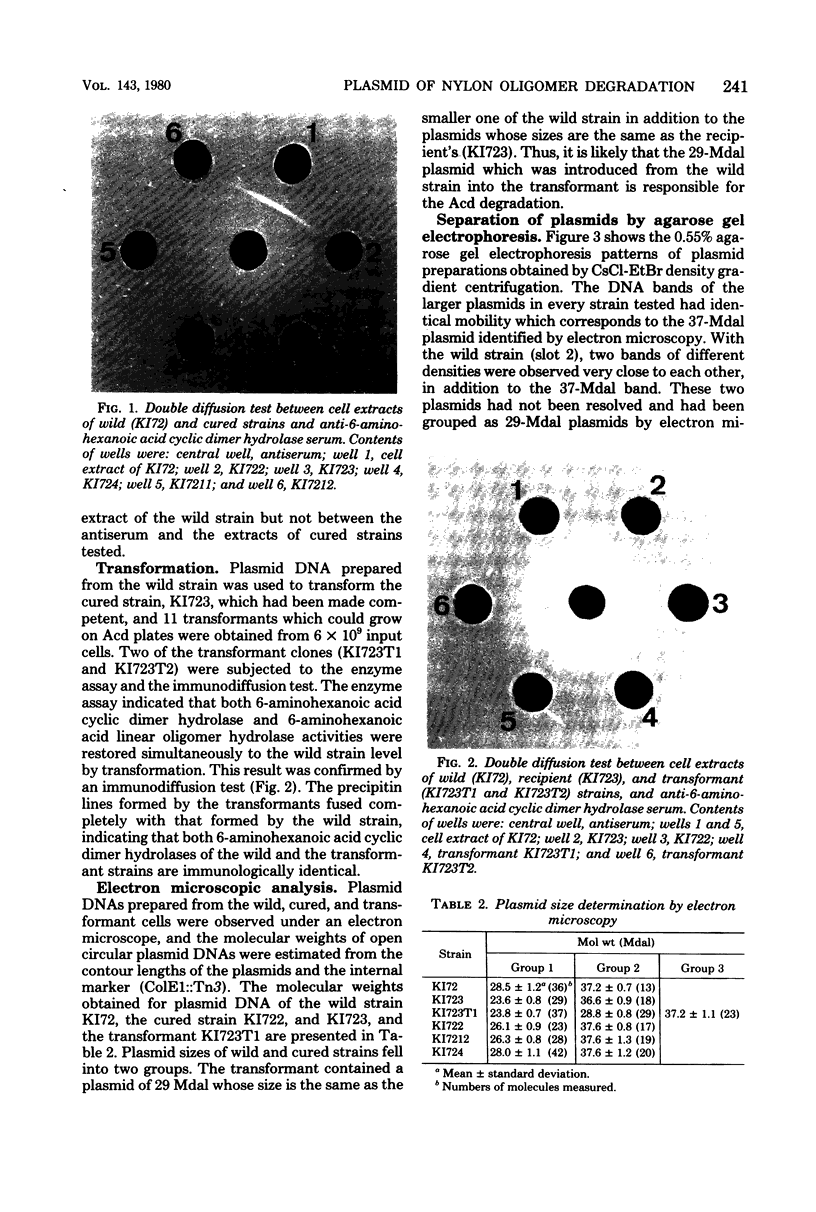
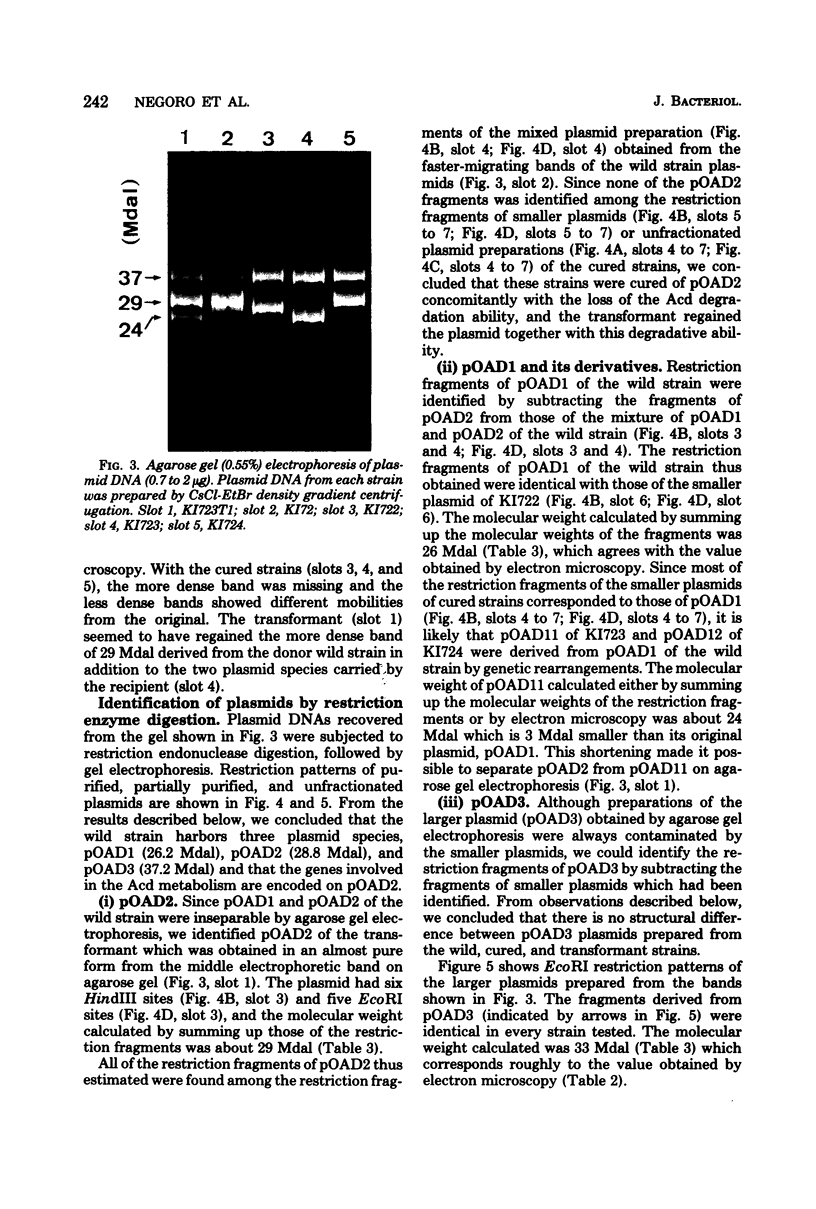
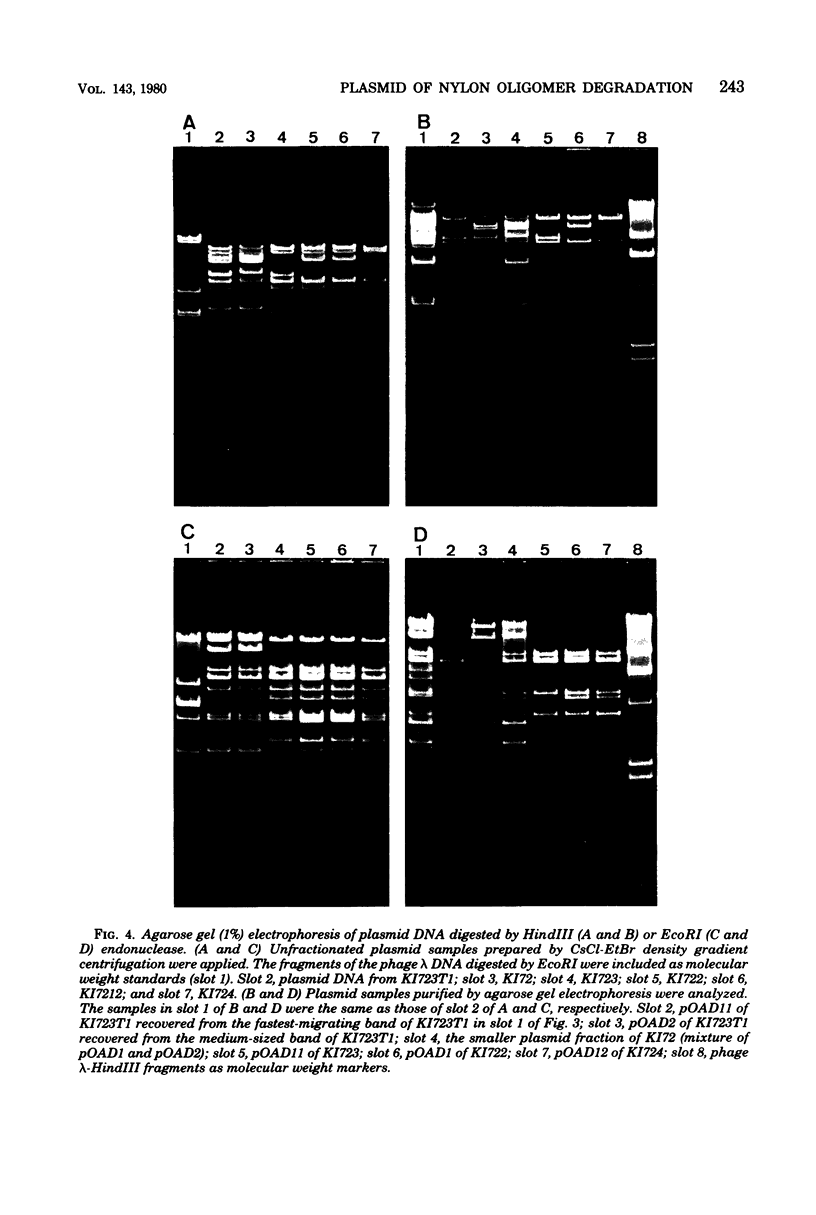
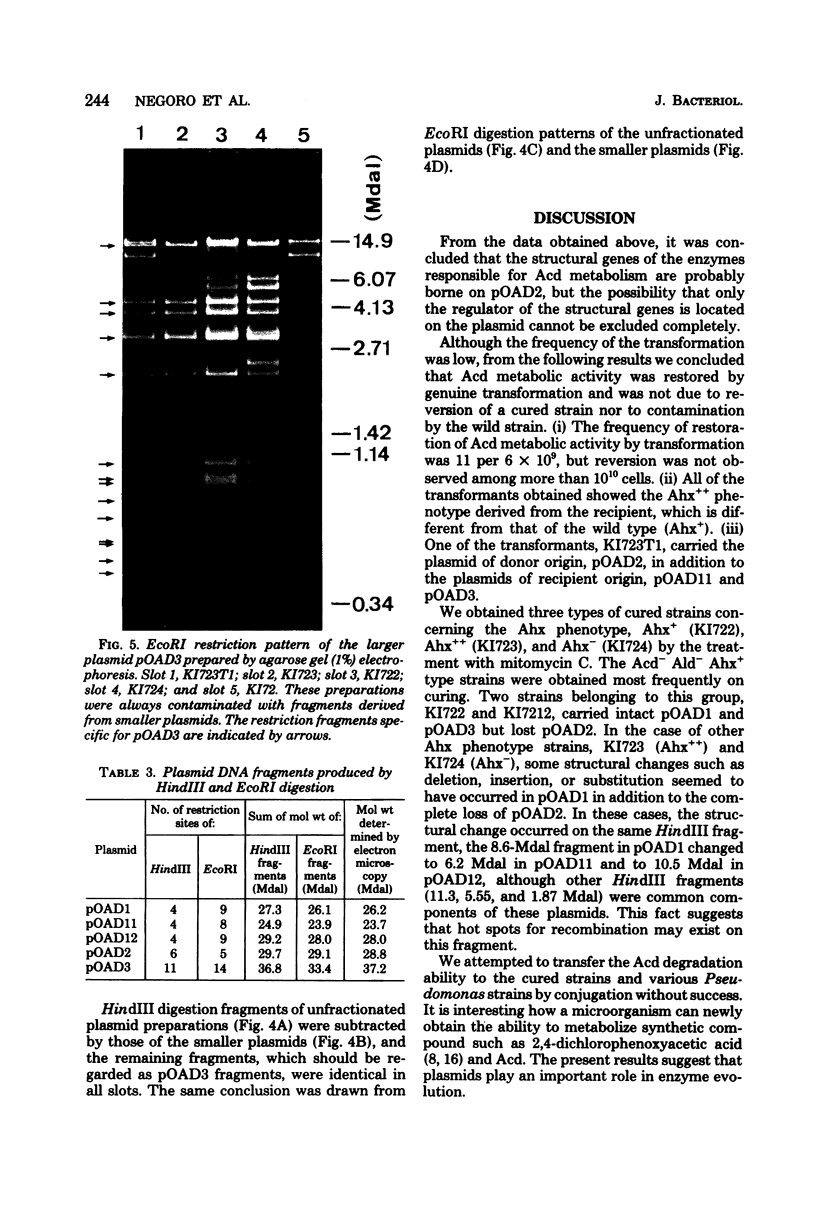
Flavobacterium sp. K172, which is able to grow on 6-aminohexanoic acid cyclic dimer as the sole source of carbon and nitrogen, and plasmid control of the responsible enzymes, 6-aminohexanoic acid cyclic dimer hydrolase and 6-aminohexanoic acid linear oligomer hydrolase, were studied. The wild strain of K172 harbors three kinds of plasmid, pOAD1 (26.2 megadaltons), pOAD2 (28.8 megadaltons), and pOAD3 (37.2 megadaltons). The wild strain K172 was readily cured of its ability to grow on the cyclic dimer by mitomycin C, and the cyclic dimer hydrolase could not be detected either as catalytic activity or by antibody precipitation. No reversion of the cured strains was detected. pOAD2 was not detected in every cured strain tested but was restored in a transformant. The transformant recovered both of the enzyme activities, and the cyclic dimer hydrolase of the transformant was immunologically identical with that of the wild strain. All of the strains tested, including the wild, cured, and transformant ones, possessed identical pOAD3 irrespective of the metabolizing activity. Some of the cured strains possessed pOAD1 identical with the wild strain, but the others harbored plasmids with partially altered structures which were likely to be derived from pOAD1 by genetic rearrangements such as deletion, insertion, or substitution. These results suggested that the genes of the enzymes were borne on pOAD2.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Bazaral M., Helinski D. R. Circular DNA forms of colicinogenic factors E1, E2 and E3 from Escherichia coli. J Mol Biol. 1968 Sep 14;36(2):185–194. doi: 10.1016/0022-2836(68)90374-4. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic fusion of incompatible plasmids in Pseudomonas. Proc Natl Acad Sci U S A. 1973 Jun;70(6):1641–1644. doi: 10.1073/pnas.70.6.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty A. M. Plasmids in Pseudomonas. Annu Rev Genet. 1976;10:7–30. doi: 10.1146/annurev.ge.10.120176.000255. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. R., Appleton J., Pemberton J. M. Isolation and characterization of the pesticide-degrading plasmid pJP1 from Alcaligenes paradoxus. J Bacteriol. 1978 Sep;135(3):798–804. doi: 10.1128/jb.135.3.798-804.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B. Some factors affecting electron microscopic length of deoxyribonucleic acid. J Mol Biol. 1967 Apr 28;25(2):209–216. doi: 10.1016/0022-2836(67)90138-6. [DOI] [PubMed] [Google Scholar]
- Kinoshita S., Negoro S., Muramatsu M., Bisaria V. S., Sawada S., Okada H. 6-Aminohexanoic acid cyclic dimer hydrolase. A new cyclic amide hydrolase produced by Achromobacter guttatus KI74. Eur J Biochem. 1977 Nov 1;80(2):489–495. doi: 10.1111/j.1432-1033.1977.tb11904.x. [DOI] [PubMed] [Google Scholar]
- Palchaudhuri S., Chakrabarty A. Isolation of plasmid deoxyribonucleic acid from Pseudomonas putida. J Bacteriol. 1976 Apr;126(1):410–416. doi: 10.1128/jb.126.1.410-416.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Fisher P. R. 2,4-D plasmids and persistence. Nature. 1977 Aug 25;268(5622):732–733. doi: 10.1038/268732a0. [DOI] [PubMed] [Google Scholar]
- TOSA T., CHIBATA I. UTILIZATION OF CYCLIC AMIDES AND FORMATION OF OMEGA-AMINO ACIDS BY MICROORGANISMS. J Bacteriol. 1965 Mar;89:919–920. doi: 10.1128/jb.89.3.919-920.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]